Transcriptomic Analysis of the Photosynthetic, Respiration, and Aerenchyma Adaptation Strategies in Bermudagrass (Cynodon dactylon) under Different Submergence Stress
Abstract
1. Introduction
2. Results
2.1. Sequencing Quality Evaluation and Assembly
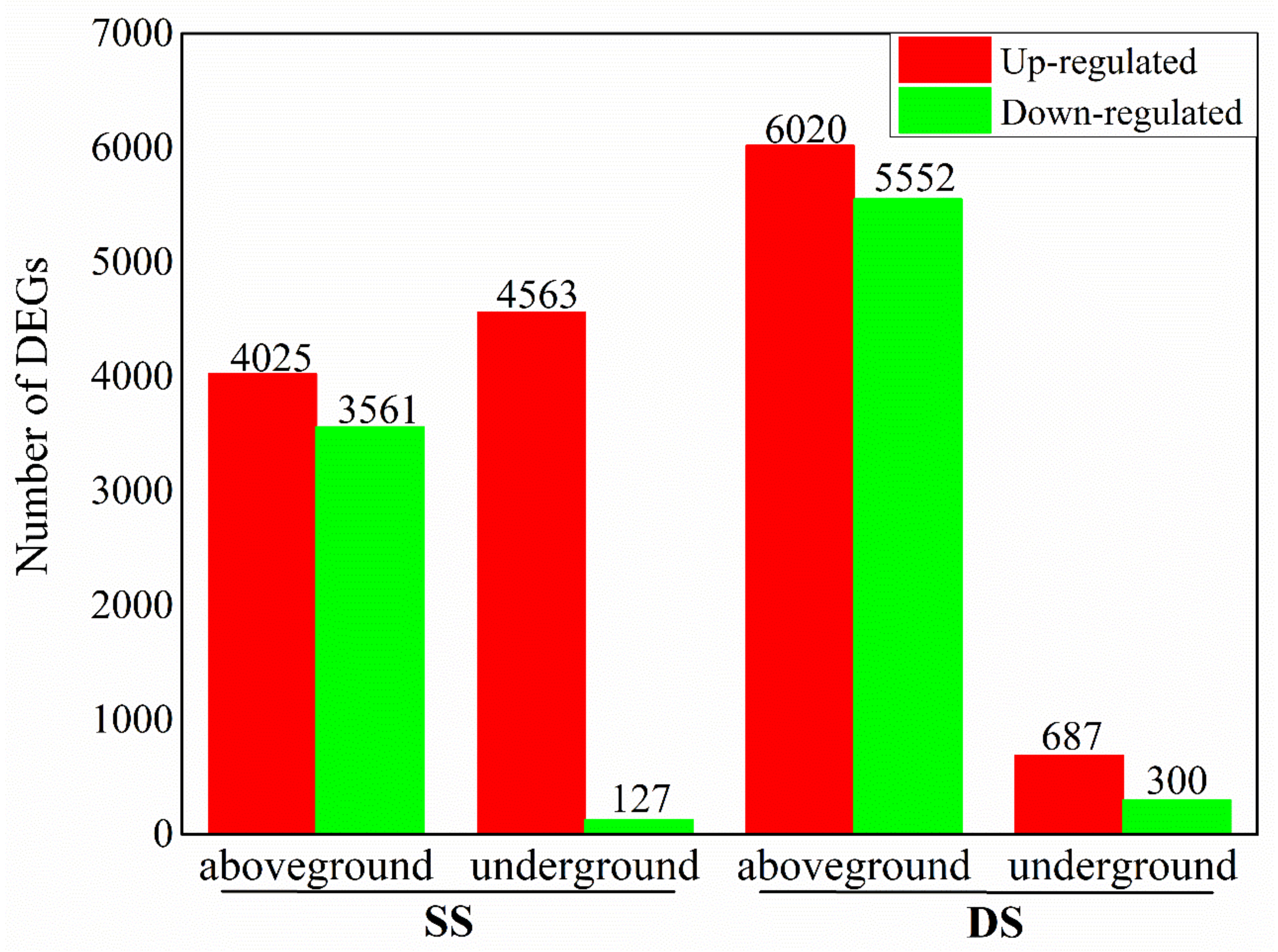
2.2. Identification of DEGs under Different Water Regimes
2.3. DEGs Related to Photosynthesis
2.4. DEGs Related to Energy Metabolism
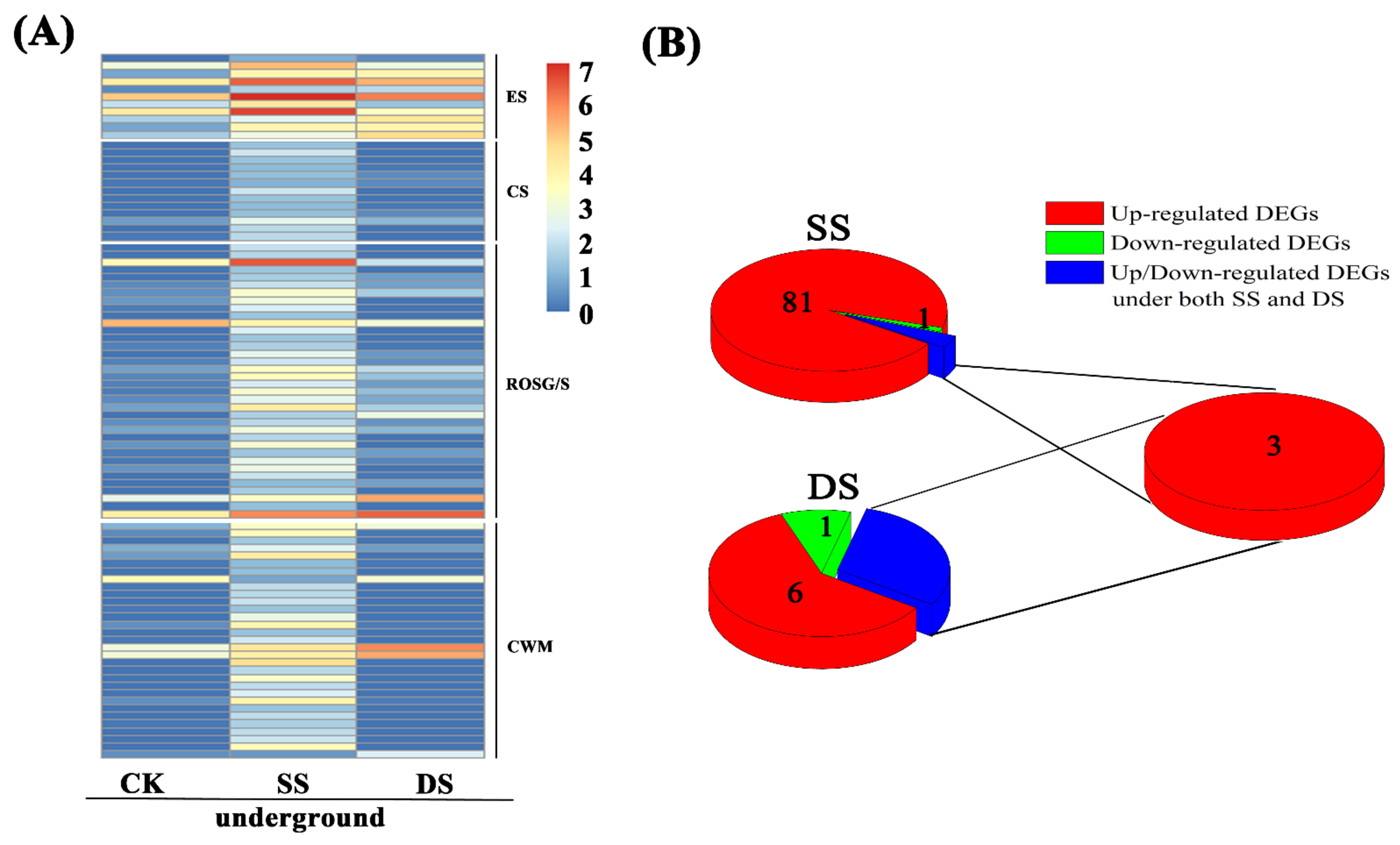
2.5. DEGs Related to Internal Gas Diffusion: Aerenchyma
3. Discussion
3.1. Modulation of Photosynthetic Pathways under SS and DS
3.2. Modulation of Energy Metabolism Pathways under SS and DS
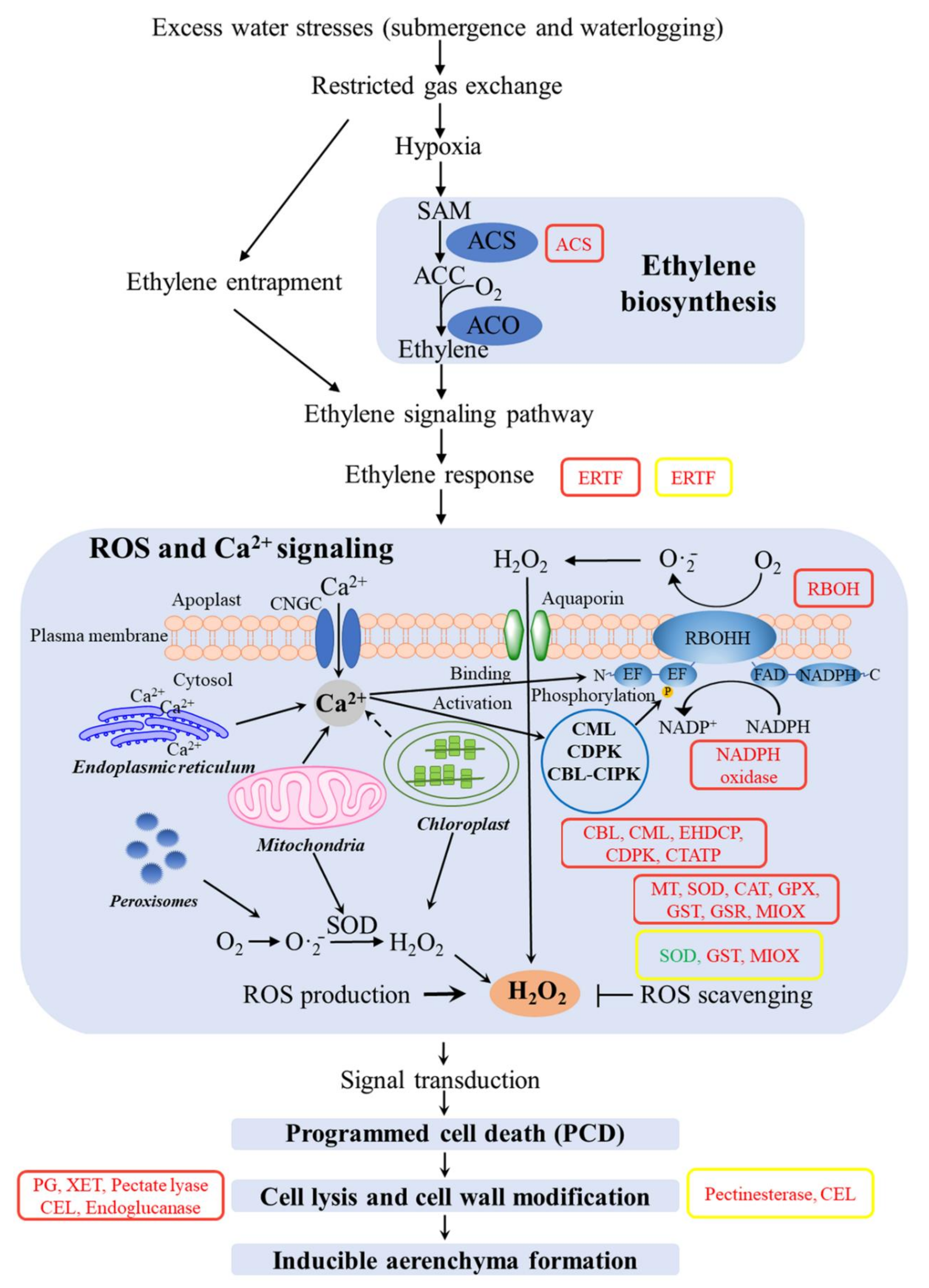
3.3. Modulation of Aerenchyma Pathways under SS and DS
4. Materials and Methods
4.1. Plant Materials and Experimental Treatments
4.2. RNA Isolation and Sequencing/RNA-seq Library Preparation and Sequencing
4.3. Identification and Functional Annotation of DEGs
4.4. The KEGG Pathway Enrichment Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC: | 1-aminocyclopropane-1-carboxylic acid |
| ACS: | ACC synthase |
| AMY: | α-amylase |
| ATP: | Adenosine triphosphate |
| CAT: | Catalase |
| CBL: | Calcineurin B-like protein |
| CDPK: | Calcium/calmodulin-dependent protein kinases |
| CEL: | Cellulase |
| CIPK: | CBL interacting protein kinases |
| CK: | Control |
| CML: | Calcium-binding proteins and calmodulin-like protein |
| CO2: | Carbon dioxide |
| CTATP: | Ca2+-transporting ATPase |
| DEGs: | Differentially expressed genes |
| DS: | Deep submergence |
| EHDCP: | EH domain-containing protein |
| EMP: | Embdem–Meyerhof–Parnas |
| ENO: | Enolase |
| ERTF: | Ethylene-responsive transcription factor |
| FBA: | D-fructose-1,6-diphosphate |
| GAPDH: | Glyceraldehyde-3-phosphate dehydrogenase |
| GPX: | Glutathione peroxidase |
| GPI: | Glucose-6-phosphate isomerase |
| GSR: | Glutathione reductase |
| GST: | Glutathione s-transferase |
| H2O2: | Hydrogen peroxide |
| HXK: | Hexokinase |
| INV: | Beta-fructofuranosidase |
| LHC: | Light-harvesting chlorophyll protein complex |
| MIOX: | Myo-inositol oxygenase |
| MT: | Metallothionein |
| NADPH: | Nicotinamide adenine dinucleotide phosphate |
| O2 | Superoxide anion radical |
| PCD: | Programmed cell death |
| PFK: | Phosphofructokinase |
| PG: | Polygalacturonase |
| PGAM: | Phosphoglyceratemutase |
| PGK: | Phosphoglycerate kinase |
| PK: | Pyruvate kinase |
| RBOH: | NADPH oxidase respiratory burst oxidase homolog |
| ROS: | Reactive oxygen species |
| SOD: | Superoxide dismutase |
| SS: | Shallow submergence |
| TCA cycle: | Tricarboxylic acid cycle |
| TGDR: | Three Gorges Dam Reservoir |
| TPI: | Triose-phosphate isomerase |
| XET: | Xyloglucan endotransglucosylase |
References
- Gill, M.B.; Zeng, F.; Shabala, L.; Zhang, G.P.; Yu, M.; Demidchik, V.; Shabala, S.; Zhou, M.X. Identification of QTL related to ROS formation under hypoxia and their association with waterlogging and salt tolerance in barley. Int. J. Mol. Sci. 2019, 20, 699. [Google Scholar] [CrossRef] [PubMed]
- Candan, N.; Tarhan, L. Tolerance or sensitivity responses of Mentha pulegium to osmotic and waterlogging stress in terms of antioxidant defense systems and membrane lipid peroxidation. Environ. Exp. Bot. 2012, 75, 83–88. [Google Scholar] [CrossRef]
- Chen, Z.T.; Chen, X.M.; Wang, C.Y.; Li, C.X. Foliar cellulose and lignin degradation of two dominant tree species in a riparian zone of the Three Gorges Dam Reservoir, China. Front. Plant Sci. 2020, 11, 569871. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.; Zheng, J.; Wokadala, C.; Zhang, S.L.; Yuan, Z.X.; Chen, Z.T.; Dong, Z.; He, X.R.; Li, C.X. Assessing riparian zone changes under the influence of stress factors in higher-order streams and tributaries: Implications for the management of massive dams and reservoirs. Sci. Total Environ. 2021, 776, 146011. [Google Scholar] [CrossRef]
- Zheng, J.; Arif, M.; Zhang, S.L.; Yuan, Z.X.; Zhang, L.M.; Dong, Z.; Tan, X.; Wokadala, C.; Li, C.X. The convergence of species composition along the drawdown zone of the Three Gorges Dam Reservoir, China: Implications for restoration. Environ. Sci. Pollut. Res. Int. 2021, 33818726. [Google Scholar]
- Fan, D.Y.; Xiong, G.M.; Zhang, A.Y.; Liu, X.; Xie, Z.Q.; Li, Z.J. Effect of water-lever regulation on species selection for ecological restoration practice in the water-level fluctuation zone of Three Gorges Reservoir. Chin. J. Plant Ecol. 2015, 39, 416–432. [Google Scholar]
- Arif, M.; Zhang, S.L.; Zheng, J.; Wokadala, C.; Sanelisiwe Mzondi, P.; Li, C.X. Evaluating the effects of pressure indicators on riparian zone health conditions in the Three Gorges Dam Reservoir, China. Forests 2020, 11, 214. [Google Scholar] [CrossRef]
- Li, Y.J.; Jia, H.Y.; Li, Q.T.; Bai, Y.X.; Liu, Y.S. Analysis of SNPs and SSRs of suitable Cynodon dactylon in fluctuating zone of Three Gorges Reservoir Area based on transcriptome data. Southwest China J. Agric. Sci. 2020, 33, 524–528. [Google Scholar]
- Tufail, A.; Ahmad, F.; Hameed, M.; Ahmad, R. growth performance and stomatal behavior in relation to ecotypic adaptations in cynodon dactylon (L.) pers. Pak. J. Bot. 2017, 49, 1395–1403. [Google Scholar]
- Xie, C.C.; Pu, S.Y.; Xiong, X.; Chen, S.Y.; Peng, L.L.; Fu, J.Y.; Sun, L.X.; Guo, B.M.; Jiang, M.Y.; Li, X. Melatonin-assisted phytoremediation of Pb-contaminated soil using bermudagrass. Environ. Sci. Pollut. R 2021, 33846924. [Google Scholar] [CrossRef]
- Zhu, Z.H.; Chen, Z.L.; Li, L.; Shao, Y. Response of dominant plant species to periodic flooding in the riparian zone of the Three Gorges Reservoir (TGR), China. Sci. Total Environ. 2020, 747, 141101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Peng, J.H.; Wan, C.Y.; Zheng, Z.W.; Qiu, D.G. Distribution and decomposition dynamics of herb plants in Pengxihe water-fluctuation-zone of the Three Gorges Reservoir. Acta Prataculturae Sin. 2010, 19, 146–152. [Google Scholar]
- Ke, Z.Y.; Wang, Q.; Shen, Q.Y.; Xie, M.T.; Xiao, H.L.; Liu, Y. Characteristics of plant community in the hydro-fluctuation belt of the Three Gorges Reservoir at the Zhong to Zigui section. Res. Environ. Yangtze Basin 2020, 29, 1975–1985. [Google Scholar]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Han, W.J.; Bai, L.L.; Li, C.X. Effects of flooding on photosynthesis, growth and nutrient content of Cynodon dactylon. Acta Prataculturae Sin. 2016, 25, 49–59. [Google Scholar]
- Li, X.X.; Li, C.X. Carbohydrate consumption of Hemarthria altissima and Cynodon dactylon is affected by flooding and plant density. Pratacultural Sci. 2018, 35, 2593–2601. [Google Scholar]
- Bailey-Serres, J.; Voesenek, L.A. Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef]
- Pedersen, O.; Colmer, T.D. Underwater photosynthesis and internal aeration of submerged terrestrial wetland plants. In Low-Oxygen Stress in Plants; VanDongen, J.T., Licausi, F., Eds.; Springer: London, UK, 2014; Volume 21, pp. 315–327. [Google Scholar]
- Basu, S.; Kumari, S.; Kumar, P.; Kumar, G.; Rajwanshi, R. Redox imbalance impedes photosynthetic activity in rice by disrupting cellular membrane integrity and induces programmed cell death under submergence. Physiol. Plant. 2021, 172, 1764–1778. [Google Scholar] [CrossRef]
- Boru, G.; Vantoai, T.; Alves, J.; Hua, D.; Knee, M. Responses of soybean to oxygen deficiency and elevated root-zone carbon dioxide concentration. Ann. Bot. 2003, 91, 447–453. [Google Scholar] [CrossRef]
- Li, Y.J.; Liu, R.H.; Yang, J.N.; Zhou, D.X.; Qin, H.W. Dynamics of enzyme activities of Cynodon dactulon roots from hydro-fluctuation belt in the Three Gorges Reservoir area during flooding. Res. Soil Water Conserv. 2014, 21, 288–292. [Google Scholar]
- Hu, Z.Y.; Qi, X.H.; Zhang, M.F.; Chen, X.H.; Nakazono, M. Roles of phytohormones in morphological and anatomical responses of plants to flooding stress. In Plant Hormones under Challenging Environmental Factors; Ahammed, G.J., Yu, J.Q., Eds.; Springer: Dordrecht, The Netherlands, 2016; pp. 117–132. [Google Scholar]
- Yamauchi, T.; Tanaka, A.; Inahashi, H.; Nishizawa, N.K.; Tsutsumi, N.; Inukai, Y.; Nakazono, M. Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 20770–20775. [Google Scholar] [CrossRef]
- Bartoli, G.; Forino, L.M.C.; Durante, M.; Tagliasacchi, A.M. A lysigenic programmed cell death-dependent process shapes schizogenously formed aerenchyma in the stems of the waterweed Egeria densa. Ann. Bot. 2015, 116, 91–99. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kordyum, E.; Kozeko, L.; Ovcharenko, Y.; Brykov, V. Assessment of alcohol dehydrogenase synthesis and aerenchyma formation in the tolerance of Sium L. species (Apiaceae) to water-logging. Aquat. Bot. 2017, 142, 71–77. [Google Scholar] [CrossRef]
- Visser, E.J.; Bogemann, G.M. Aerenchyma formation in the wetland plant Juncus effusus is independent of ethylene. New Phytol. 2006, 171, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Yamauchi, T.; Colmer, T.D.; Nakazono, M. Aerenchyma Formation in Plants. In Plant Cell Monographs; Van Dongen, J.T., Licausi, F., Eds.; Springer: London, UK, 2014; Volume 21, pp. 247–265. [Google Scholar]
- Evans, D.E. Aerenchyma Formation. New Phytol. 2004, 161, 35–49. [Google Scholar] [CrossRef]
- Rajhi, I.; Yamauchi, T.; Takahashi, H.; Nishiuchi, S.; Shiono, K.; Watanabe, R.; Mliki, A.; Nagamura, Y.; Tsutsumi, N.; Nishizawa, N.K.; et al. Identification of genes expressed in maize root cortical cells during lysigenous aerenchyma formation using laser microdissection and microarray analyses. New Phytol. 2011, 190, 351–368. [Google Scholar] [CrossRef]
- Chen, L.; Fan, J.B.; Hu, L.X.; Hu, Z.R.; Xie, Y.; Zhang, Y.Z.; Lou, Y.H.; Nevo, E.; Fu, J.M. A transcriptomic analysis of bermudagrass (Cynodon dactylon) provides novel insights into the basis of low temperature tolerance. BMC Plant Biol. 2015, 15, 216. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.T.; Shi, H.T.; Wang, Y.P.; Chan, Z.L. Contrasting changes caused by drought and submergence stresses in bermudagrass (Cynodon dactylon). Front. Plant Sci. 2015, 6, 951. [Google Scholar] [CrossRef]
- Zhu, H.S.; Yu, X.J.; Xu, T.; Wang, T.L.; Du, L.X.; Ren, G.H.; Dong, K.H. Transcriptome profiling of cold acclimation in bermudagrass (Cynodon dactylon). Sci. Hortic. 2015, 194, 230–236. [Google Scholar] [CrossRef]
- Dassanayake, M.; Larkin, J.C. Making plants break a sweat: The structure, function, and evolution of plant salt glands. Front. Plant Sci. 2017, 8, 406. [Google Scholar] [CrossRef]
- Li, Y.J.; Liu, R.H.; Zhou, D.X.; Yang, J.N.; Jiang, M.Y.; Wu, W.; Guo, S.H.; Gao, X. Transcriptome analysis of the adaptability of wild Cynodon dactylon in the fluctuating belt of Three Gorges Reservoir during flooding. Acta Ecol. Sin. 2018, 38, 8434–8441. [Google Scholar]
- Li, Y.J.; Yang, J.N.; Liu, R.H.; Zhou, D.X.; Gan, L.P.; Wu, Y.M. Transcriptome analysis of transcription factor of suitable cynodon dactylon in Three Gorges Reservoir Area under waterlogging. Southwest China J. Agric. Sci. 2018, 31, 265–269. [Google Scholar]
- Ma, Q.; Bao, A.K.; Chai, W.W.; Wang, W.Y.; Zhang, J.L.; Li, Y.X.; Wang, S.M. Transcriptomic analysis of the succulent xerophyte Zygophyllum xanthoxylum in response to salt treatment and osmotic stress. Plant Soil 2016, 402, 343–361. [Google Scholar] [CrossRef]
- Yu, L.P.; Tian, L.R.; Zhang, C.L.; Ma, N.; Li, J. Effects of low phosphorus stress on photosynthesis of rapeseed leaves in different periods. Chin. Agric. Sci. Bull. 2008, 24, 232–236. [Google Scholar]
- Beale, S.I. Green genes gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Shpilyov, A.V.; Zinchenko, V.V.; Shestakov, S.V.; Grimm, B.; Lokstein, H. Inactivation of the geranylgeranyl reductase (ChlP) gene in the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 2005, 1706, 195–203. [Google Scholar] [CrossRef]
- Xiang, R.; Shi, J.Q.; Zhang, H.B.; Dong, C.C.; Liu, L.; Fu, J.K.; He, X.Y.; Yan, Y.J.; Wu, Z.X. Chlorophyll a fluorescence and transcriptome reveal the toxicological effects of bisphenol A on an invasive cyanobacterium, Cylindrospermopsis raciborskii. Aquat. Toxicol. 2018, 200, 188–196. [Google Scholar] [CrossRef]
- Debus, R.J. The manganese and calcium ions of photosynthetic oxygen evolution. Biochim. Biophys. Acta 1992, 1102, 269–352. [Google Scholar] [CrossRef]
- Kondo, M.; Matsuda, H.; Noji, T.; Nango, M.; Dewa, T. Photocatalytic activity of the light-harvesting complex of photosystem II (LHCII) monomer. J. Photochem. Photobiol. A Chem. 2021, 406, 112926. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Lee, S.C.; Brinton, E. Waterproofing crops: Effective flooding survival strategies. Plant Physiol. 2012, 160, 1698–1709. [Google Scholar] [CrossRef]
- Locke, A.M.; Barding, G.J.; Sathnur, S.; Larive, C.K.; Bailey-Serres, J. Rice SUB1A constrains remodelling of the transcriptome and metabolome during submergence to facilitate post-submergence recovery. Plant Cell Environ. 2018, 41, 721–736. [Google Scholar] [CrossRef]
- Pan, Q.M.; Han, X.G.; Bai, Y.F.; Yang, J.C. Advances in physiology and ecology studies on stored non-structure carbohydrates in plants. Chin. Bull. Bot. 2002, 19, 30–38. [Google Scholar]
- Li, M.H.; Xiao, W.F.; Shi, P.; Wang, S.G.; Zhong, Y.D.; Liu, X.L.; Wang, X.D.; Cai, X.H.; Shi, Z.M. Nitrogen and carbon source-sink relationships in trees at the Himalayan treelines compared with lower elevations. Plant Cell Environ. 2008, 31, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.J.; Huang, X.P.; Zhang, J.P. Effect of environmental stress on non-structural carbohydrates reserves and transfer in seagrasses. Acta Ecol. Sin. 2012, 32, 6242–6250. [Google Scholar] [CrossRef][Green Version]
- Ye, X.Q.; Zeng, B. survival and carbohydrate storage in two tolerant plant species exposed to prolonged flooding in the Three Gorges Reservoir Region. Acta Hydrobiol. Sin. 2013, 37, 450–457. [Google Scholar]
- Patton, A.J.; Cunningham, S.M.; Volenec, J.J.; Reicher, Z.J. Differences in freeze tolerance of zoysiagrasses: II. carbohydrate and proline Accumulation. Crop. Sci. 2007, 47, 2170–2181. [Google Scholar] [CrossRef]
- Zhou, C.P.; Bai, T.; Wang, Y.; Wu, T.; Zhang, X.Z.; Xu, X.F.; Han, Z.H. Morpholoical and enzymatic responses to waterlogging in three Prunus species. Sci. Hortic. 2017, 221, 62–67. [Google Scholar] [CrossRef]
- Zeng, Y.L. Analysis of Glycolysis Way and Study on the Function of Aldolase Family Genes in Camellia oleifera Seed; Central South University of Forestry and Technology: Changsha, China, 2013. [Google Scholar]
- Jia, L.T. The Metabolic Characteristics and Physiological Regulation of GABA in Cerasus sachalinensis Roots under Waterlogging Stress; Shenyang Agricultural University: Shenyang, China, 2018. [Google Scholar]
- Yang, Y.J.; Shi, J.Q.; Jia, Y.L.; Bai, F.; Yang, S.Q.; Mi, W.M.; He, S.H.; Wu, Z.X. Unveiling the impact of glycerol phosphate (DOP) in the dinoflagellate, Peridinium bipes by photosynthesis and transcriptomic analysis. Environ. Sci. Eur. 2020, 32, 38. [Google Scholar] [CrossRef]
- Kogawara, S.; Yamanoshita, T.; Norisada, M.; Kojima, K. Steady sucrose degradation is a prerequisite for tolerance to root hypoxia. Tree Physiol. 2014, 34, 229–240. [Google Scholar] [CrossRef][Green Version]
- Colmer, T.D. Long-distance transport of gases in plants: A perspective on internal aeration and radial oxygen loss from roots. Plant Cell Environ. 2003, 26, 17–36. [Google Scholar] [CrossRef]
- Yamauchi, T.; Shiono, K.; Nagano, M.; Fukazawa, A.; Ando, M.; Takamure, I.; Mori, H.; Nishizawa, N.K.; Kawai-Yamada, M.; Tsutsumi, N.; et al. Ethylene biosynthesis is promoted by very-long-chain fatty acids during lysigenous aerenchyma formation in rice roots. Plant Physiol. 2015, 169, 180–193. [Google Scholar] [CrossRef]
- Zarei, A.; Korbes, A.P.; Younessi, P.; Montiel, G.; Champion, A.; Memelink, J. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 2011, 75, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Yoshioka, M.; Fukazawa, A.; Mori, H.; Nishizawa, N.K.; Tsutsumi, N.; Yoshioka, H.; Nakazono, M. An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 2017, 29, 775–790. [Google Scholar] [CrossRef] [PubMed]
- Beffagna, N.; Buffoli, B.; Busi, C. Modulation of reactive oxygen species production during osmotic stress in Arabidopsis thaliana cultured cells: Involvement of the plasma membrane Ca2+-ATPase and H+-ATPase. Plant Cell Physiol. 2005, 46, 1326–1339. [Google Scholar] [CrossRef]
- Kurusu, T.; Kuchitsu, K.; Tada, Y. Plant signaling networks involving Ca2+ and Rboh/Nox-mediated ROS production under salinity stress. Front. Plant Sci. 2015, 6, 427. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, Y.; Kaya, H.; Hiraoka, G.; Yumoto, F.; Kimura, S.; Kadota, Y.; Hishinuma, H.; Senzaki, E.; Yamagoe, S.; Nagata, K.; et al. Synergistic activation of the Arabidopsis NADPH oxidase atrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 2008, 283, 8885–8892. [Google Scholar] [CrossRef]
- Drerup, M.M.; Schlücking, K.; Hashimoto, K.; Manishankar, P.; Steinhorst, L.; Kuchitsu, K.; Kudla, J. The calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant 2013, 6, 559–569. [Google Scholar] [CrossRef]
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.P.; Schulze, W.X.; Romeis, T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749. [Google Scholar] [CrossRef]
- Overmyer, K.; Brosché, M.; Kangasjärvi, J. Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 2003, 8, 335–342. [Google Scholar] [CrossRef]
- Marti, M.C.; Stancombe, M.A.; Webb, A.A. Cell- and stimulus type-specific intracellular free Ca2+ signals in Arabidopsis. Plant Physiol. 2013, 163, 625–634. [Google Scholar] [CrossRef]
- Choi, W.G.; Toyota, M.; Kim, S.H.; Hilleary, R.; Gilroy, S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA 2014, 111, 6497–6502. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Watanabe, K.; Fukazawa, A.; Mori, H.; Abe, F.; Kawaguchi, K.; Oyanagi, A.; Nakazono, M. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J. Exp. Bot. 2014, 65, 261–273. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Harir, Y.; Mittler, R. The ROS Signaling Network of Cells. In Reactive Oxygen Species in Plant Signaling; DelRio, L.A., Puppo, A., Eds.; Springer: London, UK, 2009; pp. 165–174. [Google Scholar]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef]
- Hassinen, V.H.; Tervahauta, A.I.; Schat, H.; Karenlampi, S.O. Plant metallothioneins--metal chelators with ROS scavenging activity? Plant Biol. 2011, 13, 225–232. [Google Scholar] [CrossRef]
- Kong, Y.; Wang, Z.; Gu, Y.J.; Wang, Y.X. Research progress on aerenchyma formation in plant roots. Chin. Bull. Bot. 2008, 25, 248–253. [Google Scholar]
- Eric, J.W.V.; Laurentius, A.C.J.V. Acclimation to soil flooding-sensing and signal-transduction. Plant Soil 2005, 254, 197–214. [Google Scholar]
- Khan, M.I.R.; Trivellini, A.; Chhillar, H.; Chopra, P.; Ferrante, A.; Khan, N.A.; Ismail, A.M. The significance and functions of ethylene in flooding stress tolerance in plants. Environ. Exp. Bot. 2020, 179, 104188. [Google Scholar] [CrossRef]
- Arif, M.; Tahir, M.; Jie, Z.; Changxiao, L. Impacts of riparian width and stream channel width on ecological networks in main waterways and tributaries. Sci. Total Environ. 2021, 792, 148457. [Google Scholar] [CrossRef]
- Chen, Z.; Arif, M.; Wang, C.; Chen, X.; Li, C. Effects of hydrological regime on foliar decomposition and nutrient release in the riparian zone of the Three Gorges Reservoir, China. Front. Plant Sci. 2021, 12, 661865. [Google Scholar] [CrossRef]
- He, X.; Wang, T.; Wu, K.; Wang, P.; Qi, Y.; Arif, M.; Wei, H. Responses of swamp cypress (Taxodium distichum) and Chinese willow (Salix matsudana) roots to periodic submergence in mega-reservoir: Changes in organic acid concentration. Forests 2021, 12, 203. [Google Scholar] [CrossRef]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009, 37, e123. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]







| Sample | Base Number | GC Content | % ≥ Q30 | Clean Reads | Mapped Reads | Mapped Ratio |
|---|---|---|---|---|---|---|
| CK aboveground | 7,708,624,970 | 53.06% | 88.32% | 25,745,275 | 19,292,852 | 74.94% |
| CK underground | 8,084,087,134 | 52.92% | 88.99% | 26,992,331 | 20,142,196 | 74.62% |
| SS aboveground | 6,837,998,294 | 53.65% | 89.31% | 22,842,105 | 16,743,522 | 73.30% |
| SS underground | 7,941,119,312 | 53.13% | 88.97% | 26,520,179 | 19,451,568 | 73.35% |
| DS aboveground | 6,968,619,394 | 55.04% | 89.19% | 23,287,321 | 16,828,567 | 72.26% |
| DS underground | 6,834,848,554 | 52.71% | 89.00% | 22,825,956 | 16,885,731 | 73.98% |
| Name | Definition | Expression | Number |
|---|---|---|---|
| bglB | beta-glucosidase | down-regulated | 14 |
| egl | endoglucanase | down-regulated | 2 |
| glgA | starch synthase | down-regulated | 3 |
| glgC | glucose-1-phosphate adenylyl transferase | down-regulated | 4 |
| PYG | glycogen phosphorylase | down-regulated | 3 |
| bam | beta-amylase | down-regulated | 1 |
| Pgm | phosphoglucomutase | down-regulated | 1 |
| malQ | 4-alpha-glucanotransferase | down-regulated | 2 |
| GBE1 | 1,4-alpha-glucan branching enzyme | down-regulated | 3 |
| INV | beta-fructofuranosidase | down-regulated | 3 |
| AMY | alpha-amylase | down-regulated | 1 |
| ALDO | fructose-bisphosphate aldolase | down-regulated | 4 |
| PGK | phosphoglycerate kinase | down-regulated | 1 |
| gpmB | probable phosphoglycerate mutase | down-regulated | 1 |
| PK | pyruvate kinase | down-regulated | 1 |
| PDHA | pyruvate dehydrogenase E1 component alpha subunit | down-regulated | 1 |
| ACLY | ATP citrate (pro-S)-lyase | down-regulated | 2 |
| MDH | malate dehydrogenase | down-regulated | 2 |
| NdhJ | NAD(P)H-quinone oxidoreductase subunit J | down-regulated | 1 |
| NdhF | NAD(P)H-quinone oxidoreductase subunit 5 | down-regulated | 1 |
| CYTB | ubiquinol-cytochrome c reductase cytochrome b subunit | down-regulated | 2 |
| COX1 | cytochrome c oxidase subunit 1 | down-regulated | 1 |
| COX2 | cytochrome c oxidase subunit 2 | down-regulated | 1 |
| COX3 | cytochrome c oxidase subunit 3 | down-regulated | 1 |
| ATPF1G | F-type H+-transporting ATPase subunit gamma | down-regulated | 1 |
| ATPF0B | F-type H+-transporting ATPase subunit b | down-regulated | 1 |
| ATPF1D | F-type H+-transporting ATPase subunit delta | down-regulated | 1 |
| ATP1 | F-type H+-transporting ATPase subunit alpha | down-regulated | 1 |
| ATP2 | F-type H+-transporting ATPase subunit beta | down-regulated | 1 |
| ATP6 | F-type H+-transporting ATPase subunit a | down-regulated | 1 |
| ATP6B | V-type H+-transporting ATPase subunit B | down-regulated | 2 |
| AMY | alpha-amylase | up-regulated | 1 |
| INV | beta-fructofuranosidase | up-regulated | 1 |
| TPS | trehalose 6-phosphate synthase/phosphatase | up-regulated | 6 |
| GPI | glucose-6-phosphate isomerase | up-regulated | 1 |
| PFK | 6-phosphofructokinase 1 | up-regulated | 2 |
| ND4 | NADH-ubiquinone oxidoreductase chain 4 | up-regulated | 1 |
| Sum | 76 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Z.; Ni, X.; Arif, M.; Dong, Z.; Zhang, L.; Tan, X.; Li, J.; Li, C. Transcriptomic Analysis of the Photosynthetic, Respiration, and Aerenchyma Adaptation Strategies in Bermudagrass (Cynodon dactylon) under Different Submergence Stress. Int. J. Mol. Sci. 2021, 22, 7905. https://doi.org/10.3390/ijms22157905
Yuan Z, Ni X, Arif M, Dong Z, Zhang L, Tan X, Li J, Li C. Transcriptomic Analysis of the Photosynthetic, Respiration, and Aerenchyma Adaptation Strategies in Bermudagrass (Cynodon dactylon) under Different Submergence Stress. International Journal of Molecular Sciences. 2021; 22(15):7905. https://doi.org/10.3390/ijms22157905
Chicago/Turabian StyleYuan, Zhongxun, Xilu Ni, Muhammad Arif, Zhi Dong, Limiao Zhang, Xue Tan, Jiajia Li, and Changxiao Li. 2021. "Transcriptomic Analysis of the Photosynthetic, Respiration, and Aerenchyma Adaptation Strategies in Bermudagrass (Cynodon dactylon) under Different Submergence Stress" International Journal of Molecular Sciences 22, no. 15: 7905. https://doi.org/10.3390/ijms22157905
APA StyleYuan, Z., Ni, X., Arif, M., Dong, Z., Zhang, L., Tan, X., Li, J., & Li, C. (2021). Transcriptomic Analysis of the Photosynthetic, Respiration, and Aerenchyma Adaptation Strategies in Bermudagrass (Cynodon dactylon) under Different Submergence Stress. International Journal of Molecular Sciences, 22(15), 7905. https://doi.org/10.3390/ijms22157905







