Proteins Binding to the Carbohydrate HNK-1: Common Origins?
Abstract
1. Introduction
2. Results
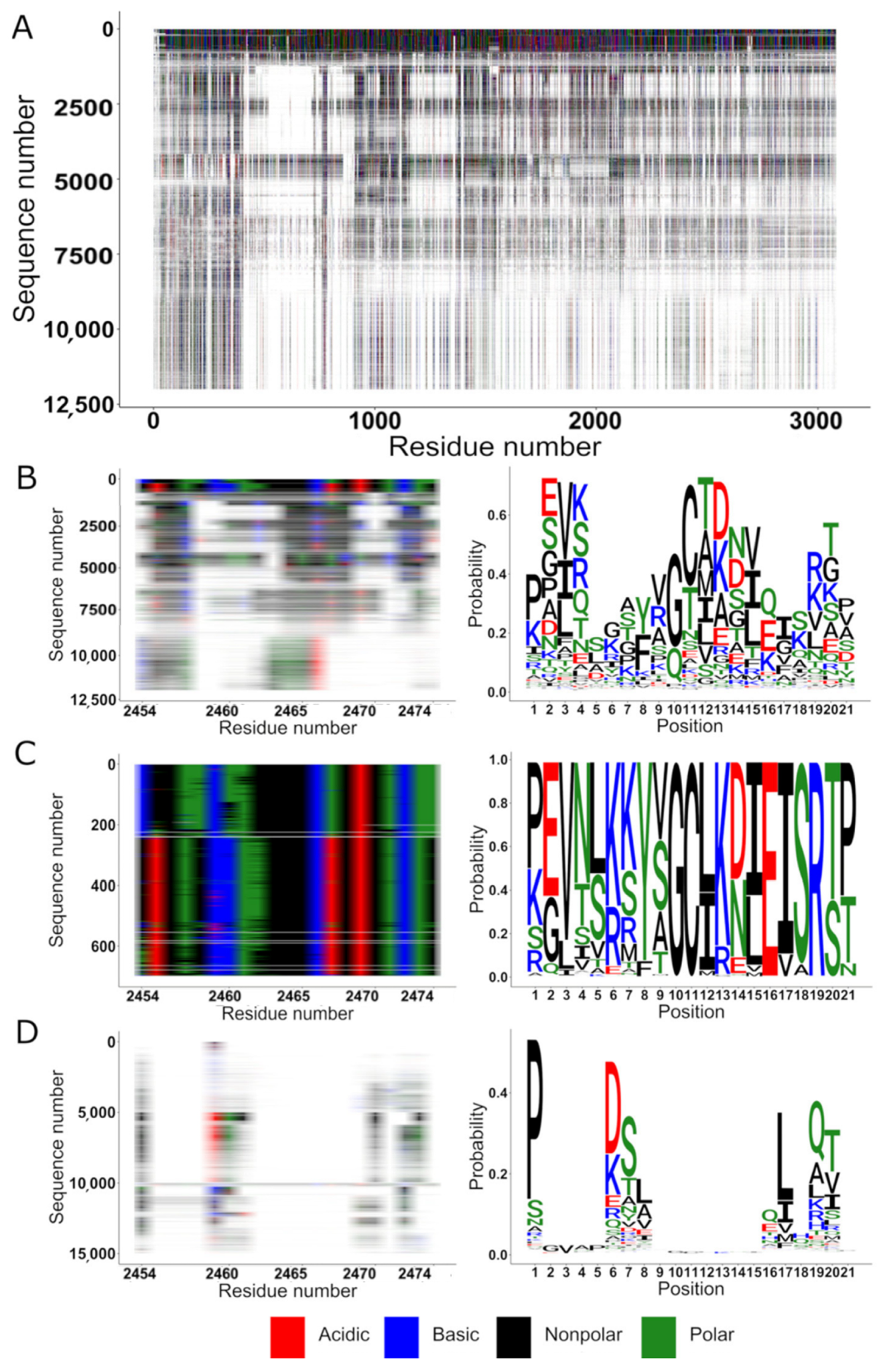
2.1. Alignment and Conservation of HNK-1 Receptors
2.2. Motifs Common to All Families
3. Discussion
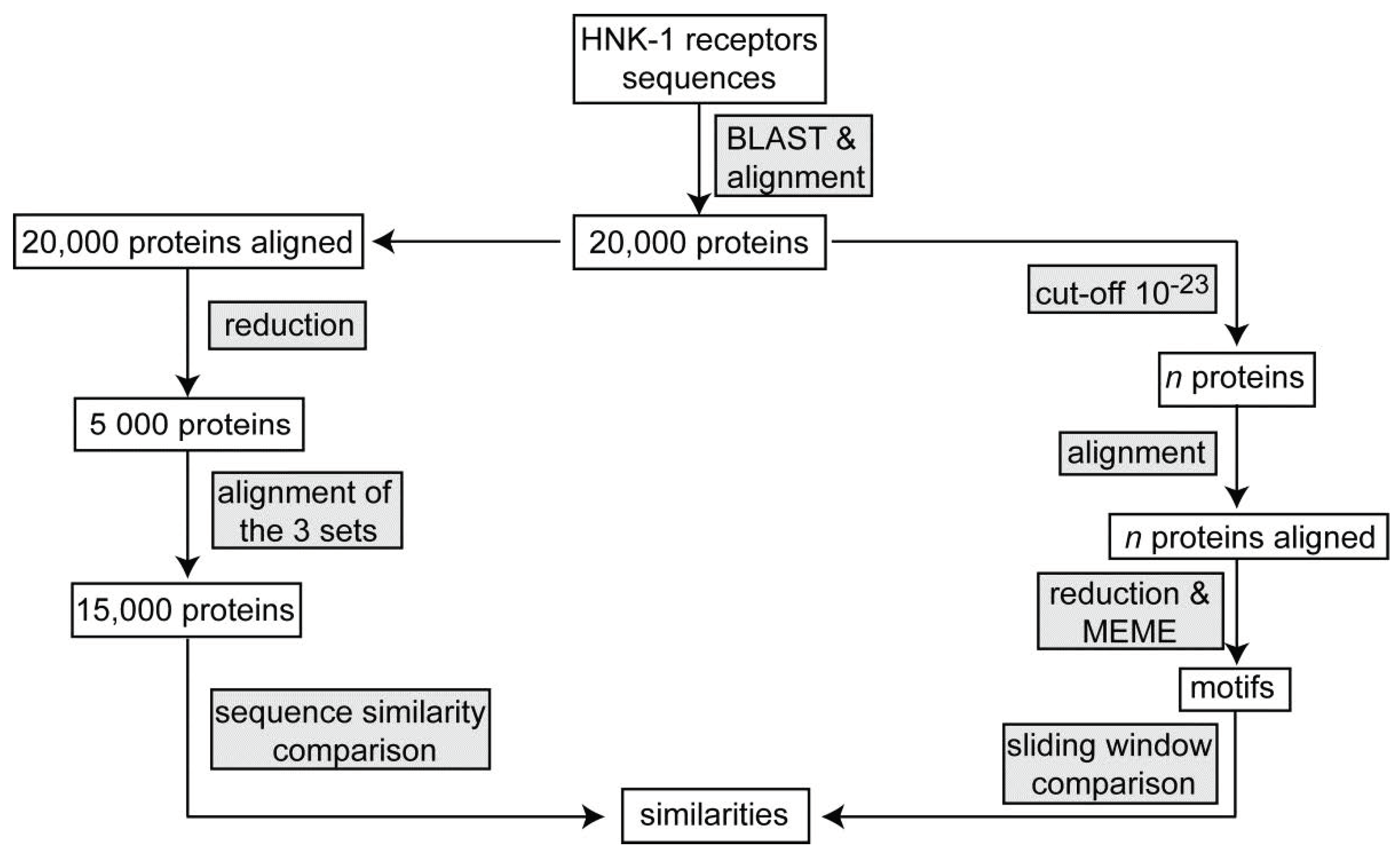
4. Materials and Methods
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMPA | α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| GluR2 | glutamate receptor 2 |
| HMGB | high-mobility group box |
| HNK | human natural killer |
References
- Sheikh, M.; Venzke, D.; Anderson, M.E.; Yoshida-Moriguchi, T.; Glushka, J.N.; Nairn, A.V.; Galizzi, M.; Moremen, K.W.; Campbell, K.P.; Wells, L. HNK-1 sulfotransferase modulates α-dystroglycan glycosylation by 3-O-sulfation of glucuronic acid on matriglycan. Glycobiology 2020, 30, 817–829. [Google Scholar] [CrossRef]
- Schwarting, G.A.; Jungalwala, F.B.; Chou, D.K.; Boyer, A.M.; Yamamoto, M. Sulfated glucuronic acid-containing glycoconjugates are temporally and spatially regulated antigens in the developing mammalian nervous system. Dev. Biol. 1987, 120, 65–76. [Google Scholar] [CrossRef]
- Chou, D.K.H.; Schwarting, G.A.; Evans, J.E.; Jungalwala, F.B. Sulfoglucuronyl-neolacto series of glycolipids in peripheral nerves reacting with HNK-1 antibody. J. Neurochem. 1987, 49, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Chou, D.K.; Ilyas, A.A.; Evans, J.E.; Costello, C.; Quarles, R.H.; Jungalwala, F.B. Structure of sulfated glucuronyl glycolipids in the nervous system reacting with HNK-1 antibody and some IgM paraproteins in neuropathy. J. Biol. Chem. 1986, 261, 11717–11725. [Google Scholar] [CrossRef]
- Sytnyk, V.; Leshchyns’Ka, I.; Schachner, M. Neural glycomics: The sweet side of nervous system functions. Cell. Mol. Life Sci. 2020, 78, 93–116. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Oka, S. Regulated expression and neural functions of human natural killer-1 (HNK-1) carbohydrate. Cell. Mol. Life Sci. 2012, 69, 4135–4147. [Google Scholar] [CrossRef]
- Poltorak, M.; Sadoul, R.; Keilhauer, G.; Landa, C.; Fahrig, T.; Schachner, M. Myelin-associated glycoprotein, a member of the L2/HNK-1 family of neural cell adhesion molecules, is involved in neuron-oligodendrocyte and oligodendrocyte-oligodendrocyte interaction. J. Cell Biol. 1987, 105, 1893–1899. [Google Scholar] [CrossRef] [PubMed]
- Martini, R.; Schachner, M. Immunoelectron microscopic localization of neural cell adhesion molecules (L1, N-CAM, and MAG) and their shared carbohydrate epitope and myelin basic protein in developing sciatic nerve. J. Cell Biol. 1986, 103, 2439–2448. [Google Scholar] [CrossRef] [PubMed]
- Kruse, J.; Mailhammer, R.; Wernecke, H.; Faissner, A.; Sommer, I.; Goridis, C.; Schachner, M.; Kruse, J.; Mailhammer, R.; Wernecke, H.; et al. Neural cell adhesion molecules and myelin-associated glycoprotein share a common carbohydrate moiety recognized by monoclonal antibodies L2 and HNK-1. Nat. Cell Biol. 1984, 311, 153–155. [Google Scholar] [CrossRef]
- Kruse, J.; Keilhauer, G.; Faissner, A.; Timpl, R.; Schachner, M. The J1 glycoprotein—A novel nervous system cell adhesion molecule of the L2/HNK-1 family. Nature 1985, 316, 146–148. [Google Scholar] [CrossRef]
- Morise, J.; Takematsu, H.; Oka, S. The role of human natural killer-1 (HNK-1) carbohydrate in neuronal plasticity and disease. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 2455–2461. [Google Scholar] [CrossRef] [PubMed]
- Morita, I.; Kakuda, S.; Takeuchi, Y.; Itoh, S.; Kawasaki, N.; Kizuka, Y.; Kawasaki, T.; Oka, S. HNK-1 Glyco-epitope regulates the stability of the glutamate receptor subunit GluR2 on the neuronal cell surface. J. Biol. Chem. 2009, 284, 30209–30217. [Google Scholar] [CrossRef] [PubMed]
- Schachner, M.; Martini, R.; Hall, H.; Orberger, G. Functions of the L2/HNK-1 carbohydrate in the nervous system. Prog. Brain. Res. 1995, 105, 183–188. [Google Scholar] [CrossRef]
- Jungalwala, F.B. Expression and biological functions of sulfoglucuronyl glycolipids (SGGLs) in the nervous system? A review. Neurochem. Res. 1994, 19, 945–957. [Google Scholar] [CrossRef]
- Hall, H.; Liu, L.; Schachner, M.; Schmitz, B. The L2/HNK-1 carbohydrate mediates adhesion of neural cells to laminin. Eur. J. Neurosci. 1993, 5, 34–42. [Google Scholar] [CrossRef]
- Yabuno, K.; Morise, J.; Kizuka, Y.; Hashii, N.; Kawasaki, N.; Takahashi, S.; Miyata, S.; Izumikawa, T.; Kitagawa, H.; Takematsu, H.; et al. A sulfated glycosaminoglycan linkage region is a novel type of human natural killer-1 (HNK-1) epitope expressed on aggrecan in perineuronal nets. PLoS ONE 2015, 10, e0144560. [Google Scholar] [CrossRef]
- Saghatelyan, A.; Gorissen, S.; Albert, M.; Hertlein, B.; Schachner, M.; Dityatev, A. The extracellular matrix molecule tenascin-R and its HNK-1 carbohydrate modulate perisomatic inhibition and long-term potentiation in the CA1 region of the hippocampus. Eur. J. Neurosci. 2000, 12, 3331–3342. [Google Scholar] [CrossRef]
- Martini, R.; Schachner, M.; Brushart, T.M. The L2/HNK-1 carbohydrate is preferentially expressed by previously motor axon-associated Schwann cells in reinnervated peripheral nerves. J. Neurosci. 1994, 14, 7180–7191. [Google Scholar] [CrossRef]
- Low, K.; Orberger, G.; Schmitz, B.; Martini, R.; Schachner, M. The L2/HNK-1 carbohydrate is carried by the myelin associated glycoprotein and sulphated glucuronyl glycolipids in muscle but not cutaneous nerves of adult mice. Eur. J. Neurosci. 1994, 6, 1773–1781. [Google Scholar] [CrossRef] [PubMed]
- Simova, O.; Irintchev, A.; Mehanna, A.; Liu, J.; Dihné, M.; Bächle, D.; Sewald, N.; Loers, G.; Schachner, M. Carbohydrate mimics promote functional recovery after peripheral nerve repair. Ann. Neurol. 2006, 60, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.; Li, R.; Kadeyala, P.K.; Liu, S.; Schachner, M. The human natural killer-1 (HNK-1) glycan mimetic ursolic acid promotes functional recovery after spinal cord injury in mouse. J. Nutr. Biochem. 2018, 55, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Kared, H.; Martelli, S.; Ng, T.P.; Pender, S.L.; Larbi, A. CD57 in human natural killer cells and T-lymphocytes. Cancer Immunol. Immunother. 2016, 65, 441–452. [Google Scholar] [CrossRef]
- Hall, H.; Vorherr, T.; Schachner, M. Characterization of a 21 amino acid peptide sequence of the laminin G2 domain that is involved in HNK-1 carbohydrate binding and cell adhesion. Glycobiology 1995, 5, 435–441. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chou, D.K.H.; Zhang, J.; Smith, F.I.; McCaffery, P.; Jungalwala, F.B. Developmental expression of receptor for advanced glycation end products (RAGE), amphoterin and sulfoglucuronyl (HNK-1) carbohydrate in mouse cerebellum and their role in neurite outgrowth and cell migration. J. Neurochem. 2004, 90, 1389–1401. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Schachner, M.; Shen, Y.-Q. HMGB1 in development and diseases of the central nervous system. Mol. Neurobiol. 2012, 45, 499–506. [Google Scholar] [CrossRef]
- Rauvala, H.; Rouhiainen, A. Physiological and pathophysiological outcomes of the interactions of HMGB1 with cell surface receptors. Biochim. Biophys. Acta Gene Regul. Mech. 2010, 1799, 164–170. [Google Scholar] [CrossRef]
- Schmidt, J.T.; Schachner, M. Role for cell adhesion and glycosyl (HNK-1 and oligomannoside) recognition in the sharpening of the regenerating retinotectal projection in goldfish. J. Neurobiol. 1998, 37, 659–671. [Google Scholar] [CrossRef]
- Hall, H.; Deutzmann, R.; Timpl, R.; Vaughan, L.; Schmitz, B.; Schachner, M. HNK-1 carbohydrate-mediated cell adhesion to laminin-1 is different from heparin-mediated and sulfatide-mediated cell adhesion. JBIC J. Biol. Inorg. Chem. 1997, 246, 233–242. [Google Scholar] [CrossRef]
- Schmitz, B.; Schachner, M.; Ito, Y.; Nakano, T.; Ogawa, T. Determination of structural elements of the L2/HNK-1 carbohydrate epitope required for its function. Glycoconj. J. 1994, 11, 345–352. [Google Scholar] [CrossRef]
- Chou, D.K.; Evans, J.E.; Jungalwala, F.B. Identity of nuclear high-mobility-group protein, HMG-1, and sulfoglucuronyl carbohydrate-binding protein, SBP-1, in brain. J. Neurochem. 2001, 77, 120–131. [Google Scholar] [CrossRef]
- Chou, D.K.H.; Henion, T.R.; Jungalwala, F.B. Regulation of expression of sulfoglucuronyl carbohydrate (HNK-1), Amphoterin and RAGE in retinoic acid-differentiated P19 embryonal carcinoma cells. J. Neurochem. 2003, 86, 917–931. [Google Scholar] [CrossRef]
- Morita, I.; Kizuka, Y.; Kakuda, S.; Oka, S. Expression and function of the HNK-1 carbohydrate. J. Biochem. 2007, 143, 719–724. [Google Scholar] [CrossRef]
- Ma, L.; Shen, H.-F.; Shen, Y.-Q.; Schachner, M. The adhesion molecule-characteristic HNK-1 carbohydrate contributes to functional recovery after spinal cord injury in adult zebrafish. Mol. Neurobiol. 2016, 54, 3253–3263. [Google Scholar] [CrossRef]
- Lang, D.M.; Stuermer, C.A. Adaptive plasticity of Xenopus glial cells in vitro and after CNS fiber tract lesions in vivo. Glia 1996, 18, 92–106. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; McWilliam, H.; Goujon, M.; Cowley, A.; Lopez, R.; Pearson, W.R. PSI-Search: Iterative HOE-reduced profile SSEARCH searching. Bioinformatics 2012, 28, 1650–1651. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W.; Li, W.; López, R. Query-seeded iterative sequence similarity searching improves selectivity 5–20-fold. Nucleic Acids Res. 2016, 45, e46. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Mach. Learn. 1995, 21, 51–80. [Google Scholar] [CrossRef]
- Shapiro, L.; Kwong, P.D.; Fannon, A.M.; Colman, D.R.; Hendrickson, W.A. Considerations on the folding topology and evolutionary origin of cadherin domains. Proc. Natl. Acad. Sci. USA 1995, 92, 6793–6797. [Google Scholar] [CrossRef]
- Reeves, R.; Nissen, M.S. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 1990, 265, 8573–8582. [Google Scholar] [CrossRef]
- Beckmann, G.; Hanke, J.; Bork, P.; Reich, J.G. Merging extracellular domains: Fold prediction for laminin G-like and amino-terminal thrombospondin-like modules based on homology to pentraxins. J. Mol. Biol. 1998, 275, 725–730. [Google Scholar] [CrossRef]
- Hohenester, E. Structural biology of laminins. Essays Biochem. 2019, 63, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Briggs, D.; Yoshida-Moriguchi, T.; Zheng, T.; Venzke, D.; Anderson, M.E.; Strazzulli, A.; Moracci, M.; Yu, L.; Hohenester, E.; Campbell, K.P. Structural basis of laminin binding to the LARGE glycans on dystroglycan. Nat. Chem. Biol. 2016, 12, 810–814. [Google Scholar] [CrossRef]
- Zhang, R.; Loers, G.; Schachner, M.; Boelens, R.; Wienk, H.; Siebert, S.; Eckert, T.; Kraan, S.; Rojas-Macias, M.A.; Lütteke, T.; et al. Molecular basis of the receptor interactions of polysialic acid (polySia), polySia mimetics, and sulfated polysaccharides. ChemMedChem 2016, 11, 990–1002. [Google Scholar] [CrossRef] [PubMed]
- Spiwok, V. CH/π Interactions in carbohydrate recognition. Molecules 2017, 22, 1038. [Google Scholar] [CrossRef]
- Hudson, K.L.; Bartlett, G.J.; Diehl, R.C.; Agirre, J.; Gallagher, T.; Kiessling, L.L.; Woolfson, D.N. Carbohydrate–aromatic interactions in proteins. J. Am. Chem. Soc. 2015, 137, 15152–15160. [Google Scholar] [CrossRef]
- Tisi, D.; Talts, J.F.; Timpl, R.; Hohenester, E. Structure of the C-terminal laminin G-like domain pair of the laminin alpha 2 chain harbouring binding sites for alpha -dystroglycan and heparin. EMBO J. 2000, 19, 1432–1440. [Google Scholar] [CrossRef]
- Ichikawa, N.; Kasai, S.; Suzuki, N.; Nishi, N.; Oishi, S.; Fujii, N.; Kadoya, Y.; Hatori, K.; Mizuno, Y.; Nomizu, M.; et al. Identification of neurite outgrowth active sites on the laminin alpha4 chain G domain. Biochemistry 2005, 44, 5755–5762. [Google Scholar] [CrossRef]
- Bhunia, A.; Vivekanandan, S.; Eckert, T.; Burg-Roderfeld, M.; Wechselberger, R.; Romanuka, J.; Bächle, D.; Kornilov, A.V.; von der Lieth, C.W.; Jiménez-Barbero, J.; et al. Why structurally different cyclic peptides can be glycomimetics of the HNK-1 carbohydrate antigen. J. Am. Chem. Soc. 2010, 132, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Abo, T.; Balch, C.M. A differentiation antigen of human NK and K cells identified by a monoclonal antibody (HNK-1). J. Immunol. 1981, 127, 1024–1029. [Google Scholar] [PubMed]
- Noronha, A.B.; Ilyas, A.; Antonicek, H.; Schachner, M.; Quarles, R.H. Molecular specificity of L2 monoclonal antibodies that bind to carbohydrate determinants of neural cell adhesion molecules and their resemblance to other monoclonal antibodies recognizing the myelin-associated glycoprotein. Brain Res. 1986, 385, 237–244. [Google Scholar] [CrossRef]
- Tamura, M.; Natori, K.; Kobayashi, M.; Miyamura, T.; Takeda, N. Genogroup II noroviruses efficiently bind to heparan sulfate proteoglycan associated with the cellular membrane. J. Virol. 2004, 78, 3817–3826. [Google Scholar] [CrossRef] [PubMed]
- Taube, S.; Perry, J.W.; Yetming, K.; Patel, S.P.; Auble, H.; Shu, L.; Nawar, H.F.; Lee, C.H.; Connell, T.D.; Shayman, J.A.; et al. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J. Virol. 2009, 83, 4092–4101. [Google Scholar] [CrossRef]
- Ji, X.; Gewurz, H.; Spear, G.T. Mannose binding lectin (MBL) and HIV. Mol. Immunol. 2005, 42, 145–152. [Google Scholar] [CrossRef]
- Marzi, A.; Mitchell, D.A.; Chaipan, C.; Fisch, T.; Doms, R.W.; Carrington, M.; Desrosiers, R.C.; Pöhlmann, S. Modulation of HIV and SIV neutralization sensitivity by DC-SIGN and mannose-binding lectin. Virology 2007, 368, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Neu, U.; Bauer, J.; Stehle, T. Viruses and sialic acids: Rules of engagement. Curr. Opin. Struct. Biol. 2011, 21, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wu, L.; Eckert, T.; Burg-Roderfeld, M.; Rojas-Macias, M.A.; Lütteke, T.; Krylov, V.B.; Argunov, D.A.; Datta, A.; Markart, P.; et al. Lysozyme’s lectin-like characteristics facilitates its immune defense function. Q. Rev. Biophys. 2017, 50, e9. [Google Scholar] [CrossRef]
- Battistel, M.D.; Azurmendi, H.F.; Yu, B.; Freedberg, D.I. NMR of glycans: Shedding new light on old problems. Prog. Nucl. Magn. Reson. Spectrosc. 2014, 79, 48–68. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, Y.E.; Burg-Roderfeld, M.; Loers, G.; Arda, A.; Sukhova, E.V.; Khatuntseva, E.A.; Grachev, A.A.; Chizhov, A.O.; Siebert, H.-C.; Schachner, M.; et al. Synthesis and molecular recognition studies of the HNK-1 trisaccharide and related oligosaccharides. The specificity of monoclonal anti-HNK-1 antibodies as assessed by surface plasmon resonance and STD NMR. J. Am. Chem. Soc. 2011, 134, 426–435. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Torda, A.E. Reduce ACM. Sigsam Bull. 2020, 24, 14–15. [Google Scholar] [CrossRef]
- Strait, B.; Dewey, T.; Strait, B.; Dewey, T. The Shannon information entropy of protein sequences. Biophys. J. 1996, 71, 148–155. [Google Scholar] [CrossRef]
- Torda, A.E. Entropy.pl. 2019. Available online: https://zenodo.org/record/4334283#.YQICgI4zZPY (accessed on 17 December 2020).
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2017, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Pagès, H.A.P.; Gentleman, R.; DebRoy, S. Biostrings: Efficient manipulation of biological strings. R Package Version 2019, 2, 10–18129. [Google Scholar]
- Available online: https://nam02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.nist.gov%2Fpml%2Fspecial-publication-811%2Fnist-guide-si-chapter-10-more-printing-and-using-symbols-and-numbers%231053&data=04%7C01%7Cschachner%40dls.rutgers.edu%7Cf4bf2bb292e443e2fd0008d95029d375%7Cb92d2b234d35447093ff69aca6632ffe%7C1%7C1%7C637628962842219163%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000&sdata=Dnky8qDcpBI2rPm0OhZChlDaBWbETQN740X2Uun%2BdXg%3D&reserved=0 (accessed on 27 July 2021).
- Available online: https://nam02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.bipm.org%2Fen%2Fcommittees%2Fcg%2Fcgpm%2F22-2003%2Fresolution-10&data=04%7C01%7Cschachner%40dls.rutgers.edu%7Cf4bf2bb292e443e2fd0008d95029d375%7Cb92d2b234d35447093ff69aca6632ffe%7C1%7C1%7C637628962842219163%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000&sdata=LWQDiDaVZyeynw9lqgDClv%2FVCpoWgjkJL7Vkye0x9Bg%3D&reserved=0 (accessed on 27 July 2021).

| ID | Sequence Length | n Homologues | |
|---|---|---|---|
| Cadherin-2 | NP_031690.3 | 906 | 14,128 |
| HMGB-1 | NP_034569.1 | 215 | 1590 |
| HMGB-2 | NP_032278.1 | 199 | 1573 |
| Laminin (subunit α-1) | NP_032506.2 | 3083 | 11,999 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castillo, G.; Kleene, R.; Schachner, M.; Loers, G.; Torda, A.E. Proteins Binding to the Carbohydrate HNK-1: Common Origins? Int. J. Mol. Sci. 2021, 22, 8116. https://doi.org/10.3390/ijms22158116
Castillo G, Kleene R, Schachner M, Loers G, Torda AE. Proteins Binding to the Carbohydrate HNK-1: Common Origins? International Journal of Molecular Sciences. 2021; 22(15):8116. https://doi.org/10.3390/ijms22158116
Chicago/Turabian StyleCastillo, Gaston, Ralf Kleene, Melitta Schachner, Gabriele Loers, and Andrew E. Torda. 2021. "Proteins Binding to the Carbohydrate HNK-1: Common Origins?" International Journal of Molecular Sciences 22, no. 15: 8116. https://doi.org/10.3390/ijms22158116
APA StyleCastillo, G., Kleene, R., Schachner, M., Loers, G., & Torda, A. E. (2021). Proteins Binding to the Carbohydrate HNK-1: Common Origins? International Journal of Molecular Sciences, 22(15), 8116. https://doi.org/10.3390/ijms22158116








