Targeting the Highly Expressed microRNA miR-146b with CRISPR/Cas9n Gene Editing System in Thyroid Cancer
Abstract
:1. Introduction
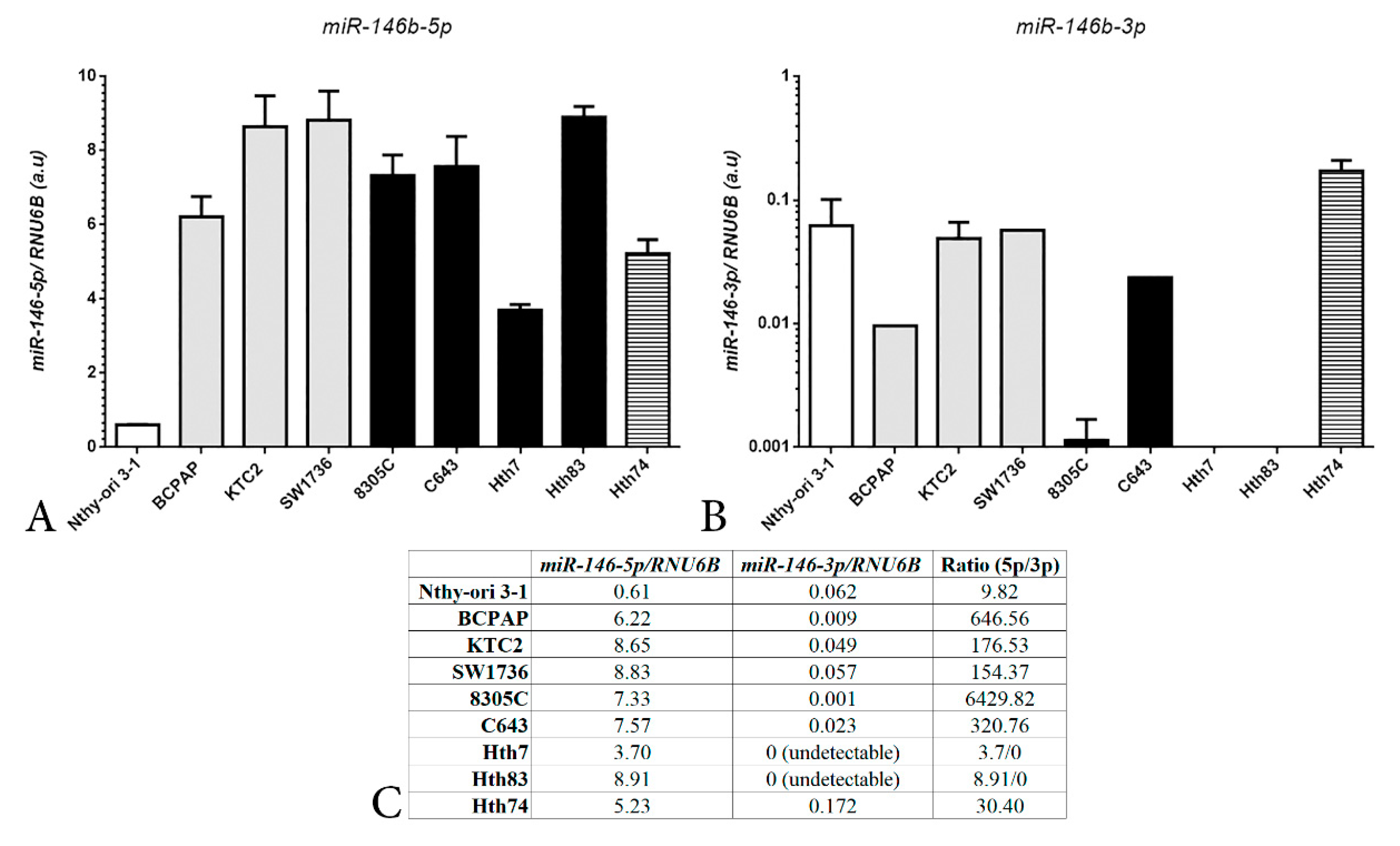
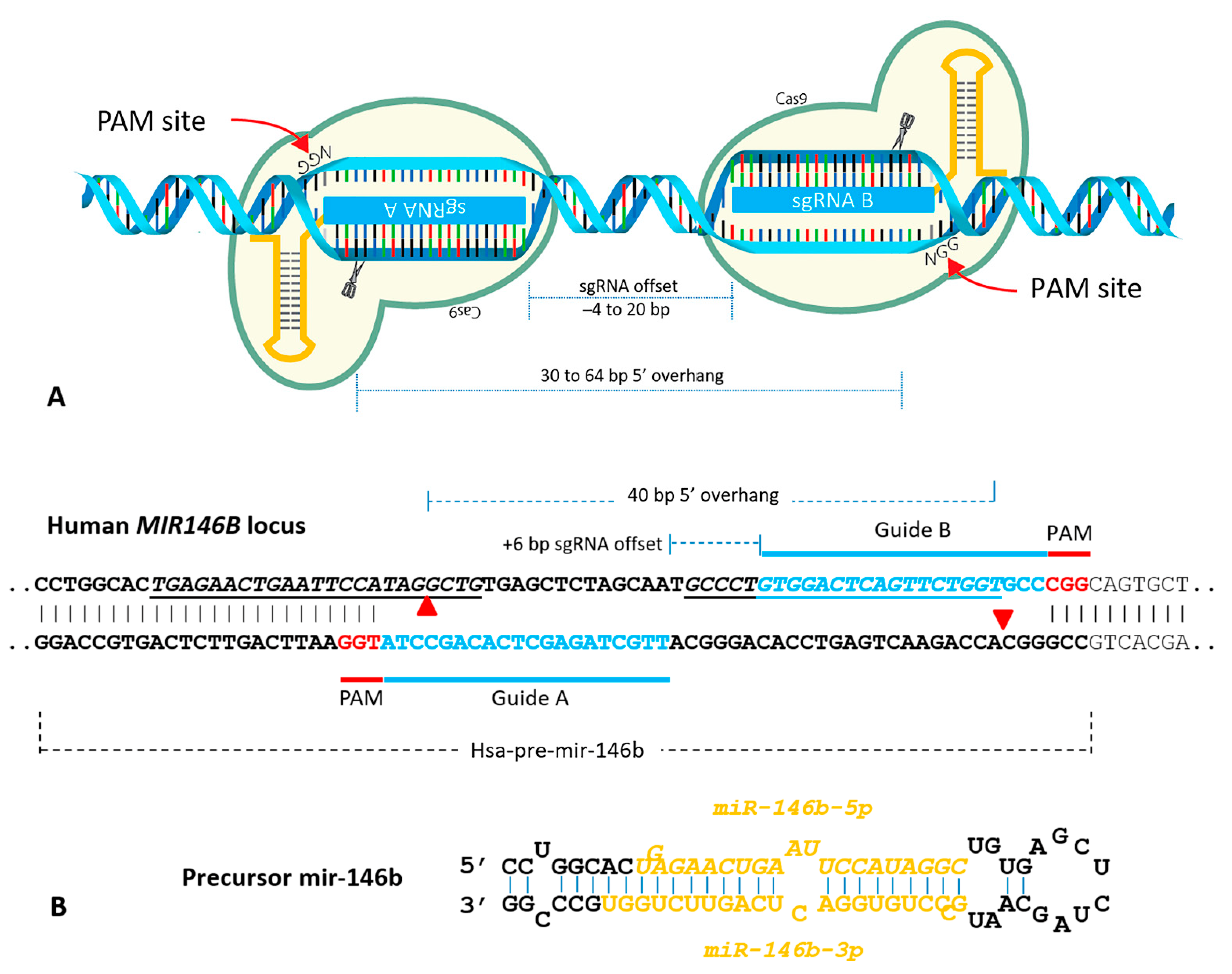
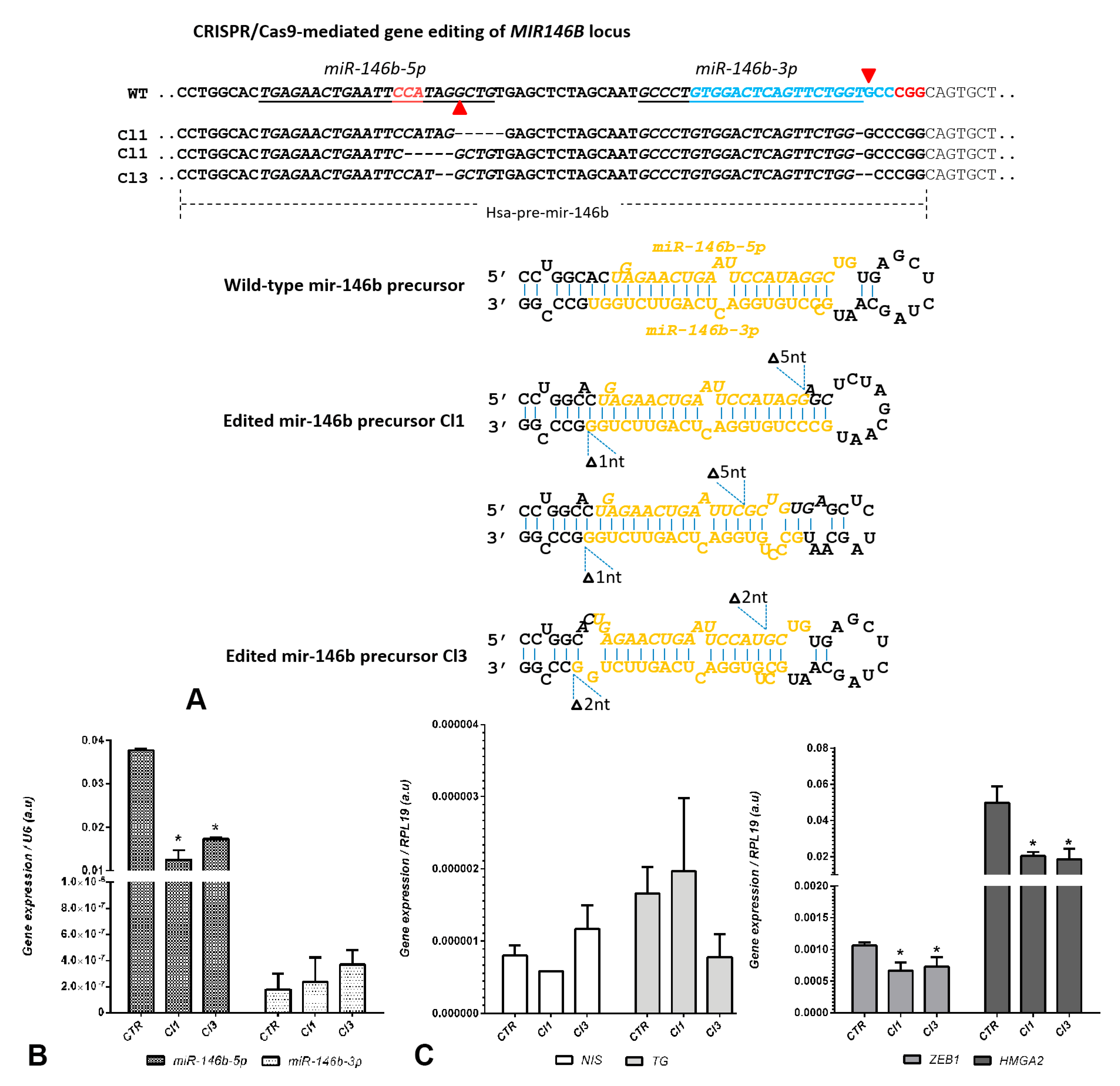
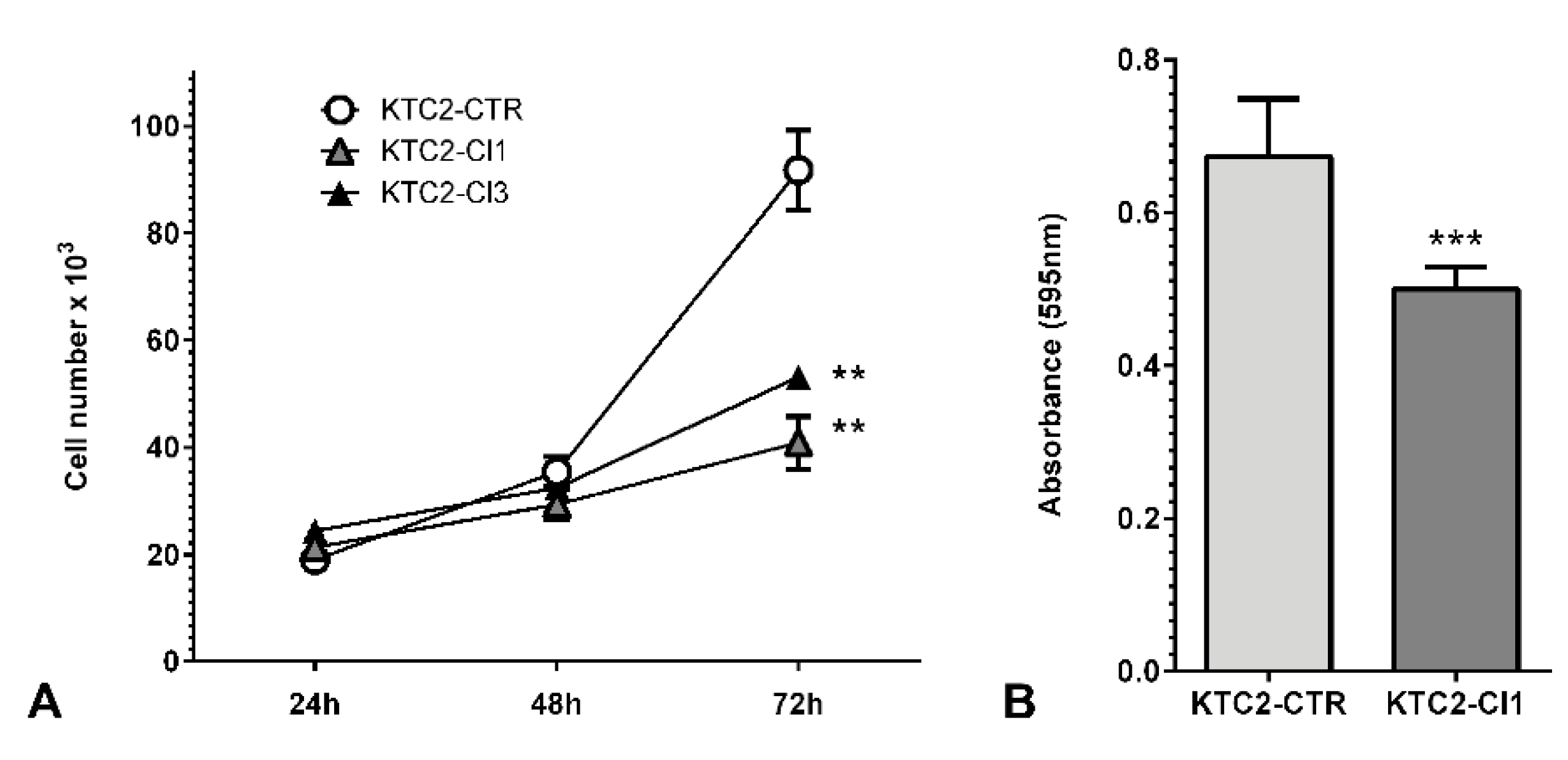
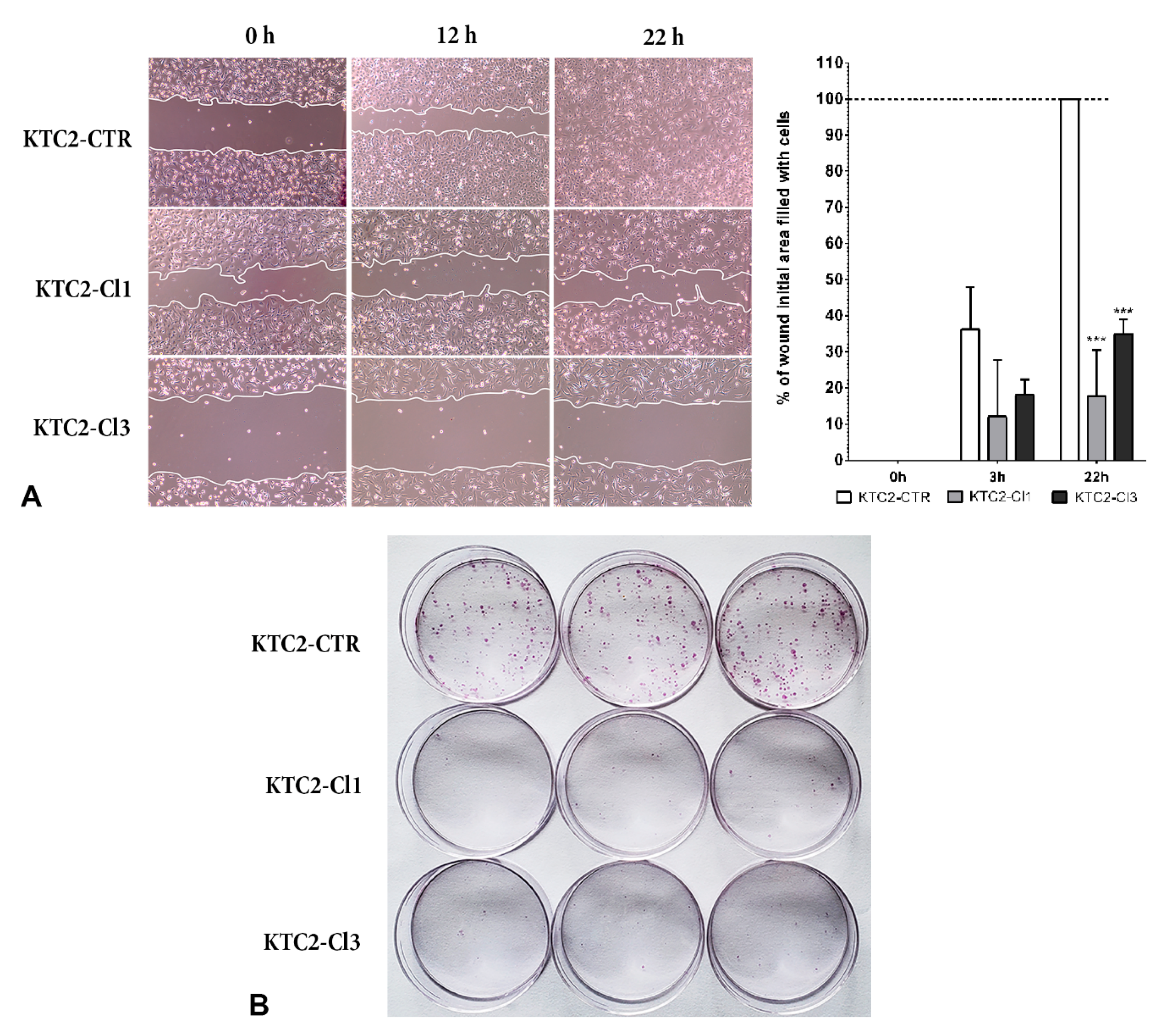
2. Results and Discussion
Construction of a CRISPR/Cas9n Gene Editing to Target MIR146B Gene
3. Materials and Methods
3.1. MIR146B Gene Editing Using CRISPR/Cas9n
3.2. Cell Culture
3.3. Genomic DNA Sequencing for Gene Editing Validation
3.4. Gene Expression Analysis
3.5. Cell Function Assays
3.5.1. Cell Counting
3.5.2. Cell Viability Assay (MTT)
3.5.3. Wound Healing Assay
3.5.4. Colony Formation
3.5.5. In Vivo Xenotransplant in Nude Mice
3.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Yeh, C.D.; Richardson, C.D.; Corn, J.E. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 2019, 21, 1468–1478. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, E.T.; Nikiforova, M.N.; Zhu, Z.; Knauf, J.A.; Nikiforov, Y.E.; Fagin, J.A. High prevalence of BRAF mutations in thyroid cancer: Genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003, 63, 1454–1457. [Google Scholar]
- Xing, M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat. Rev. Cancer 2013, 13, 184–199. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [Green Version]
- Fuziwara, C.S.; Saito, K.C.; Kimura, E.T. Thyroid Follicular Cell Loss of Differentiation Induced by MicroRNA miR-17-92 Cluster Is Attenuated by CRISPR/Cas9n Gene Silencing in Anaplastic Thyroid Cancer. Thyroid 2020, 30, 81–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, P.; Wang, L.; Sliz, P.; Gregory, R.I. A Biogenesis Step Upstream of Microprocessor Controls miR-17 approximately 92 Expression. Cell 2015, 162, 885–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Jazdzewski, K.; Li, W.; Liyanarachchi, S.; Nagy, R.; Volinia, S.; Calin, G.A.; Liu, C.G.; Franssila, K.; Suster, S.; et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 2005, 102, 19075–19080. [Google Scholar] [CrossRef] [Green Version]
- Pallante, P.; Visone, R.; Ferracin, M.; Ferraro, A.; Berlingieri, M.T.; Troncone, G.; Chiappetta, G.; Liu, C.G.; Santoro, M.; Negrini, M.; et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr. Relat. Cancer 2006, 13, 497–508. [Google Scholar] [CrossRef] [Green Version]
- Visone, R.; Pallante, P.; Vecchione, A.; Cirombella, R.; Ferracin, M.; Ferraro, A.; Volinia, S.; Coluzzi, S.; Leone, V.; Borbone, E.; et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene 2007, 26, 7590–7595. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Tseng, G.C.; Steward, D.; Diorio, D.; Nikiforov, Y.E. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J. Clin. Endocrinol. Metab. 2008, 93, 1600–1608. [Google Scholar] [CrossRef]
- Swierniak, M.; Wojcicka, A.; Czetwertynska, M.; Stachlewska, E.; Maciag, M.; Wiechno, W.; Gornicka, B.; Bogdanska, M.; Koperski, L.; de la Chapelle, A.; et al. In-depth characterization of the microRNA transcriptome in normal thyroid and papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2013, 98, E1401–E1409. [Google Scholar] [CrossRef]
- Lima, C.R.; Geraldo, M.V.; Fuziwara, C.S.; Kimura, E.T.; Santos, M.F. MiRNA-146b-5p upregulates migration and invasion of different Papillary Thyroid Carcinoma cells. BMC Cancer 2016, 16, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riesco-Eizaguirre, G.; Wert-Lamas, L.; Perales-Paton, J.; Sastre-Perona, A.; Fernandez, L.P.; Santisteban, P. The miR-146b-3p/PAX8/NIS Regulatory Circuit Modulates the Differentiation Phenotype and Function of Thyroid Cells during Carcinogenesis. Cancer Res. 2015, 75, 4119–4130. [Google Scholar] [CrossRef] [Green Version]
- Geraldo, M.V.; Yamashita, A.S.; Kimura, E.T. MicroRNA miR-146b-5p regulates signal transduction of TGF-beta by repressing SMAD4 in thyroid cancer. Oncogene 2012, 31, 1910–1922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yip, L.; Kelly, L.; Shuai, Y.; Armstrong, M.J.; Nikiforov, Y.E.; Carty, S.E.; Nikiforova, M.N. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann. Surg. Oncol. 2011, 18, 2035–2041. [Google Scholar] [CrossRef] [Green Version]
- Chou, C.K.; Chen, R.F.; Chou, F.F.; Chang, H.W.; Chen, Y.J.; Lee, Y.F.; Yang, K.D.; Cheng, J.T.; Huang, C.C.; Liu, R.T. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid 2010, 20, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Zhao, J.T.; Clifton-Bligh, R.J.; Gill, A.; Gundara, J.S.; Ip, J.C.; Glover, A.; Sywak, M.S.; Delbridge, L.W.; Robinson, B.G.; et al. MicroRNA-222 and microRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer 2013, 119, 4358–4365. [Google Scholar] [CrossRef]
- Jiang, K.; Li, G.; Chen, W.; Song, L.; Wei, T.; Li, Z.; Gong, R.; Lei, J.; Shi, H.; Zhu, J. Plasma Exosomal miR-146b-5p and miR-222-3p are Potential Biomarkers for Lymph Node Metastasis in Papillary Thyroid Carcinomas. OncoTargets Ther. 2020, 13, 1311–1319. [Google Scholar] [CrossRef] [Green Version]
- Chou, C.K.; Yang, K.D.; Chou, F.F.; Huang, C.C.; Lan, Y.W.; Lee, Y.F.; Kang, H.Y.; Liu, R.T. Prognostic implications of miR-146b expression and its functional role in papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2013, 98, E196–E205. [Google Scholar] [CrossRef]
- Bible, K.C.; Kebebew, E.; Brierley, J.; Brito, J.P.; Cabanillas, M.E.; Clark, T.J., Jr.; Di Cristofano, A.; Foote, R.; Giordano, T.; Kasperbauer, J.; et al. 2021 American Thyroid Association Guidelines for Management of Patients with Anaplastic Thyroid Cancer. Thyroid 2021, 31, 337–386. [Google Scholar] [CrossRef]
- Cabanillas, M.E.; Ferrarotto, R.; Garden, A.S.; Ahmed, S.; Busaidy, N.L.; Dadu, R.; Williams, M.D.; Skinner, H.; Gunn, G.B.; Grosu, H.; et al. Neoadjuvant BRAF- and Immune-Directed Therapy for Anaplastic Thyroid Carcinoma. Thyroid 2018, 28, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, T.W.; le Sage, C.; Larrieu, D.; Demir, M.; Jackson, S.P. CRISPR-Cas9(D10A) nickase-based genotypic and phenotypic screening to enhance genome editing. Sci. Rep. 2016, 6, 24356. [Google Scholar] [CrossRef]
- El Fatimy, R.; Subramanian, S.; Uhlmann, E.J.; Krichevsky, A.M. Genome Editing Reveals Glioblastoma Addiction to MicroRNA-10b. Mol. Ther. 2017, 25, 368–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.S. Functional Anatomy of the Human Microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, S.C.; Nguyen, T.A.; Choi, Y.G.; Jo, M.H.; Hohng, S.; Kim, V.N.; Woo, J.S. Structure of Human DROSHA. Cell 2016, 164, 81–90. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Z.; Li, H.; Wang, J.; Sun, C. miR-146a and miR-146b in the diagnosis and prognosis of papillary thyroid carcinoma. Oncol. Rep. 2017, 38, 2735–2740. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Moya, J.; Wert-Lamas, L.; Riesco-Eizaguirre, G.; Santisteban, P. Impaired microRNA processing by DICER1 downregulation endows thyroid cancer with increased aggressiveness. Oncogene 2019, 38, 5486–5499. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Moya, J.; Wert-Lamas, L.; Santisteban, P. MicroRNA-146b promotes PI3K/AKT pathway hyperactivation and thyroid cancer progression by targeting PTEN. Oncogene 2018, 37, 3369–3383. [Google Scholar] [CrossRef]
- Czajka, A.A.; Wojcicka, A.; Kubiak, A.; Kotlarek, M.; Bakula-Zalewska, E.; Koperski, L.; Wiechno, W.; Jazdzewski, K. Family of microRNA-146 Regulates RARbeta in Papillary Thyroid Carcinoma. PLoS ONE 2016, 11, e0151968. [Google Scholar] [CrossRef]
- Yu, C.; Zhang, L.; Luo, D.; Yan, F.; Liu, J.; Shao, S.; Zhao, L.; Jin, T.; Zhao, J.; Gao, L. MicroRNA-146b-3p Promotes Cell Metastasis by Directly Targeting NF2 in Human Papillary Thyroid Cancer. Thyroid 2018, 28, 1627–1641. [Google Scholar] [CrossRef]
- Li, L.; Lv, B.; Chen, B.; Guan, M.; Sun, Y.; Li, H.; Zhang, B.; Ding, C.; He, S.; Zeng, Q. Inhibition of miR-146b expression increases radioiodine-sensitivity in poorly differential thyroid carcinoma via positively regulating NIS expression. Biochem. Biophys. Res. Commun. 2015, 462, 314–321. [Google Scholar] [CrossRef]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef]
- Wen, L.; Zhao, C.; Song, J.; Ma, L.; Ruan, J.; Xia, X.; Chen, Y.E.; Zhang, J.; Ma, P.X.; Xu, J. CRISPR/Cas9-Mediated TERT Disruption in Cancer Cells. Int. J. Mol. Sci. 2020, 21, 653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 2019, 38, 6172–6183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labun, K.; Montague, T.G.; Krause, M.; Torres Cleuren, Y.N.; Tjeldnes, H.; Valen, E. CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019, 47, W171–W174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Simon, P. Q-Gene: Processing quantitative real-time RT-PCR data. Bioinformatics 2003, 19, 1439–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Sequence 5′-3′ | Experiment | |
|---|---|---|
| GuideA Fw | CACCGTTGCTAGAGCTCACAGCCTATGG | cloning in PX462 plasmid |
| GuideA Rv | AAACCCATAGGCTGTGAGCTCTAGCAAC | cloning in PX462 plasmid |
| GuideB Fw | CACCGGTGGACTCAGTTCTGGTGCCCGG | cloning in PX461 plasmid |
| GuideB Rv | AAACCCGGGCACCAGAACTGAGTCCACC | cloning in PX461 plasmid |
| NIS Fw | AGTACATTGTAAGCCACGATGCTGTA | qPCR |
| NIS Rv | CGGTCACTTGGTTCAGGATGA | qPCR |
| TG Fw | CCTGCTGGCTCCACCTTGTTT | qPCR |
| TG Rv | CCTTGTTCTGAGCCTCCCATCGTT | qPCR |
| ZEB1 Fw | GATGACCTGCCAACAGACCA | qPCR |
| ZEB1 Rv | GCCCTTCCTTTCCTGTGTCA | qPCR |
| HMGA2 Fw | AAAGCAGCTCAAAAGAAAGCA | qPCR |
| HMGA2 Rv | TGTTGTGGCCATTTCCTAGGT | qPCR |
| RPL19 Fw | TCTCATGGAACACATCCACAA | qPCR |
| RPL19 Rv | TGGTCAGCCAGGAGCTTCTT | qPCR |
| Seq Fw | CTGGGAACGGGAGACGATTC | sequencing of target region |
| Seq Rv | GAAAGCTAAGTGGAGGCCGT | sequencing of target region |
| Histology | Genetic Driver | Culture Medium | |
|---|---|---|---|
| Nthy-ori 3-1 | Non-malignant | none | RPMI1640 + 10% SFB + 2 mM Glutamine |
| BCPAP | PTC | BRAFV600E | DMEM + 10% SFB |
| KTC2 | ATC | BRAFV600E | RPMI1640 + 5% SFB |
| 8305C | ATC | BRAFV600E | RPMI1640 + 10% SFB |
| C643 | ATC | HRASG13R | RPMI1640 + 10% SFB |
| Hth7 | ATC | NRASQ61R | DMEM + 10%SFB + 2 mM Glutamine |
| Hth83 | ATC | HRASQ61R | RPMI1640 + 10% SFB |
| Hth74 | ATC | NF1L732F | RPMI1640 + 10% SFB + 2 mM Glutamine |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santa-Inez, D.C.d.; Fuziwara, C.S.; Saito, K.C.; Kimura, E.T. Targeting the Highly Expressed microRNA miR-146b with CRISPR/Cas9n Gene Editing System in Thyroid Cancer. Int. J. Mol. Sci. 2021, 22, 7992. https://doi.org/10.3390/ijms22157992
Santa-Inez DCd, Fuziwara CS, Saito KC, Kimura ET. Targeting the Highly Expressed microRNA miR-146b with CRISPR/Cas9n Gene Editing System in Thyroid Cancer. International Journal of Molecular Sciences. 2021; 22(15):7992. https://doi.org/10.3390/ijms22157992
Chicago/Turabian StyleSanta-Inez, Daniel Casartelli de, Cesar Seigi Fuziwara, Kelly Cristina Saito, and Edna Teruko Kimura. 2021. "Targeting the Highly Expressed microRNA miR-146b with CRISPR/Cas9n Gene Editing System in Thyroid Cancer" International Journal of Molecular Sciences 22, no. 15: 7992. https://doi.org/10.3390/ijms22157992
APA StyleSanta-Inez, D. C. d., Fuziwara, C. S., Saito, K. C., & Kimura, E. T. (2021). Targeting the Highly Expressed microRNA miR-146b with CRISPR/Cas9n Gene Editing System in Thyroid Cancer. International Journal of Molecular Sciences, 22(15), 7992. https://doi.org/10.3390/ijms22157992







