Abstract
Hybrid varieties can provide the boost needed to increase stagnant wheat yields through heterosis. The lack of an efficient hybridization system, which can lower the cost of goods of hybrid seed production, has been a major impediment to commercialization of hybrid wheat varieties. In this review, we discuss the progress made in characterization of nuclear genetic male sterility (NGMS) in wheat and its advantages over two widely referenced hybridization systems, i.e., chemical hybridizing agents (CHAs) and cytoplasmic male sterility (CMS). We have characterized four wheat genes, i.e., Ms1, Ms5, TaMs26 and TaMs45, that sporophytically contribute to male fertility and yield recessive male sterility when mutated. While Ms1 and Ms5 are Triticeae specific genes, analysis of TaMs26 and TaMs45 demonstrated conservation of function across plant species. The main features of each of these genes is discussed with respect to the functional contribution of three sub-genomes and requirements for complementation of their respective mutants. Three seed production systems based on three genes, MS1, TaMS26 and TaMS45, were developed and a proof of concept was demonstrated for each system. The Tams26 and ms1 mutants were maintained through a TDNA cassette in a Seed Production Technology-like system, whereas Tams45 male sterility was maintained through creation of a telosome addition line. These genes represent different options for hybridization systems utilizing NGMS in wheat, which can potentially be utilized for commercial-scale hybrid seed production.
1. Introduction
Wheat is one of the most important and widely grown crops in the world. It is the third largest produced cereal crop, providing 20% of the world’s calories [1]. With the predicted growth in world population to over nine billion by 2050, the Food and Agriculture Organization (FAO) set a target of 60% increased food production by 2050 [2]. A continued improvement in the yield of major food crops, including wheat, is required to meet this target [3]. Breeding for hybrid varieties to exploit hybrid vigor is one way to increase stagnant yields, particularly in wheat [4,5]. Heterosis has the potential to increase yield up to 20% in wheat, highlighting the prospects for hybrid varieties [4,5,6]. Yield stability of hybrids is another major advantage of cultivating hybrid wheat. Hybrids have demonstrated superior performance compared with traditional varieties under biotic and abiotic stress conditions, thus providing opportunity to expand wheat cultivation under marginal growing conditions [7,8].
The major hindrance to large-scale commercialization of hybrid varieties in wheat is the lack of a hybridization system to economically produce hybrid seed. Wheat is an autogamous species with a perfect flower, thus excluding the option of mechanical emasculation techniques such as detasseling in maize. A biological or a chemical hybridization system, therefore, is a prerequisite for hybrid seed production in wheat. Importantly, such a system should be reliable and efficient to lower the cost of goods to produce hybrid seed. Thus far two main methods have been deployed on a small-scale in wheat to produce hybrid seed: CHAs and CMS [9,10]. Each of these systems has limitations that make large-scale deployment for commercial hybrid seed production challenging [9,10,11,12,13].
In this review we focus on a third type of male sterility system in wheat, NGMS, and its utilization for hybrid seed production in wheat. NGMS-based hybridization systems can offer several advantages over CHAs and the CMS system because of simpler genetics of male sterility and fertility restoration [2,14]. Over the past several decades many male sterile mutants have been identified in wheat and male sterility genes cloned, including four genes that have been characterized by our group at CortevaTM Agriscience in partnership with external collaborators. Based on these and other male fertility genes, several NGMS hybridization systems have been proposed and are discussed in this review.
2. Sporophytic Genes Involved in Anther and Pollen Development in Wheat
Flowering plants have developed specialized structures to produce male and female gametes to accomplish sexual reproduction. Successful production of male gametes relies on proper formation of male reproductive organs. Pollen grains or microgametophytes are formed in the anthers, the male reproductive organ, and deliver male gametes to organs bearing female gametes (reviewed in [15]). Pollen grains are surrounded by protective pollen walls, intine and exine to enable survival of pollen in diverse environmental conditions. The intine is composed of cellulosic material, whereas sporopollenin is the major component of the exine (reviewed in [16]). Sporopollenin fortifies the exine through formation of a skeletal structure and a durable covering. The components of the exine are synthesized by the surrounding tapetum and deposited on the surface of developing microspores [17].
NGMS arises due to mutations in the genes involved in anther and sporophytic stages of pollen development, including sporopollenin biosynthesis pathway. Spontaneous male-sterile mutants have been reported in more than 175 species of angiosperms [18]. Of the mutants that have been characterized, more than 60% are attributed to a single recessive gene [18]. In addition to the spontaneous mutants, several sporophytic male fertility genes have been identified through mutant screens (reviewed in [19,20,21]) (Table 1).

Table 1.
Mutants and temperature- or photoperiod-variants of the male fertility genes in wheat.
2.1. Male Sterile Mutants in Wheat Identified through Forward Genetics
In hexaploid bread wheat, the three genomes A, B and D exhibit extensive functional redundancy, with all three homeologs contributing towards many traits. Therefore, a triple knockout of all three genomes is often required to uncover a mutant phenotype [32,33,47]. Due to this functional redundancy, few male sterile mutants have been identified in wheat compared with single genome diploid species such as maize, rice and barley. Nevertheless, a few male sterile mutants have been identified through forward genetics, either as spontaneous mutations or through mutagenesis (http://www.shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp) (accessed on 6 May 2021). Genetic studies and subsequent cloning of genes for many of these mutants has revealed these as either dominant mutations or mutations in genes with a predominant functional homeolog (Table 1). ms1 and ms5 are two such recessive mutants where one genome predominantly contributes to gene function [25,27,31]. ms1a mutant was the first male sterile mutant reported in wheat [48]. Subsequently, various forward genetic screens have identified seven ms1 mutant alleles (ms1a-g). Dominant male sterility can arise due to mutations in a single homeolog; therefore, more dominant mutants, such as Ms2, Ms3 and Ms4, have been identified as compared with recessive mutants. Of the dominant mutants, MS2 encodes an orphan protein that is specifically activated in the Ms2 mutants through the insertion of a retro-element in the promoter of the gene [28]. Tang et al. [49] utilized CRISPR/Cas9 to edit Ms2 to restore male fertility to Ms2 mutant. The genes for the Ms3 and Ms4 have not been identified yet.
2.2. Male Sterile Mutants in Wheat Revealed through Reverse Genetics
The role of an increasing number of wheat genes in male fertility is now being revealed through reverse genetics (Table 1). The availability of more efficient gene editing technologies, such as CRISPR/Cas, has accelerated the functional analysis of wheat genes, as it is possible to obtain concurrent mutations in three homeologs [33]. It is not surprising that all the genes investigated through reverse genetics exhibit functional redundancy of the three homeologs. Utilizing the new gene editing tools, wheat homologs of the maize MS26 and MS45 genes were characterized for their role in male fertility [32,33,50]. For both these genes, knockouts of all three homeologs are required for complete loss of function, which resulted in male sterility. Similarly, it has been observed that for TaNP1, the wheat homolog of the rice OsNP1 and maize ZmIPE1 genes encoding a putative glucose-methanol-choline oxidoreductase, knockouts in all three homeologs are required for male sterility [34]. Milner et al. [36] identified two male fertility genes in wheat, CALS5- and RPG1-like, through gene expression analysis. Functional analysis of these genes through CRISPR/Cas showed that knockouts in all three homeologs are required to obtain male sterile plants [36]. Similarly, triple knockouts of all the three homeologs for TaTDF, the wheat homolog of the Arabidopsis TDF1 (Tapetal Development and Function1), are required to completely abolish function and achieve male sterility [35].
2.3. Analysis of Male Fertility Genes Ms1, Ms5, TaMs26 and TaMs45
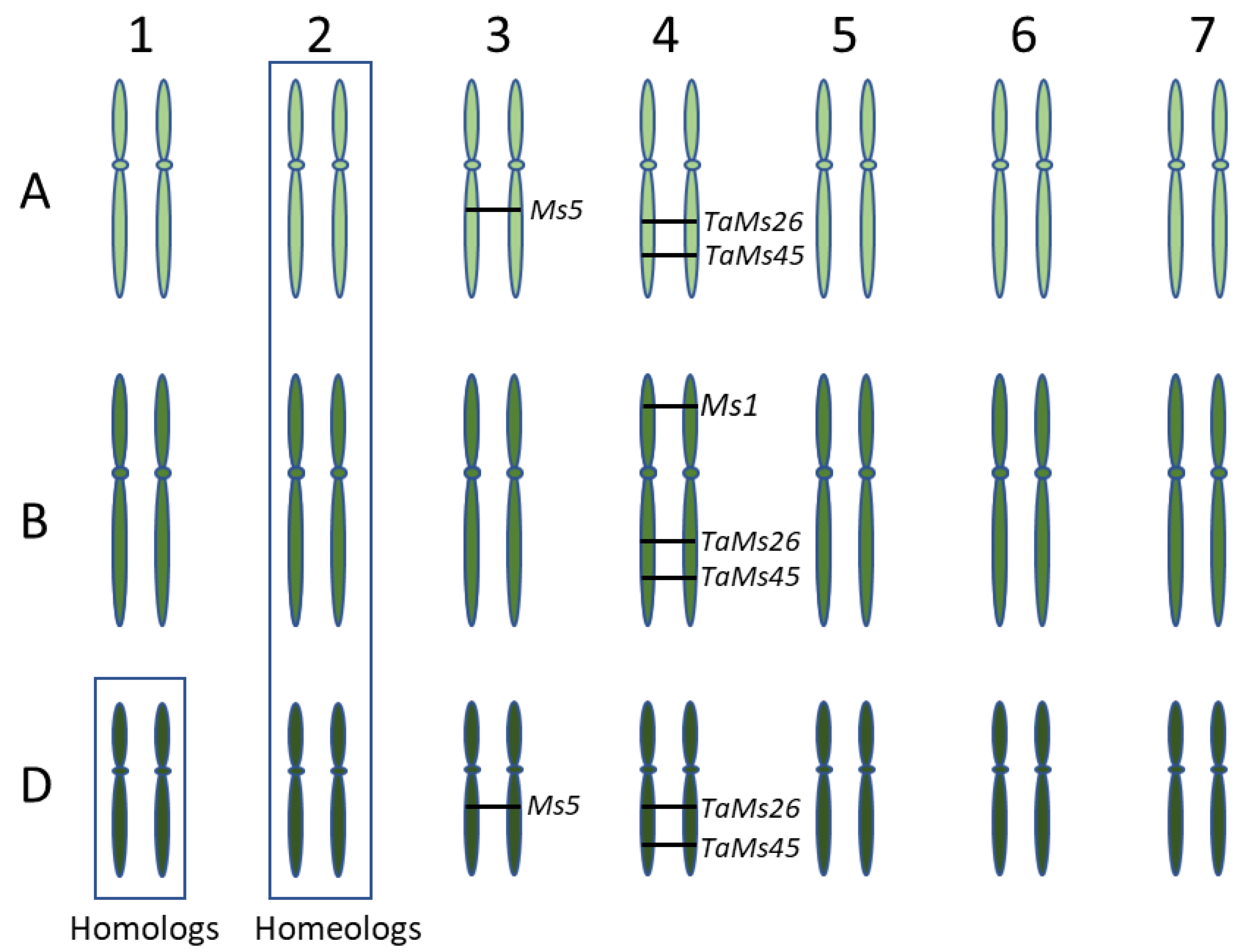
To understand the genetic basis of male fertility in wheat, we analyzed four genes, MS1, MS5, TaMS26 and TaMS45, that sporophytically contribute to male fertility in wheat (Table 2). The putative function of these genes suggests a role in tapetum function, and thus pollen development, and their loss of function causes defective tapetum and pollen development. MS1 and MS5 genes were identified through positional cloning of ms1 and ms5 mutants. TaMS26 and TaMS45 are homologs of the maize male fertility genes MS26 and MS45. MS1, TaMS26 and TaMS45 are located on the group 4 chromosomes, while MS5 is located on group 3 chromosomes (Figure 1). The number of functional homeologs varied from one to three for each the four wheat fertility genes (Figure 1).

Table 2.
Chromosomal location, functional homeologs and alleles, and number of alleles required for complementation of four male fertility genes in wheat.
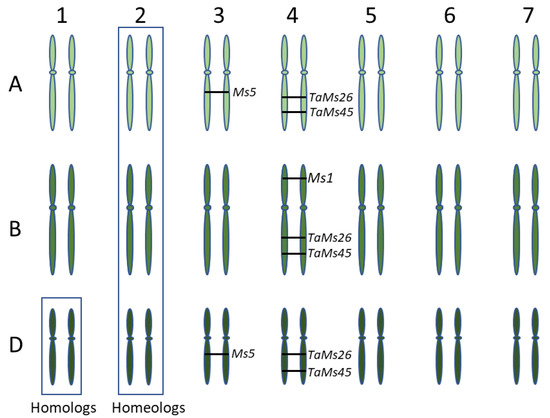
Figure 1.
Diagrammatic representation of the three wheat sub-genomes illustrating homologs and homeologs. The numbers at the top indicate the homeologous chromosome group, while alphabets on the left indicate the three sub-genomes. Position of functional homeologs of four wheat fertility genes, MS1, MS5, TaMS26 and TaMS45 is indicated.
Genetic studies with several ms1 alleles showed monogenic recessive segregation of male sterility attributed to chromosome 4 BS [2]. MS1 was cloned through positional cloning and was determined to encode a glycosylphosphatidylinositol-anchored lipid transfer protein (LTP), necessary for pollen exine development [27].
Gene expression analysis revealed that only the B-genome-derived TaMS1 homeolog is expressed during microspore development. Microscopic analysis and metabolomic profiling of anthers from ms1 mutants suggested the requirement of TaMS1 for either biosynthesis or transport of sporopollenin, and hence pollen exine formation. It was also demonstrated that an exogenous copy of TaMS1 completely restored fertility to the ms1d mutant, confirming TaMS1 as the causal gene for ms1 mutation [27]. Similarly, TaMS5-A, the gene responsible for ms5 phenotype, was also cloned using positional cloning, and like TaMS1, encodes a glycosylphosphatidylinositol-anchored LTP required for pollen exine development [31]. Phylogenetic analysis showed that MS1 and MS5 proteins are distinct from the other members of the wheat LTP super-family [51]. Sequence homology and gene expression pattern of MS5 were also unique to Triticeae and highly distinct from the homologs outside the Triticeae family [31]. Like ms1d, ms5 was also defective in exine formation of the microspores, although an irregular nexine structure was unique to the ms5 mutants. Like TaMS1, an extraneous copy of TaMS5-A homeolog was sufficient for complementation of ms5 mutants to restore male fertility [31].
The maize MALE STERILE26 (MS26) gene encodes a cytochrome P450 mono-oxygenase enzyme, CYP704B1 [52]. Subfamily CYP704 of the cytochrome P450s has an essential role in male fertility through hydroxylation of the fatty acid constituents of predicted sporopollenin precursors [21]. ms26 mutants are male sterile due to defective tapetum and microspores that lack sporopollenin deposition on the exine [52,53]. Gene expression analysis in wheat revealed that all three TaMS26 homeologs are expressed in the anthers from tetrad to early uninucleate microspore stages at comparable levels [32]. Mutations in the A, B and D homeologs of the putative MS26/CYP704B wheat gene were obtained utilizing a meganuclease [50]. Similar to the maize ms26 mutants, triple homozygous wheat mutants were male sterile due to defective pollen and anther development [32]. Expression and functional analysis confirmed the contribution of all three homeologs of TaMS26 towards male fertility in wheat. Interestingly, a single heterologous MS26/CYP704B gene was unable to restore fertility to a triple homozygous mutant background, but two heterologous genes completely restored male fertility [32].
The maize MALE STERILE45 (MS45) gene encodes a strictosidine synthase-like enzyme required for male fertility [54,55]. Analysis of CRISPR-Cas derived TaMS45 mutations demonstrated that all three homeologs contribute to male fertility and that triple homozygous mutants are required to abort pollen development and achieve male sterility [33]. Further, it was demonstrated that a single wild-type copy of the MS45 gene from rice or maize was able to restore fertility to the male sterile wheat mutants, suggesting a conservation of function between these species [33]. This conservation in expression and function was also utilized to complement the TaMS45 promoter inverted repeat (pIR)-based dominant male sterility through the expression of maize MS45 with maize or rice MS45 promoters [56].
2.4. Ms1, Ms5, TaMs26 and TaMs45 Exhibit Different Levels of Functional Redundancy
Bread wheat is an allohexaploid (2n = 6× = 42) consisting of three ancestral genomes (AABBDD), formed following two hybridization events between three progenitor species [57,58,59]. As a consequence of polyploidization and sub-functionalization, asymmetrical genetic contribution of the three subgenomes is an important characteristic in wheat [1,60,61]. Interestingly, the four male fertility genes we analyzed represented a complete spectrum of dosage of the three sub-genomes towards male fertility (Table 2). While MS1 and MS5 have one and two functional homeologs, respectively, TaMS26 and TaMS45 both have three functional homologs. Similarly, these genes also displayed a difference in the minimum number of wild-type alleles required for male fertility and the number of transformed wild-type genes required for complementation of their respective mutants (Table 2).
ms1 and ms5 both show monogenic inheritance; however, the underlying basis for this inheritance is different for both these genes, with ms5 presenting a more complex control of male fertility. In the case of MS1, sub-functionalization occurred due to inactivation of the A and D genome homeologs, with B genome being the only functional homeolog [27,62]. DNA methylation analysis of MS1 promoter of A, B and D homeologs, along with orthologs from allotetraploid and diploid wheat species, showed that the A and D homeologs are epigenetically silenced with only the B homeolog expressed in the anthers [62]. Therefore, a knockout of the B-genome homeolog is sufficient to abolish MS1 function and result in male sterility. However, variable penetrance of male sterility has been reported in various ms1 mutants [27]. This variation could be due to activation of the A- and D-genome homeologs, although this needs to be further investigated. For MS5, the predominant functional homeolog is TaMS5-A [31]. TaMS5-D exhibits has two allelic forms, one of which is non-functional, while the second form, although functional, displays incomplete dominance, suggesting reduced functionality [31]. Consistent with observed differences in function, the transcript abundance in developing anthers is lower for TaMS5-D than TaMS5-A. The non-functional form confers monogenic inheritance of male sterility, while the second form displays a two-gene inheritance [31]. Therefore, depending upon the TaMS5-D allele present, knockout of either one or two TaMS5 homeologs is required for achieving male sterility. For the TaMS5-B homeolog, two different non-functional alleles were observed in the 178 diverse hexaploid and tetraploid wheats analyzed, including several landraces and Triticum dicoccoides [31]. The prevalence of the non-functional TaMS5-B alleles in diverse hexaploid and tetraploid wheats suggests an early elimination of function of this homeolog during evolution. Thus, TaMS5 homeologs appear to have undergone two subsequent steps of sub-functionalization: an ancestral inactivation of TaMS5-B and a more recent reduced functionality of TaMS5-D.
Both TaMS26 and TaMS45 genes have three functional homeologs, as demonstrated by the requirement of triple knockouts to achieve a complete loss of function. However, for TaMS26, an in-depth characterization revealed differences in functional contribution of A, B and D homeologs [32]. In addition to the triple mutants, it was observed that double homozygous-single heterozygous mutants also exhibited male sterility but with varying levels of residual fertility (Figure 2). The fertility of these triple mutants was dependent upon the homeolog contributing the wild-type allele. This residual fertility was the highest in plants heterozygous for D genome and lowest in plants heterozygous for B genome (Figure 2). Genome A heterozygotes were intermediate for residual male fertility. Therefore, the B homeolog contributes the least towards male fertility, whereas the D homeolog contributes the most [32]. Thus, when a single wild-type allele of each homeolog was analyzed against five mutant alleles, differences appear in contribution of each genome. This also suggested that two wild-type alleles out of six TaMS26 alleles are required for complete male fertility, with the two alleles contributed by one genome or a combination of any two genomes [32]. Compared with TaMS26, only one wild-type allele is required for complete fertility TaMS45 [33]. This was evident from the analysis of fertility in double homozygous-single heterozygous TaMS45 mutant plants and from complementation analysis of triple mutants. This difference in the function of TaMS45 and TaMS26 highlights the functional differences between the homeologs of these genes, which could be ascribed to transcriptional, translational or enzymatic differences. Thus, MS1, MS5, TaMS26 and TaMS45 represent different aspects of polyploid biology, which is important to understanding how to create hybridization control systems.
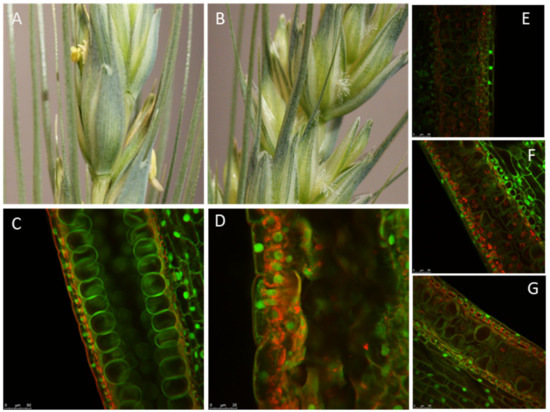
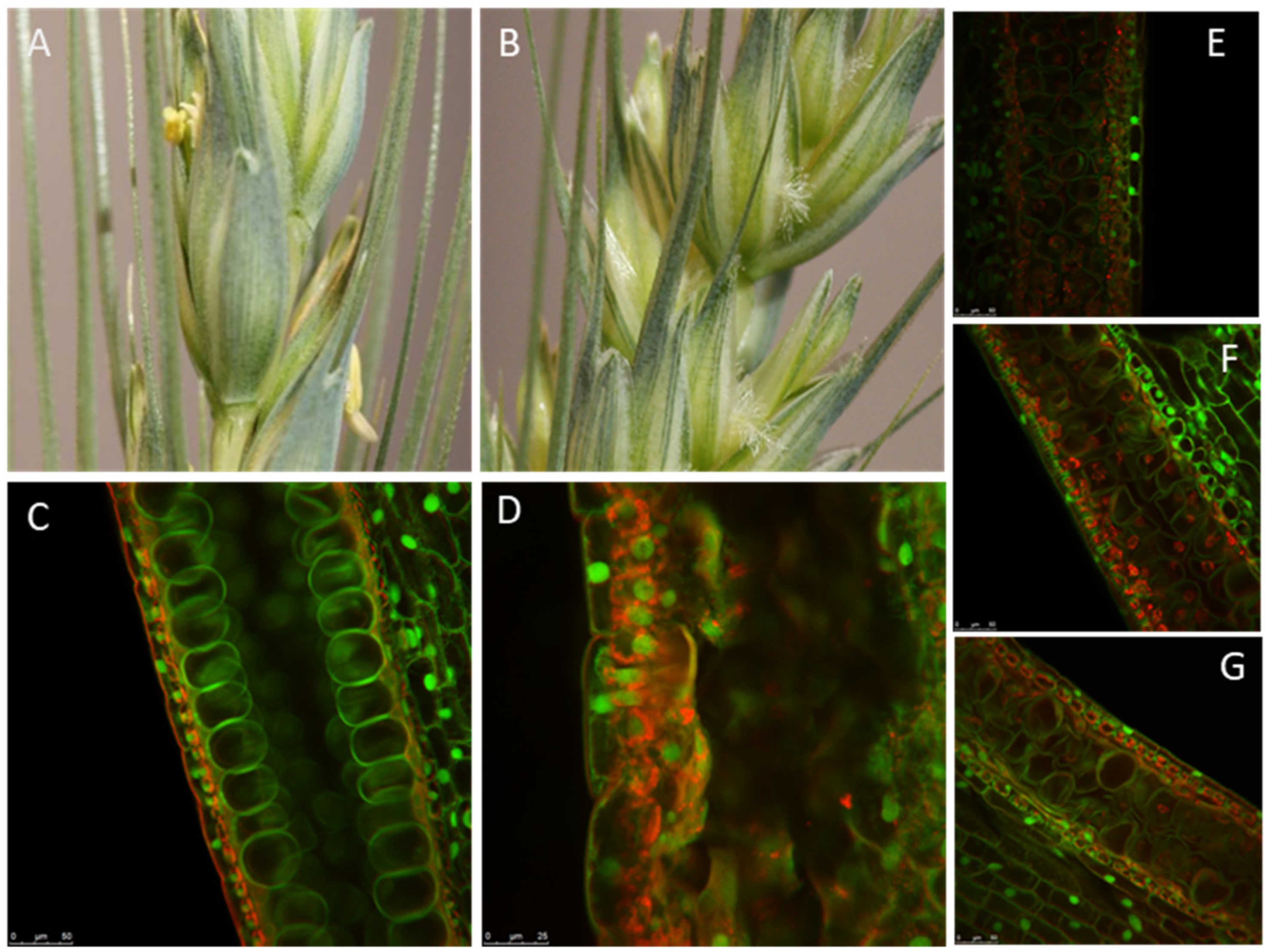
Figure 2.
Functional analysis of TaMS26 mutant wheat plants. Spikes from (A) wild-type and (B) Tams26-abd mutant plants. Microspores at late vacuolate stage from (C) wild-type TaMS26-ABD; (D) triple-recessive Tams26-abd. Double homozygous-single heterozygous: (E) Tams26-Aabd, (F) Tams26-aBbd and (G) Tams26-abDd plants. (E), (F) and (G) illustrate the differences in pollen morphology of double homozygous-single heterozygous mutants that are heterozygous for A-, B- and D-genomes, respectively. Scale bars = 25 μm. (Adapted from Singh et al. [32].)
2.5. Temperature and Photoperiod Influenced Male Fertility
In addition to the non-conditional genetic mutants described above, two types of environmentally influenced nuclear genes have also been reported in wheat that affect male fertility (Table 1). Thermo-sensitive genetic male sterility (TGMS) is controlled by genes that are influenced by temperature, while photoperiod- and temperature-sensitive genetic male sterility (PTGMS) is influenced both by temperature and photoperiod. TGMS and PTGMS variants have been mainly identified by wheat researchers in China. Four mutants have been reported thus far that can be classified in the TGMS category. YanZhan 4110S is one such line where male fertility is sensitive to temperature during late uninucleate stage of microspore development, with complete male sterility at temperature above 20 °C [38]. Two genes, TaMUT11 and TaSF3, have been reported to be associated with TGMS in YanZhan 4110S [63]. BNY-S is another TGMS line identified as a spontaneous mutant of the fertile wheat line BNY-F [39]. BNY-S exhibits male sterility at temperatures less than 10 °C during spikelet differentiation stage but is male fertile at temperatures higher than 10 °C. Genetic analysis indicated that sterility in BNY-S was controlled by a single recessive gene, wtms1 [39]. TGMS line tmsBS20T has also been used for hybrid wheat breeding in northern China [41]. Male sterility in tmsBS20T segregates as a single gene that was mapped to chromosome 2BL. BNS, another TGMS line identified in wheat, exhibits male sterility in the temperature range of 7.4 °C to 11.4 °C but is male fertile at temperature higher than 11.4 °C. [40,64].
In addition to TGMS mutants, several PTGMS lines have also been identified in wheat. BS366 is one such line that is the female parent of several hybrid varieties developed through two-line breeding system [44,65]. BS366 exhibits male sterility under a sterile condition of 10 °C with 12–14 h daytime during pollen development but fertility at 20 °C with 12–14 h daytime [44]. C49S, another PTGMS line, is also being utilized in two-line hybrid wheat development in China [45]. The critical temperature to induce complete male sterility is less than 13.5 °C, with near normal male fertility at 15 °C or higher. Two improved PTGMS lines, K78S and C412S, were subsequently derived from C49S [66,67]. BS210 is a PTGMS line that is male sterile between 10–12 °C and a photoperiod of 10–12.5 h during the critical stages of anther development. It exhibits up to 70% fertility at temperatures of 8–10 °C and 14 h photoperiod [43]. Male sterility in Xinong 291S (XN291S), another PTGMS line, is controlled by one or two recessive major genes [46].
3. Hybridization Systems for Hybrid Seed Production in Wheat
In the past six decades of hybrid wheat research, two main systems for hybridization have been the focus of wheat breeders, i.e., CHAs and CMS. However, NGMS-based hybridization systems can provide certain advantages over these two systems, which makes NGMS systems very attractive. The unique characteristics and the major challenges for each of these systems are discussed below.
3.1. CHA-Based Hybridization System
Chemical hybridizing agents are a class of chemical compounds, also known as gametocides, that can selectively induce male sterility without affecting female fertility (Figure 3). The major advantage of CHAs for hybrid seed production is the elimination of the need of any genetic manipulation for creating male sterile lines, which can effectively reduce the cost of goods to produce hybrid seed. The research for utilizing CHAs in wheat started with the first generation of CHAs, which included many commonly used growth hormones. However, these chemicals were highly variable in efficacy and exhibited phytotoxic effects [2]. This led to the discovery, mainly by the chemical industry, of much improved second generation CHAs [68]. Gametocide Hybrex (active substance RH 007 CHA) was invented by Rohm & Haas, compound SD 84811 was invented by Shell/Nickerson, Genesis® (active substance - MON 21200, Clofencet) was discovered by Monsanto, and Croisor® (active substance—Sintofen) was discovered by DuPont. These second generation CHAs, which are effective across a broad range of genotypes and have reduced phytotoxicity, have been reasonably utilized for hybrid seed production in wheat. This is evident from registration of seventy-four hybrid wheat varieties that were produced through the use of CHAs from 1996 to 2016 [68].

Figure 3.
Utilization of CHAs for hybrid seed production in wheat. The rows with white bags represent the male-sterile female parent treated with CHA. The unbagged rows represent the male-fertile male parent. (Photograph courtesy of Bill Curran, CortevaTM Agriscience).
Still, the use of CHAs is limited due to a number of factors that restrict their large-scale adoption. The narrow window of CHA application, which can be severely affected by prevailing environmental conditions, is the major limitation to utilizing CHAs. This can make the application of CHAs challenging and therefore risky for seed production. Emergence of late tillers that can escape application of CHAs adds to that risk of contamination with self-pollinated seed. Phytotoxicity associated with CHAs also necessitates dosage optimization for diverse inbreds. Another key drawback of a CHA-based system is the inability to utilize blend planting of male and female parents for hybrid seed production (Figure 3). With the current available technologies, strip planting of male and female lines is the only option for a CHA-based hybridization system, as opposed to male and female blend planting proposed for genetic hybridization systems to maximize cross pollination [69,70]. The restriction of using strip planting necessitates the development of super males with exceptional male characteristics, to get optimal hybrid seed production. Consequently, current hybrid wheat breeding programs utilizing CHAs rely on a few male inbreds [68]. However, with the advent of advanced technologies for pollen preservation and application, it may be possible to increase cross pollination even for CHA-based hybridization systems [70]. Nevertheless, even with the most effective CHAs, any variation due to plant growth or application makes utilization of CHAs for hybrid seed production challenging.
3.2. Genetic Hybridization Systems
The genetic hybridization systems rely on genetic factors for the induction of male sterility, without the need for any external manipulation or application. Two main types of genetic male sterility systems have been utilized for creating hybridization systems: CMS and NGMS.
3.2.1. CMS Hybridization System
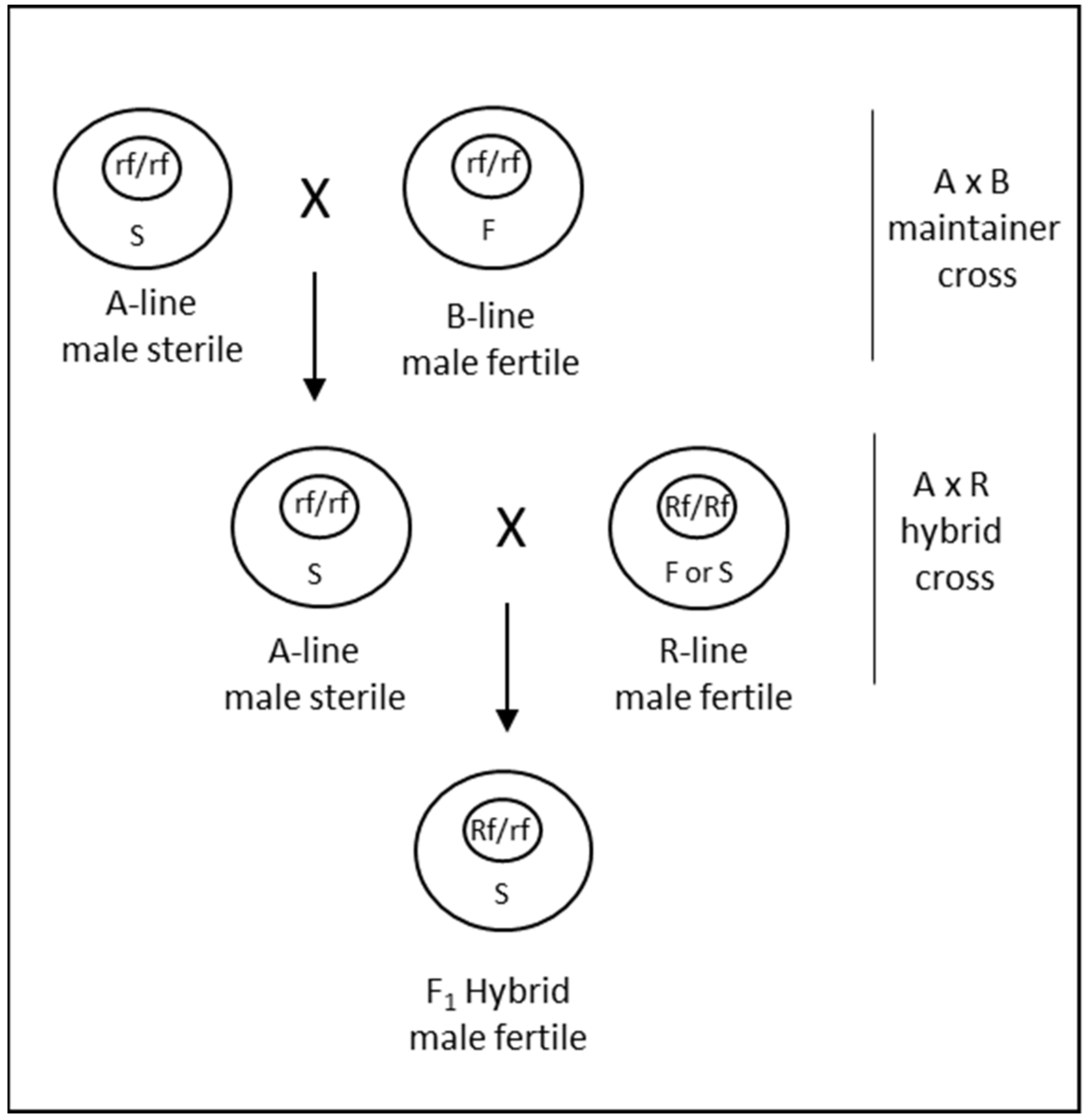
Cytoplasmic male sterility occurs due to the interaction of nuclear and cytoplasmic genetic factors [71]. This generally occurs when cytoplasm from a distantly related species is combined with the nucleus of another species, resulting in an alloplasmic line. The male sterility can be reversed with restorer of fertility (Rf) genes, which in many instances are derived from the species that contributed the cytoplasm. In addition to the male sterile A-line and the male fertile R-line, an additional B-line is also required for the maintenance of male sterility through A- × B-line cross (Figure 4). Thus, a CMS system is a 3-line system that requires cross-pollination at two steps to produce hybrid seed. Since its discovery in wheat in 1951 [72], CMS has been extensively researched with the aim of utilizing it for commercial seed production. The cytoplasms of various wheat-related species have been combined with the nucleus of T. aestivum to investigate CMS. Tsunewaki et al. [73] analyzed 46 cytoplasms transferred to wheat from Triticum and Aegilops species, of which 31 could induce partial to complete male sterility. Other studies have also demonstrated the ability of cytoplasm from many species to induce CMS when combined with the T. aestivum nucleus [74]. Due to the stability of male sterility, predominance of recessive alleles for restoration and identification of corresponding dominant restoration factors, CMS induced by T. timopheevii Zhuk. has been primarily considered for commercial hybrid seed production in wheat [69]. Several hybrid varieties were commercialized from the late 1970s to the 1990s utilizing a timopheevii CMS system [74].
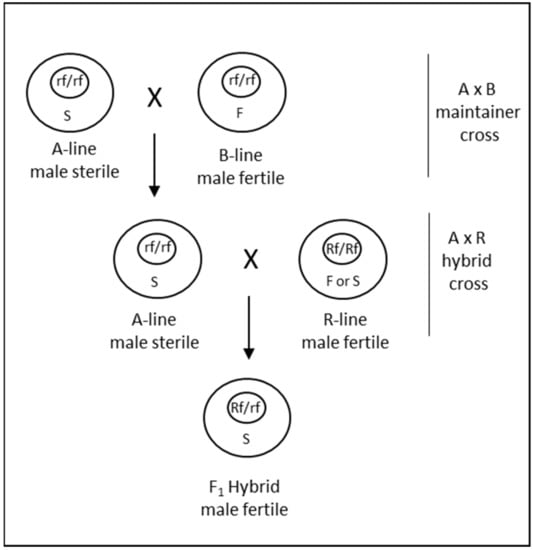
Figure 4.
Diagrammatic representation of the CMS system for hybrid seed production in wheat. The bigger outer and the smaller internal circles represent the cytoplasm and the nucleus, respectively. S and F denote the sterile and fertile cytoplasms; Rf and rf denote the fertility restoring and non-restoring alleles of the restorer gene, respectively. A, B and R lines represent the male-sterile female parent, the male-fertile female isogenic maintainer and the restorer male parent, respectively.
However, there are certain aspects of CMS systems, including timopheevii CMS, that render these systems more complex for hybrid seed production in wheat. First, various studies have revealed that combinations of two or three and perhaps more Rf genes are needed for complete fertility restoration of CMS in wheat [75,76,77]. To add to that complexity, epistatic interactions among Rfs and between Rfs and modifier loci have also been highlighted by several studies [78]. Consequently, it is required to stack multiple Rf genes in a genotype-dependent combination to achieve complete fertility of F1 hybrids. In the past, stacking and breeding of restorer genes has proved challenging, although with contemporary genomics and molecular breeding techniques it is now possible to precisely track Rf genes for breeding [79]. Another major limitation for utilizing a CMS for hybrid seed production is the requirement for A × B crossing for female parent seed increase (Figure 4). The strip plots used for A × B female-increase limit the efficiency of female inbred multiplication. For such a maintainer crossing, it is necessary that the B line should also have good male characteristics. This adds another layer of complexity to breeding for female parents. Several undesirable pleiotropic effects that are environment-dependent have also been reported for timopheevii cytoplasm, including shriveled F1 seed [13]. All these factors can lead to the increased cost of goods for hybrid seed production, which is likely the reason that currently no hybrid wheat varieties that were produced utilizing a CMS system are commercially available.
3.2.2. Hybridization Systems Based on NGMS
An alternative to the utilization of CHAs or CMS are the systems based on NGMS (Table 3). There are two major advantages of hybridization systems based on the recessive NGMS. First, no genetic manipulation, trait introgression or breeding is required of the male inbreds for fertility restoration. Since most of the mutants or variants of the nuclear genes are recessive in nature, restoration of fertility can be achieved in the heterozygous F1 hybrid. This is also true in cases where triple homeolog mutants are required to generate male sterile lines. Therefore, in terms of restoration of male sterility, NGMS is much simpler compared with CMS. Dominant or semi-dominant NGMS systems, however, require a more complex strategy for restoration of male fertility. The second advantage of NGMS based hybridization systems is the propagation or maintenance of the male-sterile female lines through self-pollination of the maintainer lines. As mentioned previously, maintenance of male sterile plants in an A × B crossing scheme for CMS adds complexity to the hybridization system and increases the cost of goods. Thus NGMS, including both conditional and non-conditional, can offer an advantage for a hybridization system in wheat.
TGMS- and PTGMS-Based Hybridization Systems
While several TGMS and PTGMS lines have been characterized in wheat (Table 1), few are being utilized for hybrid seed production [70]. BS366 is an important PTGMS line for a hybrid wheat breeding program in China and is the female parent of Jingmai-series hybrid wheat varieties that include Jingmai 7 (JM7), Jingmai 8 (JM8) and Jingmai 9 (JM9) [44,65]. A major concern for large-scale implementation of TGMS and PTGMS is the effect of sudden changes in temperature and/or photoperiod on male sterility and fertility of the female lines. Any leakiness of sterility due to temperature or photoperiod fluctuations will result in contamination of hybrid seed with female parent seed. Similarly, any variation from the required conditions can reduce the seed set of female inbreds during maintenance, resulting in supply chain issues. Spreading seed production over a number of locations can alleviate the risks associated with temperature or photoperiod variation. However, the number of such locations may be limited due the narrow range of climatic zones where the hybrid seed production and female maintenance can occur. Penetrance of sterility in TGMS/PTGMS variants may also be a concern for their utilization for hybrid seed production. It is reported that under sterile conditions PTGMS lines BS366 and BS210 exhibit 95–100% male sterility, suggesting up to 5% fertility under sterile conditions [43,44], which can impact purity of hybrid seed. Another issue, which appears to be wheat specific, is the manifestation of TGMS and PTGMS, in general, at lower temperature, with fertility occurring at higher temperatures. This contrasts with the classical TGMS mutants in other crops, where TGMS manifests at higher temperatures [80]. Observations similar to wheat have also been reported in barley [81], suggesting that this may be a common feature of the Triticeae family. This feature can be an obstacle to utilizing TGMS and PTGMS for hybrid seed production in wheat since both the spring and winter wheat flower in spring when the daily temperatures are rising. This can decrease the penetrance of male sterility and enhance reversion to fertility of female lines. Restoration of sterility in F1 hybrids generated through TGMS and PTGMS can also be complex [44,82]. Thus, the TGMS and PTGMS systems have several drawbacks that may limit their application for hybrid seed production in wheat.
Hybridization Systems Based on Non-Conditional Recessive NGMS
Following the discovery of the first nuclear male sterile mutant in wheat [48], the first NGMS hybridization system was proposed in 1972 [83]. This system, known as the XYZ, can be described as the first-generation concept for utilizing NGMS for hybrid seed production. It was based on a recessive male-sterility mutant, ms, that could be complemented with a homeologous chromosome derived from a wheat-related species to render it male fertile. X, Y and Z, the three lines required for maintenance for male sterile females, are all homozygous for the male sterile mutation (ms/ms) but carry 2, 1 and 0 doses of the alien chromosome, respectively [83]. In a hypothetical example, Driscoll considered a male sterile mutant of homeologous group 5 for wheat that could be complemented with the addition of rye chromosome 5R, which also carries a marker gene for hairy peduncle [83]. While this concept was a major step forward for utilizing NGMS for hybrid wheat seed production, the selection of ms/ms plants at a vegetative stage would be difficult to implement in the field. A modified XYZ was later proposed that did away with the need for the X component [84]. However, this system also required a Z × Y crossing step, similar to the A × B cross in the CMS system, for the maintenance of the male sterile female, thus making it a 3-line system (Table 3).

Table 3.
Hybridization system concepts for wheat based on non-conditional NGMS.
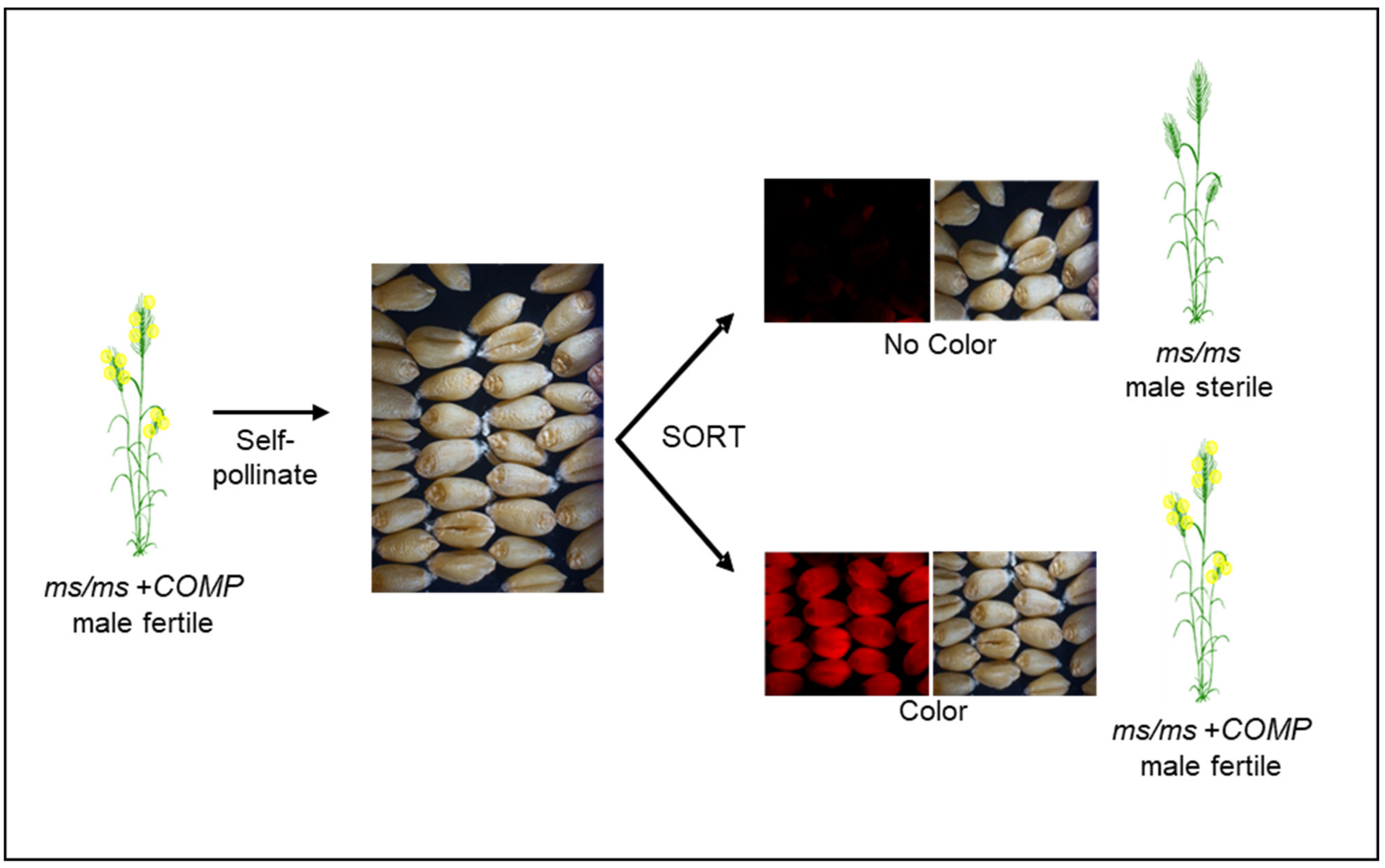
Since the mid-2000s, several groups have refocused attention on the development of NGMS-based systems that can be described as second-generation concepts for utilizing NGMS. Zhou et al. [85] devised the 4E-ms system, which is a further modification the XYZ system (Table 3). The selection for ms/ms plants is based on seed phenotype, which predicts the genotype of the plant derived from that seed (Figure 5). In this system, the ms component, a mutant allele of the MS1 gene, was combined with the complementation component, 4E chromosome from Agropyron elongatum ssp. Ruthenicum, essentially creating a monosomic addition line [85]. Chromosome 4E also has the BLUE ALEURONE (BA) gene, which imparts a light blue color to the seed containing the 4E monosomic. In a working system, the background is always ms1/ms1 for the ms component, but the plants are fertile due to presence of the 4E chromosome. A plant derived from a light blue-colored seed, upon self-pollination, segregates for three categories of seed—red, light blue and blue carrying, 0, 1 and 2 doses of the 4E chromosome [85]. The proportion of red and blue seeds is about 65% and 35%, respectively, where the red seed represents the male sterile females for hybrid seed production, while the light blue seed serves as the female maintainer line.
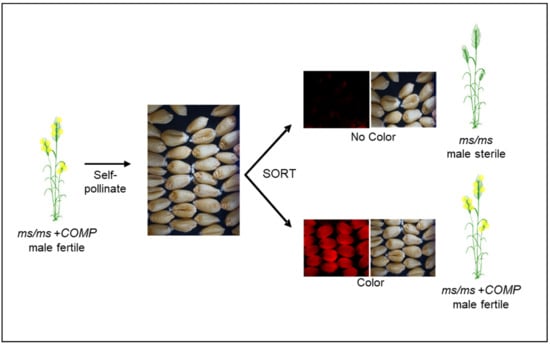
Figure 5.
Diagrammatic representation of a NGMS system based on seed color marker for hybrid seed production in wheat. The ms/ms and COMP denote the male sterility and complementing components, respectively. The male sterility and complementation components including seed color can originate from different sources that can be combined through genetic manipulation.
Although an excellent concept, the commercial use of the 4E-ms system has not been reported. This may be due to a few inherent shortcomings of this system that relate both to the ms and the 4E complementation components. The first issue can be attributed to the location of MS1 and BA components on the 4E chromosome. If the synteny of genes holds true between the wheat group 4 chromosomes and 4E, the MS1 and BA gene are likely to be on opposite arms. It is known that the monosomics form univalent chromosomes during meiosis and tend to break at centromeres during meiosis. Therefore, it is possible that due to breakage at the centromere, MS1 and BA can be unlinked, resulting in segregation and thus misclassification of red and blue seed. This can result in red seeds producing male fertile plants and male sterile plants derived from blue seeds. Several studies have also suggested variation in the penetrance of ms1 mutations [27,90,91]. This could also lead to the presence of female inbred seed in the hybrid seed, depending on the penetrance of the ms component. Finally, the efficiency of such a system relies on the ability to sort various classes of seeds, i.e., red, light blue and dark blue seeds, particularly the ability to efficiently remove blue seeds disomic for 4E chromosome. Any environmental or genotypic variation in blue color will further increase the complexity of seed sorting, thus reducing the efficiency of this system.
Seed production technology (SPT) [55], initially developed for corn at CortevaTM Agriscience, can mitigate the linkage issue associated with the 4E-ms system (Figure 5). In the SPT system, the complementation component and the seed marker reside on the same TDNA insertion, ensuring absolute linkage of these components. The pollen-specific expression of alpha-amylase gene, another component of the TDNA, prevents the transmission of TDNA construct through pollen. Therefore, the issue of homozygous TDNA, equivalent to the disomics in 4E-ms system, is not encountered. When a ms/ms plant hemizygous for the TDNA is self-pollinated, all the seeds have a ms/ms genotype, half of which carry the TDNA, with the other half being devoid of TDNA. Each class of these seeds can be sorted based on the seed marker. ms/ms seeds without TDNA will produce a male-sterile female parent to cross with a male parent for hybrid seed production (Figure 5). An important aspect of this system is that the female parent, and thus the hybrid seed and commercial grain, is devoid of TDNA and is therefore SPT transgene-free.
Based on the maize SPT concept, we have assembled wheat hybridization systems with two male sterility mutants, ms1, and Tams26 (Table 3). Additionally, components for two more systems based on ms5 and Tams45 mutants have been determined. The first such hybridization system was reported for the TaMS26 gene [32]. Homozygous Tams26 mutants formed the ms component, whereas the complementing piece was provided by a TDNA cassette that contained a ZmMS26-OsMS26 gene combination, DSRED as a seed marker and a pollen-specific alpha-amylase gene [32]. When Tams26 plants with TDNA cassette were self-pollinated, the obtained seeds segregated approximately 50:50 for with and without a TDNA cassette (Figure 5). All Tams26 plants that carried a TDNA cassette were male fertile and set seed comparable with wild-type plants; all plants without the T-DNA cassette were male sterile [32]. The TDNA carrying seeds could be sorted by a seed sorter appropriate for the seed marker. (Figure 3). A similar system was established with ms1 as the ms component and a TDNA cassette containing TaMS1, DSRED and the pollen-specific alpha-amylase gene as the complementing piece (M. Singh, M. Albertsen, M. Cigan, unpublished results). ms1/ms1 plants with a TDNA cassette are completely fertile and upon self-pollination produce seeds that segregate approximately 50:50 for with and without TDNA. All the plants produced from seeds without the seed marker were male sterile, while those from seeds with the marker were fertile. ms1/ms1 seeds without the TDNA generated a male-sterile female parent to be crossed to a male parent for hybrid seed production (Figure 3). In addition to ms1 and Tams26 systems, the complementation component has been demonstrated for ms5 and Tams45 mutants [31,33]. These proof-of-concept studies showed that a SPT-like hybridization system is feasible in wheat.
Another hybridization system for wheat has been described by [86] through tapetum-specific expression of the phytotoxic barnase gene. The barnase gene was split into two components, each of which is placed at the same locus in separate plants, thus representing two alleles. Male sterility is induced when the two components or alleles of the barnase gene are brought together through crossing [86]. The major advantage of this system is that, like the systems based on recessive mutants, no restorer genes are required; therefore, genetic modification of male parent is not needed. However, the maintenance of this system requires an A x B like crossing scheme akin to a 3-line CMS system. The F1 hybrids still carry the transgene in a heterozygous state and therefore will be regulated as a transgenic trait.
Another hybridization system proposed for wheat is based on the chemical complementation of mutants in known biochemical pathways. Sedlacek [37] demonstrated that the inactivation of TaSPPS, a gene involved in the biosynthesis of sporopollenin, results in male sterility. This sterility can be reversed through application of 4-oxo-6-octadecane-pyran-2-olate at the tetrad stage of pollen development [37]. Like TGMS and PTGMS, such a system will be considered a classical 2-line hybridization system. However, there are no reports of this system being implemented at a commercial scale for hybrid seed production, with one of the hurdles to overcome being the registration of the chemical for commercial use.
Recently, a few hybridization systems have been proposed that seek to build up on the first- or second-generation hybridization concepts (Table 3). These concepts specifically aim to tighten the linkage between the complementation and the selection/sorting components through either native or gene editing techniques. Ms45-BA is a hybridization system based on the mutations in the TaMS45 gene as the ms component, and the telosome for long arm of 4E (t4EL) from Thinopyrum ponticum as the complementation piece [87]. It was previously shown that mutations in all three homeologs of the TaMS45 gene are required for achieving male sterility [33]. It was further shown that the Tams45 triple mutants can be complemented with heterologous MS45 genes from maize and rice, suggesting the potential of MS45 genes from a diverse range of species to complement Tams45 mutants [33]. Chromosome 4EL of Thinopyrum ponticum is homologous to chromosome 4L of wheat [92], where TaMS45 is located. Because of this homology, it was anticipated that 4EL should carry a homolog of the TaMS45 gene in addition to the BA gene for blue aleurone. Studies in our group demonstrated that the t4EL can both complement the Tams45 triple mutants and impart blue color to the seed [87]. Both these components are tightly linked due to the lack of recombination between t4EL and the wheat 4L group and the low probability of chromosome breakage that occurs with monosomic addition lines. Self-pollination of Ms45-BA plants yields red and blue seed ratios similar to those described for 4E-ms [85].
In addition to the Ms45-BA system, several concepts for hybridization systems in wheat have been proposed that rely heavily on chromosome engineering. Two hybridization concepts have been proposed to further modify 4E-ms system by moving the MS1 and BA components closer for a tighter linkage, either by conventional genetic techniques or utilizing gene editing technology [87,88]. Sedlacek and Horcicka [14] have proposed an updated XYZ hybridization system based on the TIP2 gene as the ms component and the barley 7H chromosome as the complementation component. The proposed system utilizes the amylose profiles of the wheat waxy null-mutants and the 7H monosomic addition line, which has a WT waxy allele, to sort for seed [93]. The male sterile line increase is proposed by crossing to the 7H fertile maintainer in an A × B crossing scheme, similar to a 3-line system. The self-pollination of the 7H addition line for maintainer increase, however, could pose problems, as the seed with 0, 1 and 2 doses of the monosomic will have to be sorted into each class to eliminate disomic seeds. Another hybridization concept has been recently proposed that relies heavily on gene editing technology [89]. The MFW-PV-OV Maintainer Line concept intends to create male-fertile maintainer lines that can be propagated by self-pollination without the need for seed or plant sorting [89]. However, the male sterile lines must be crossed to male-fertile maintainer plants in an A x B-like crossing scheme similar to a 3-line system. For the wheat version of this system, it is proposed to knockout 5–6 alleles of three genes, MFW, PV and OV, along with intra-chromosomal deletions.
Hybridization Systems Based on Dominant NGMS
Based on the SPT concept, two dominant male sterility systems have also been developed [56]. Both of these systems utilize the techniques of transcriptional gene silencing through promoter inverted repeats (pIR), to either create male sterility or to restore male fertility [94,95]. In the MS45-pIR system, the ms component is the pIR designed to be the promoter of the TaMS45 gene, which results in dominant male sterility [56]. This sterility can be complemented through the expression of the MS45 gene with a heterologous promoter from another plant species with an expression profile similar to the TaMS45. A second dominant system utilizes the expression of the DNA (Adenosine-N6-)-Methyltransferase (DAM) gene in a tapetum-specific manner [56]. The DAM-induced male sterility can be restored through the expression of pIRs designed to be the promoter directing the expression of the DAM gene. Both the MS45-pIR and the DAM hybridization systems can be maintained with a SPT-like TDNA that has the suppression of male sterility component as the complementation piece, along with a seed marker gene and pollen-specific alpha amylase gene. When a MS/MS plant hemizygous for the TDNA is self-pollinated, all the seeds have a MS/MS genotype but segregate approximately 50:50 for with and without TDNA, which can be sorted based on the seed marker. MS/MS seeds without TDNA will produce a male sterile female line to be crossed to a male parent for hybrid seed production, while the seeds with TDNA will produce plants in which male sterility is suppressed and thus are male fertile (Figure 3). As opposed to the recessive systems, the male parent needs to be genetically modified for restoration of fertility [56].
4. Role of Gene Editing in Developing Next Generation Wheat Hybridization Systems
Gene and genome editing utilizing the emerging tools can speed up crop improvement. Wheat, due its polyploid nature and a large genome size, can especially benefit from these technologies (reviewed in [96]). The utility of gene editing in creating hybridization systems in wheat is immediately evident from the number of triple knockouts of male fertility genes generated for TaMS26, TaMS45, TaNP1 and TaTDF. For TaMS45, the utility of the CRISPR/Cas system was effectively demonstrated through concurrent modifications in all three homeologs for two genotypes, thus allowing simultaneous testing of male sterility in two genetic backgrounds [33]. This demonstrated the utility of gene editing for quickly screening for sources of NGMS that are stable across genotypes. With these editing technologies in mind, researchers are proposing more complex NGMS systems that would not be feasible using the traditional genetics and cytogenetics techniques. The modified 4E-ms1 and MFW-PV-OV are examples of two such systems that will require chromosome engineering, with the MFW-PV-OV system requiring gene edits in three genes in addition to large chromosomal deletions [89]. For these proposed hybridization systems, including those utilizing three homeolog edits, trait introgression to different inbreds will be complex and challenging. For introgression of such hybridization systems, one approach would be to leverage the power of genome editing to introduce the hybridization systems de novo to new inbreds, as demonstrated for TaMS45 [33]. For such a large-scale trait introgression, the efficiency of gene editing will need to be substantially improved. While wheat still lags other model plant species such as Arabidopsis and rice in the utilization of CRISPR-based genome modifications, a couple of recent technological advances point to the maturing of this technology in wheat [97,98]. Next-generation monocot plant transformation systems [99] will also lead to greater efficiency of gene editing in wheat.
5. Conclusions
Even though immense progress has been made in understanding the genetic basis of male sterility in wheat, a hybridization system that combines the benefits of lower cost of goods and efficiency has not yet been implemented. NGMS offers potential advantages over CMS with respect to complexity of breeding and hybrid seed production. However, currently all NGMS based systems, particularly those based on non-conditional male sterility, are in the early stages of development, i.e., these are concepts or at proof-of-concept stages. Nevertheless, even at an early stage of development, these concepts look very promising. Some of the complex genetic manipulations that were not possible with traditional genetics or molecular techniques can now be performed through gene editing. Along with a good hybridization system, other important aspects of wheat hybrid seed production that will lower the cost of goods also need to be considered. Technologies and breeding methods to create heterotic pools and enhance cross pollination also need to be developed alongside hybridization systems.
Author Contributions
Conceptualization, M.S. and A.M.C.; writing—original draft preparation, M.S.; writing—review and editing, M.S., A.M.C. and M.C.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank Tom Greene, Sr. Research Director, Trait Discovery and Biotechnology, CortevaTM Agriscience, for his continued support and leadership. The authors would also like to thank members of the Hybrid Platforms group at CortevaTM Agriscience for helpful discussions leading to the preparation of this manuscript.
Conflicts of Interest
M.S., M.C.A. and A.M.C. are listed as inventors on one or more of the following patent applications—WO2020/056259A1, WO2014164961A3, WO2016048891A1, WO2019118342A1, WO2016100309A1.
References
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.A.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Whitford, R.; Fleury, D.; Reif, J.C.; Garcia, M.; Okada, T.; Korzun, V.; Langridge, P. Hybrid breeding in wheat: Technologies to improve hybrid wheat seed production. J. Exp. Bot. 2013, 64, 5411–5428. [Google Scholar] [CrossRef]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Longin, C.F.H.; Mühleisen, J.; Maurer, H.P.; Zhang, H.; Gowda, M.; Reif, J.C. Hybrid breeding in autogamous cereals. Theor. Appl. Genet. 2012, 125, 1087–1096. [Google Scholar] [CrossRef] [PubMed]
- Mette, M.F.; Gils, M.; Longin, C.F.H.; Reif, J.C. Hybrid Breeding in Wheat. In Advances in Wheat Genetics: From Genome to Field, Proceedings of the 12th International Wheat Genetics Symposium, Yokohama, Japan, 8–14 September 2013; Ogihara, Y., Takumi, S., Handa, H., Eds.; Springer: Tokyo, Japan, 2015; pp. 225–232. [Google Scholar]
- Gowda, M.; Longin, C.F.H.; Lein, V.; Reif, J.C. Relevance of Specific versus General Combining Ability in Winter Wheat. Crop Sci. 2012, 52, 2494–2500. [Google Scholar] [CrossRef]
- Longin, C.F.H.; Gowda, M.; Mühleisen, J.; Ebmeyer, E.; Kazman, E.; Schachschneider, R.; Schacht, J.; Kirchhoff, M.; Zhao, Y.; Reif, J.C. Hybrid wheat: Quantitative genetic parameters and consequences for the design of breeding programs. Theor. Appl. Genet. 2013, 126, 2791–2801. [Google Scholar] [CrossRef] [PubMed]
- Muhleisen, J.; Maurer, H.P.; Stiewe, G.; Bury, P.; Reif, J.C. Hybrid Breeding in Barley. Crop Sci. 2013, 53, 819–824. [Google Scholar] [CrossRef]
- McRae, D.H. Advances in chemical hybridization. Plant Breed. Rev. 1985, 3, 169–191. [Google Scholar]
- Kempe, K.; Gils, M. Pollination control technologies for hybrid breeding. Mol. Breed. 2011, 27, 417–437. [Google Scholar] [CrossRef]
- Wilson, J.A.; Ross, W.M. Male Sterility Interaction of the Triticum Aestivum Nucleus and Triticum Timopheevi Cytoplasm; Kyoto University: Kyoto, Japan, 1962. [Google Scholar]
- Mukai, Y.; Tsunewaki, K. Basic studies on hybrid wheat breeding. Theor. Appl. Genet. 1979, 54, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Adugna, A.; Nanda, G.S.; Singh, K.; Bains, N.S. A comparison of cytoplasmic and chemically-induced male sterility systems for hybrid seed production in wheat (Triticum aestivum L.). Euphytica 2004, 135, 297–304. [Google Scholar] [CrossRef]
- Sedlacek, T.; Horcicka, P. Proposal of updated XYZ system for the production of hybrid wheat seed. Czech J. Genet. Plant 2019, 55, 35–38. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, L. Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 2014, 17, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Quilichini, T.D.; Grienenberger, E.; Douglas, C.J. The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry 2015, 113, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Toriyama, K. Genetic Regulation of Sporopollenin Synthesis and Pollen Exine Development. Annu. Rev. Plant Biol. 2011, 62, 437–460. [Google Scholar] [CrossRef] [PubMed]
- Kaul, M.L.H. Genic male sterility. In Male Sterility in Higher Plants; Springer: Berlin, Germany, 1988; pp. 155–194. [Google Scholar]
- Ma, H. Molecular genetic analysis of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005, 56, 393–434. [Google Scholar] [CrossRef] [PubMed]
- Wilson, Z.A.; Zhang, D.-B. From Arabidopsis to rice: Pathways in pollen development. J. Exp. Botony 2009, 60, 1479–1492. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Cui, M.; Yang, L.; Kim, Y.-J.; Zhang, D. Genetic and Biochemical Mechanisms of Pollen Wall Development. Trends Plant Sci. 2015, 20, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Suneson, C.A. Use of Pugsley’s sterile wheat in cross breeding. Crop Sci. 1962, 2, 534–535. [Google Scholar] [CrossRef]
- Fossati, A.; Ingold, M. A male sterile mutant in Triticum aestivum. Wheat Inf. Serv. 1970, 30, 8–10. [Google Scholar]
- Driscoll, C.J. Induction and use of the ‘‘Cornerstone’’ male sterility in wheat. In Proceedings of the 5th International Wheat Genetics Symposium, New Delhi, India, 23–28 February 1978; pp. 499–502. [Google Scholar]
- Klindworth, D.L.; Williams, N.D.; Maan, S.S. Chromosomal location of genetic male sterility genes in four mutants of hexaploid wheat. Crop Sci. 2002, 42, 1447–1450. [Google Scholar] [CrossRef]
- Zhou, K.; Wang, S.; Feng, Y.; Ji, W.; Wang, G. A new male sterile mutant LZ in wheat (Triticum aestivum L.). Euphytica 2008, 159, 403–410. [Google Scholar] [CrossRef]
- Tucker, E.J.; Baumann, U.; Kouidri, A.; Suchecki, R.; Baes, M.; Garcia, M.; Okada, T.; Dong, C.; Wu, Y.; Sandhu, A.; et al. Molecular identification of the wheat male fertility gene Ms1 and its prospects for hybrid breeding. Nat. Commun. 2017, 8, 869. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.; Qi, J.; Hao, Q.Q.; Lyu, B.; Luo, M.C.; Wang, Y.; Chen, F.J.; Wang, S.Y.; Zhang, C.Z.; Epstein, L.; et al. Wheat Ms2 encodes for an orphan protein that confers male sterility in grass species. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Guttieri, M.J. Ms3 dominant genetic male sterility for wheat improvement with molecular breeding. Crop Sci. 2020, 60, 1362–1372. [Google Scholar] [CrossRef]
- Maan, S.; Kianian, S. Third dominant male sterility gene in common wheat. Wheat Inf. Serv. 2001, 93, 27–31. [Google Scholar]
- Pallotta, M.A.; Warner, P.; Kouidri, A.; Tucker, E.J.; Baes, M.; Suchecki, R.; Watson-Haigh, N.; Okada, T.; Garcia, M.; Sandhu, A.; et al. Wheat ms5 male-sterility is induced by recessive homoeologous A and D genome non-specific lipid transfer proteins. Plant J. 2019, 99, 673–685. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, M.; Thilges, K.; Cho, M.-J.; Cigan, A.M. MS26/CYP704B is required for anther and pollen wall development in bread wheat (Triticum aestivum L.) and combining mutations in all three homeologs causes male sterility. PLoS ONE 2017, 12, e0177632. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, M.; Albertsen, M.C.; Young, J.K.; Cigan, A.M. Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum Aestivum L.). Plant Mol. Biol. 2018, 97, 371–383. [Google Scholar] [CrossRef]
- Li, J.; Wang, Z.; He, G.M.; Ma, L.G.; Deng, X.W. CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterility in bread wheat. J. Genet. Genom. 2020, 47, 263–272. [Google Scholar] [CrossRef]
- Schmidt, K.; Koch, K.; Molitor, A.; Verstegen, H. Male-Sterile Plant of the Genus Triticum. International Patent Application No WO2016156583A1, 6 October 2016. [Google Scholar]
- Milner, M.J.; Craze, M.; Bowden, S.; Bates, R.; Wallington, E.J.; Keeling, A. Identification of genes involved in male sterility in wheat (Triticum aestivum L.) which could be used in a genic hybrid breeding system. Plant Direct. 2020, 4, e00201. [Google Scholar]
- Sedlacek, T. Method of Producing Hybrid F1 Seed of Angiosperm Plants. European Patent Application No. 15154573.8, 17 August 2016. [Google Scholar]
- Yang, X.T.; Geng, X.X.; Liu, Z.H.; Ye, J.L.; Xu, M.F.; Zang, L.L.; Song, X.Y. A Sterility Induction Trait in the Genic Male Sterility Wheat Line 4110S Induced by High Temperature and its Cytological Response. Crop Sci. 2018, 58, 1866–1876. [Google Scholar] [CrossRef]
- Xing, Q.H.; Ru, Z.G.; Zhou, C.J.; Xue, X.; Liang, C.Y.; Yang, D.E.; Jin, D.M.; Wang, B. Genetic analysis, molecular tagging and mapping of the thermo-sensitive genic male-sterile gene (wtms1) in wheat. Theor. Appl. Genet. 2003, 107, 1500–1504. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ru, Z.; Gao, Q.; Jiang, H.; Guo, F.; Wu, S.; Sun, Z. Male sterility and thermo-photosensitivity characteristics of BNS in wheat. Sci. Agric. Sin. 2009, 42, 3019–3027. [Google Scholar]
- Ru, Z.-G.; Zhang, L.-P.; Hu, T.-Z.; Liu, H.-Y.; Yang, Q.-K.; Weng, M.-L.; Wang, B.; Zhao, C.-P. Genetic analysis and chromosome mapping of a thermo-sensitive genic male sterile gene in wheat. Euphytica 2015, 201, 321–327. [Google Scholar] [CrossRef]
- Guo, R.X.; Sun, D.F.; Tan, Z.B.; Rong, D.F.; Li, C.D. Two recessive genes controlling thermophotoperiod-sensitive male sterility in wheat. Theor. Appl. Genet. 2006, 112, 1271–1276. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhang, F.; Wang, Y.; Ye, Z.; Qin, Z.; Bai, X.; Yang, J.; Gao, J.; Zhao, C. Fertility alteration in male sterile line BS210 of wheat. Acta Agron. Sin. 2017, 43, 171–178. [Google Scholar] [CrossRef]
- Yuan, S.H.; Bai, J.F.; Guo, H.Y.; Duan, W.J.; Liu, Z.H.; Zhang, F.T.; Ma, J.X.; Zhao, C.P.; Zhang, L.P. QTL mapping of male sterility-related traits in a photoperiod and temperature-sensitive genic male sterile wheat line BS366. Plant Breed. 2020, 139, 498–507. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, L.; He, L.; Yu, G. Thermo-sensitive period and critical temperature of fertility transition of thermo-photo-sensitive genic male sterile wheat. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2003, 14, 57–60. [Google Scholar]
- Dong, P.; Hu, Y.; Guo, G.; He, B.; Wang, L.; Yuan, J. Inheritance and chromosome location of photoperiod-thermo sensitive male sterility in wheat line Xinong 291S. Plant Breed. 2012, 131, 695–699. [Google Scholar] [CrossRef]
- Slade, A.J.; Fuerstenberg, S.I.; Loeffler, D.; Steine, M.N.; Facciotti, D. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 2005, 23, 75–81. [Google Scholar] [CrossRef]
- Pugsley, A.T.; Oram, R.N. Genic male sterility in wheat. Aust. Plant Breed. Genet. Newsl. 1959, 14, 10–11. [Google Scholar]
- Tang, H.L.; Liu, H.Y.; Zhou, Y.; Liu, H.W.; Du, L.P.; Wang, K.; Ye, X.G. Fertility recovery of wheat male sterility controlled byMs2using CRISPR/Cas9. Plant Biotechnol. J. 2021, 19, 224–226. [Google Scholar] [CrossRef] [PubMed]
- Cigan, A.M.; Singh, M.; Benn, G.; Feigenbutz, L.; Kumar, M.; Cho, M.-J.; Svitashev, S.; Young, J. Targeted mutagenesis of a conserved anther-expressed P450 gene confers male sterility in monocots. Plant Biotechol. J. 2017, 15, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Kouidri, A.; Whitford, R.; Suchecki, R.; Kalashyan, E.; Baumann, U. Genome-wide identification and analysis of non-specific Lipid Transfer Proteins in hexaploid wheat. Sci. Rep. 2018, 8, e00201. [Google Scholar] [CrossRef] [PubMed]
- Djukanovic, V.; Smith, J.; Lowe, K.; Yang, M.; Gao, H.; Jones, S.; Nicholson, M.G.; West, A.; Lape, J.; Bidney, D.; et al. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J. 2013, 76, 888–899. [Google Scholar] [CrossRef]
- Loukides, C.A.; Broadwater, A.H.; Bedinger, P.A. Two new male-sterile mutants of Zea mays (Poaceae) with abnormal tapetal cell morphology. Am. J. Bot. 1995, 82, 1017–1023. [Google Scholar] [CrossRef]
- Cigan, A.M.; Unger, E.; Xu, R.-J.; Kendall, T.L.; Fox, T.W. Phenotypic complementation of ms45 maize requires tapetal expression of MS45. Sex Plant Reprod. 2001, 14, 135–142. [Google Scholar] [CrossRef]
- Wu, Y.; Fox, T.W.; Trimnell, M.R.; Wang, L.; Xu, R.-j.; Cigan, A.M.; Huffman, G.A.; Garnaat, C.W.; Hershey, H.; Albertsen, M.C. Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 2016, 14, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Cigan, A.M. Manipulation of Dominant Male Sterility. International Patent Application No. WO2014164961A2, 9 October 2014. [Google Scholar]
- Mc, F.E.; Sears, E.R. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 1946, 37, 81–89. [Google Scholar]
- Matsuoka, Y. Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 2011, 52, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Charmet, G. Wheat domestication: Lessons for the future. C. R. Biol. 2011, 334, 212–220. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A. Genome evolution due to allopolyploidization in wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Akhunov, E.D.; Sehgal, S.; Liang, H.; Wang, S.; Akhunova, A.R.; Kaur, G.; Li, W.; Forrest, K.L.; See, D.; Šimková, H.; et al. Comparative Analysis of Syntenic Genes in Grass Genomes Reveals Accelerated Rates of Gene Structure and Coding Sequence Evolution in Polyploid Wheat. Plant Physiol. 2012, 161, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, J.; Chen, S.; Heng, Y.; Chen, Z.; Yang, J.; Zhou, K.; Pei, J.; He, H.; Deng, X.W.; et al. Poaceae-specific MS1 encodes a phospholipid-binding protein for male fertility in bread wheat. Proc. Natl. Acad. Sci. USA 2017, 114, 12614–12619. [Google Scholar] [CrossRef]
- Yang, X.T.; Ye, J.L.; Niu, F.Q.; Feng, Y.; Song, X.Y. Identification and verification of genes related to pollen development and male sterility induced by high temperature in the thermo-sensitive genic male sterile wheat line. Planta 2021, 253, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Bai, Y.X.; Liu, S.; Zhu, Q.D.; Song, Y.L.; Ma, S.C.; Ma, L.J.; Wang, X.L.; Zhang, G.S.; Wang, J.W. Microspore Abortion and Abnormal Tapetal Degeneration in BNS Thermo-sensitive Male Sterile Wheat Lines. Cereal. Res. Commun. 2019, 47, 506–517. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Gao, S.-Q.; Tang, Y.-M.; Gong, J.; Zhang, X.; Wang, Y.-B.; Zhang, L.-P.; Sun, R.-W.; Zhang, Q.; Chen, Z.-B.; et al. Transcriptome analysis of wheat seedling and spike tissues in the hybrid Jingmai 8 uncovered genes involved in heterosis. Planta 2018, 247, 1307–1321. [Google Scholar] [CrossRef]
- Mujun, Y.; Jian, G.; Kun, L. Ecological adaptability of thermo-photo-sensitive genic male sterile wheat K78S in Yunnan Province. Acta. Agron. Sin. 2006, 32, 1618–1624. [Google Scholar]
- Zhang, J.-K.; Dong, J.; Zong, X.-F.; Yu, G.-D.; Dai, X.-M.; Ruan, R.-W. Fertility Alternation of Thermo-Photo-Sensitive Genic Male Sterile (TGMS) Wheat Line C412S and its Association with Adenine Phosphoribosyltransferase Gene Expression. Acta. Agron. Sin. 2009, 35, 562–671. [Google Scholar] [CrossRef]
- Schachschneider, R. 30 years of hybrid wheat breeding in Europe: Recent developments and future prospects. In Proceedings of the Vereinigung der Pflanzenzüchter und Saatgutkaufleute Österreichs Jahrestagung, Irdning, Austria, 20–22 November 2017; BOKU-University of Natural Resources and Life Sciences: Vienna, Austria, 2018; pp. 19–26. [Google Scholar]
- Wilson, P.; Driscoll, C.J. Hybrid Wheat. In Heterosis: Reappraisal of Theory and Practice; Frankel, R., Ed.; Springer: Berlin, Germany, 1983; pp. 94–123. [Google Scholar]
- Gupta, P.K.; Balyan, H.S.; Gahlaut, V.; Saripalli, G.; Pal, B.; Basnet, B.R.; Joshi, A.K. Hybrid wheat: Past, present and future. Theor. Appl. Genet. 2019, 132, 2463–2483. [Google Scholar] [CrossRef] [PubMed]
- Kaul, M.L.H. Gene-cytoplasmic male sterility. In Male Sterility in Higher Plants; Kaul, M.L.H., Ed.; Springer: Berlin, Germany, 1988; pp. 97–192. [Google Scholar]
- Kihara, H. Substitution of Nucleus and its Effects on Genome Manifestations. Cytologia 1951, 16, 177–193. [Google Scholar] [CrossRef]
- Tsunewaki, K.; Wang, G.-Z.; Matsuoka, Y. Plasmon analysis of Triticum (wheat) and Aegilops. 1. Production of alloplasmic common wheats and their fertilities. Genes Genet. Syst. 1996, 71, 293–311. [Google Scholar] [CrossRef] [PubMed]
- Pickett, A.A.; Galwey, N.W. A further evaluation of hybrid wheat. Plant Var. Seeds 1997, 10, 15–32. [Google Scholar]
- Ahmed, T.; Tsujimoto, H.; Sasakuma, T. QTL analysis of fertility-restoration against cytoplasmic male sterility in wheat. Genes Genet. Syst. 2001, 76, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Kolb, F.L.; Domier, L.L.; Wang, S. SSR markers associated with fertility restoration genes against Triticum timopheevii cytoplasm in Triticum aestivum. Euphytica 2005, 141, 33–40. [Google Scholar] [CrossRef]
- Shahinnia, F.; Geyer, M.; Block, A.; Mohler, V.; Hartl, L. Identification of Rf9, a Gene Contributing to the Genetic Complexity of Fertility Restoration in Hybrid Wheat. Front. Plant Sci. 2020, 11, 1720. [Google Scholar] [CrossRef]
- Geyer, M.; Albrecht, T.; Hartl, L.; Mohler, V. Epistatic interactions and distribution of the fertility-restoring locus Rf1 in common wheat. Ver. Der. Pflanz. Saatgutkaufleute Osterr. 2018, 68, 29–30. [Google Scholar]
- Melonek, J.; Duarte, J.; Martin, J.; Beuf, L.; Murigneux, A.; Varenne, P.; Comadran, J.; Specel, S.; Levadoux, S.; Bernath-Levin, K.; et al. The genetic basis of cytoplasmic male sterility and fertility restoration in wheat. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.F.; Peng, G.Q.; Liu, Z.L.; Noman, A.; Alamri, S.; Hashem, M.; Qari, S.H.; al Zoubi, O.M. Molecular Control and Application of Male Fertility for Two-Line Hybrid Rice Breeding. Int. J. Mol. Sci. 2020, 21, 7868. [Google Scholar] [CrossRef]
- Fernandez-Gomez, J.; Talle, B.; Wilson, Z.A. Increased expression of the MALE STERILITY1 transcription factor gene results in temperature-sensitive male sterility in barley. J. Exp. Bot. 2020, 71, 6328–6339. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Liu, Z.; Zhang, L.; Hu, G.; Song, X. Cytological characterization of a thermo-sensitive cytoplasmic male-sterile wheat line having K-type cytoplasm of Aegilops kotschyi. Breed. Sci. 2016, 66, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, C.J. XYZ System of Producing Hybrid Wheat 1. Crop Sci. 1972, 12, 516–517. [Google Scholar] [CrossRef]
- Driscoll, C.J. Modified XYZ System of Producing Hybrid Wheat 1. Crop Sci. 1985, 25, 1115–1116. [Google Scholar] [CrossRef]
- Zhou, K.J.; Wang, S.H.; Feng, Y.Q.; Liu, Z.X.; Wang, G.X. The 4E-ms system of producing hybrid wheat. Crop Sci. 2006, 46, 250–255. [Google Scholar] [CrossRef]
- Kempe, K.; Rubtsova, M.; Gils, M. Split-gene system for hybrid wheat seed production. Proc. Natl. Acad. Sci. USA 2014, 111, 9097–9102. [Google Scholar] [CrossRef]
- Albertsen, M.; Fox, T.; Singh, M.; Williams, M. Fertility Restoration In Plants. International Patent Application No. WO 2020/056259 A1, 19 March 2020. [Google Scholar]
- Zhang, P.; Trethowan, R.; Dong, C.M.; Lage, J.; Bird, N.; Tapsell, C.; Hummel, A.; Darvey, N. Improved blue aleurone and other segregation systems. International Patent Application No. WO 2019/043082 Al, 7 March 2019. [Google Scholar]
- Keeling, A.G.; Milner, M.J. Methods And Compositions Relating To Maintainer Lines. International Patent Application No. WO/2019/165199, 29 August 2019. [Google Scholar]
- Islam, A.K.M.R.; Driscoll, C.J. Latent male fertility in ‘Cornerstone’ chromosome 4A. Can. J. Genet. Cytol. 1984, 26, 98–99. [Google Scholar] [CrossRef]
- Joshi, G.P.; Li, J.; Nasuda, S.; Endo, T.R. Development of a self-fertile ditelosomic line for the long arm of chromosome 4B and its characterization using SSR markers. Genes Genet. Syst. 2013, 88, 311–314. [Google Scholar] [CrossRef][Green Version]
- Burešová, V.; Kopecký, D.; Bartoš, J.; Martinek, P.; Watanabe, N.; Vyhnánek, T.; Doležel, J. Variation in genome composition of blue-aleurone wheat. Theor. Appl. Genet. 2015, 128, 273–282. [Google Scholar] [CrossRef]
- Dowell, F.E.; Maghirang, E.B.; Graybosch, R.A.; Berzonsky, W.A.; Delwiche, S.R. Selecting and Sorting Waxy Wheat Kernels Using Near-Infrared Spectroscopy. Cereal Chem. 2009, 86, 251–255. [Google Scholar] [CrossRef]
- Cigan, A.M.; Unger-Wallace, E.; Haug-Collet, K. Transcriptional gene silencing as a tool for uncovering gene function in maize. Plant J. 2005, 43, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Cigan, A.M.; Haug-Collet, K.; Clapp, J. Transcriptional silencing of heterologous anther promoters in maize: A genetic method to replace detasseling for seed production. Plant Reprod. 2014, 27, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kaur, A.; Pandey, A.; Mamrutha, H.M.; Singh, G.P. CRISPR-based genome editing in wheat: A comprehensive review and future prospects. Mol. Biol. Rep. 2019, 46, 3557–3569. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Su, X.; Tang, G.; Chen, Z.; Carter, J.; Wittich, P.E.; Dong, S.; Green, J.; Burch, E.; et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019, 37, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, P.; Ellison, E.; Polley, B.; Bollina, V.; Kulkarni, M.; Ghanbarnia, K.; Song, H.; Gao, C.; Voytas, D.F.; Kagale, S. Targeted mutagenesis in wheat microspores using CRISPR/Cas9. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).