Fluorescence Spectroscopic Analysis of ppGpp Binding to cAMP Receptor Protein and Histone-Like Nucleoid Structuring Protein
Abstract
1. Introduction
2. Results
2.1. Cloning, Over-Expression, and Purification of CRP and H-NS from E. coli
2.2. Binding of ppGpp and cAMP to CRP
2.3. The Binding Site for ppGpp on CRP
2.4. Binding of ppGpp and cAMP to H-NS
2.5. The Binding Site for ppGpp on H-NS
3. Discussion
4. Materials and Methods
4.1. Cloning of CRP from E. coli
4.2. Over-Expression and Isolation of H-NS and CRP
4.3. Determination of Ligand Binding by Fluorescence Spectroscopy
4.4. Computational Prediction of ppGpp-Binding to CRP
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sutherland, E.; Rall, T. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J. Biol. Chem. 1958, 232, 1077–1091. [Google Scholar] [CrossRef]
- Passner, J.; Schultz, S.; Steitz, T. Modeling the cAMP-induced Allosteric Transition Using the Crystal Structure of CAP-cAMP at 2.1Å Resolution. J. Mol. Biol. 2000, 304, 847–859. [Google Scholar] [CrossRef]
- Parkinson, G.; Wilson, C.; Gunasekera, A.; Ebright, Y.W.; Ebright, R.; Berman, H.M. Structure of the CAP-DNA Complex at 2.5 Å Resolution: A Complete Picture of the Protein-DNA Interface. J. Mol. Biol. 1996, 260, 395–408. [Google Scholar] [CrossRef]
- Hollands, K.; Busby, S.J.; Lloyd, G.S. New targets for the cyclic AMP receptor protein in theEscherichia coliK-12 genome. FEMS Microbiol. Lett. 2007, 274, 89–94. [Google Scholar] [CrossRef]
- Shimada, T.; Fujita, N.; Yamamoto, K.; Ishihama, A. Novel Roles of cAMP Receptor Protein (CRP) in Regulation of Transport and Metabolism of Carbon Sources. PLoS ONE 2011, 6, e20081. [Google Scholar] [CrossRef] [PubMed]
- Epstein, W.; Rothman-Denes, L.B.; Hesse, J. Adenosine 3’:5’-cyclic monophosphate as mediator of catabolite repression in Escherichia coli. Proc. Natl. Acad. Sci. USA 1975, 72, 2300–2304. [Google Scholar] [CrossRef]
- Pastan, I.; Adhya, S. Cyclic adenosine 5′-monophosphate in Escherichia coli. Bacteriol. Rev. 1976, 40, 527–551. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Blazy, B.; Baudras, A. An equilibrium study of the cooperative binding of adenosine cyclic 3′,5′-monophosphate and guanosine cyclic 3′,5′-monophosphate to the adenosine cyclic 3′,5′-monophosphate receptor protein from Escherichia coli. Biochemistry 1980, 19, 5124–5130. [Google Scholar] [CrossRef] [PubMed]
- Heyduk, T.; Lee, J.C. Escherichia coli cAMP receptor protein: Evidence for three protein conformational states with different promoter binding affinities. Biochemistry 1989, 28, 6914–6924. [Google Scholar] [CrossRef] [PubMed]
- Azam, T.A.; Ishihama, A. Twelve Species of the Nucleoid-associated Protein from Escherichia coli. J. Biol. Chem. 1999, 274, 33105–33113. [Google Scholar] [CrossRef]
- Bertin, P.; Terao, E.; Lee, E.H.; Lejeune, P.; Colson, C.; Danchin, A.; Collatz, E. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 1994, 176, 5537–5540. [Google Scholar] [CrossRef]
- Hommais, F.; Krin, E.; Laurent-Winter, C.; Soutourina, O.; Malpertuy, A.; Le Caer, J.-P.; Danchin, A.; Bertin, P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 2001, 40, 20–36. [Google Scholar] [CrossRef]
- Brandi, A.; Giangrossi, M.; Fabbretti, A.; Falconi, M. The hns Gene of Escherichia coli Is Transcriptionally Down-Regulated by (p)ppGpp. Microorganism 2020, 8, 1558. [Google Scholar] [CrossRef]
- Lu, P.; Vogel, C.; Wang, R.; Yao, X.; Marcotte, E.M. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2007, 25, 117–124. [Google Scholar] [CrossRef]
- Arikea, L.; Valgepeaa, K.; Peilc, L.; Nahkua, R.; Adamberga, K.; Vilua, R. Comparison and applications of label-free absolute proteome quantification methods on Escherichia coli. J. Proteom. 2012, 75, 5437–5448. [Google Scholar] [CrossRef]
- Cashel, M.; Gentry, D.R.; Hernandez, V.H.; Vinella, D. The stringent response. In Escherichia coli and Salmonella: Cellular and Molecular Biology; Neidhardt, F.C., Ed.; ASM Press: Washington, DC, USA, 1996; pp. 1458–1496. [Google Scholar]
- Dalebroux, Z.D.; Svensson, S.L.; Gaynor, E.C.; Swanson, M.S. ppGpp Conjures Bacterial Virulence. Microbiol. Mol. Biol. Rev. 2010, 74, 171–199. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, L.U.; Farewell, A.; Nyström, T. ppGpp: A global regulator in Escherichia coli. Trends Microbiol. 2005, 13, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sanders, G.M.; Grossman, A.D. Nutritional Control of Elongation of DNA Replication by (p)ppGpp. Cell 2007, 128, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Srivatsan, A.; Wang, J.D. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 2008, 11, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Traxler, M.; Summers, S.M.; Nguyen, H.-T.; Zacharia, V.M.; Hightower, G.A.; Smith, J.T.; Conway, T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 2008, 68, 1128–1148. [Google Scholar] [CrossRef]
- Wang, B.; Dai, P.; Ding, D.; Del Rosario, A.; Grant, R.A.; Pentelute, B.L.; Laub, M.T. Author Correction: Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 2019, 15, 141–150. [Google Scholar] [CrossRef]
- Harman, J.G. Allosteric regulation of the cAMP receptor protein. Biochim. Biophys. Acta 2001, 1547, 1–17. [Google Scholar] [CrossRef]
- Seok, S.-H.; Im, H.; Won, H.-S.; Seo, M.-D.; Lee, Y.-S.; Yoon, H.-J.; Cha, M.-J.; Park, J.-Y.; Lee, B.-J. Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1726–1742. [Google Scholar] [CrossRef] [PubMed]
- Wasylewski, M.; Małecki, J.; Wasylewski, Z. Fluorescence study of Escherichia coli cyclic AMP receptor protein. J. Protein Chem. 1995, 14, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Bennett, B.D.; Kimball, E.H.; Gao, M.; Osterhout, R.; Van Dien, S.J.; Rabinowitz, J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009, 5, 593–599. [Google Scholar] [CrossRef]
- Johansson, J.; Balsalobre, C.; Wang, S.-Y.; Urbonaviciene, J.; Jin, D.J.; Sondén, B.; Uhlin, B.E. Nucleoid Proteins Stimulate Stringently Controlled Bacterial Promoters: A Link between the cAMP-CRP and the (p)ppGpp Regulons in Escherichia coli. Cell 2000, 102, 475–485. [Google Scholar] [CrossRef][Green Version]
- Cashel, M. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid starved stringent strains. J. Biol. Chem. 1969, 244, 3133–3141. [Google Scholar] [CrossRef]
- Haseltine, W.A.; Block, R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. USA 1973, 70, 1564–1568. [Google Scholar] [CrossRef] [PubMed]
- Malecki, J.; Polit, A.; Wasylewski, Z. Kinetic studies of cAMP-induced allosteric changes in cyclic AMP receptor protein from Escherichia coli. J. Biol. Chem. 2000, 275, 8480–8486. [Google Scholar] [CrossRef]
- Krohn, M.; Wagner, R. Procedure for the Rapid Preparation of Guanosine Tetraphosphate (ppGpp) from Escherichia coli Ribosomes. Anal. Biochem. 1995, 225, 188–190. [Google Scholar] [CrossRef]
- Hoang, H.N.; Tran, T.T.; Jung, C. The Activation of Glycerol Dehydrogenase from Escherichia coli by ppGpp. Bull. Korean Chem. Soc. 2020, 41, 133–138. [Google Scholar] [CrossRef]
- Gill, S.C.; Von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef]
- Clark, A.R. Analysis of ligand binding by enzymes. In Enzymology Labfax; Engel, P.C., Ed.; Academic Press: San Diego, CA, USA, 1996; pp. 199–221. [Google Scholar]
- Duysak, T.; Afzal, A.R.; Jung, C.-H. Determination of glutathione-binding to proteins by fluorescence spectroscopy. Biochem. Biophys. Res. Commun. 2021, 557, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Plenum Press: New York, NY, USA, 1999; pp. 53–55. [Google Scholar]
- Shin, W.H.; Heo, L.; Lee, J.; Ko, J.; Seok, C.; Lee, J. LigDockCSA: Protein-ligand docking using conformational space annealing. J. Comput. Chem. 2011, 32, 3226–3232. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.H.; Kim, J.K.; Kim, D.S.; Seok, C. GalaxyDock2: Protein-ligand docking using beta-complex and global optimization. J. Comput. Chem. 2013, 34, 2647–2656. [Google Scholar] [CrossRef]
- Baek, M.; Shin, W.H.; Chung, H.W.; Seok, C. GalaxyDock BP2 score: A hybrid scoring function for accurate protein-ligand docking. J. Comput. Aided Mol. Des. 2017, 31, 653–666. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System, version 2.0; Schrödinger, Inc.: New York, NY, USA, 2019.
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand in-teractions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
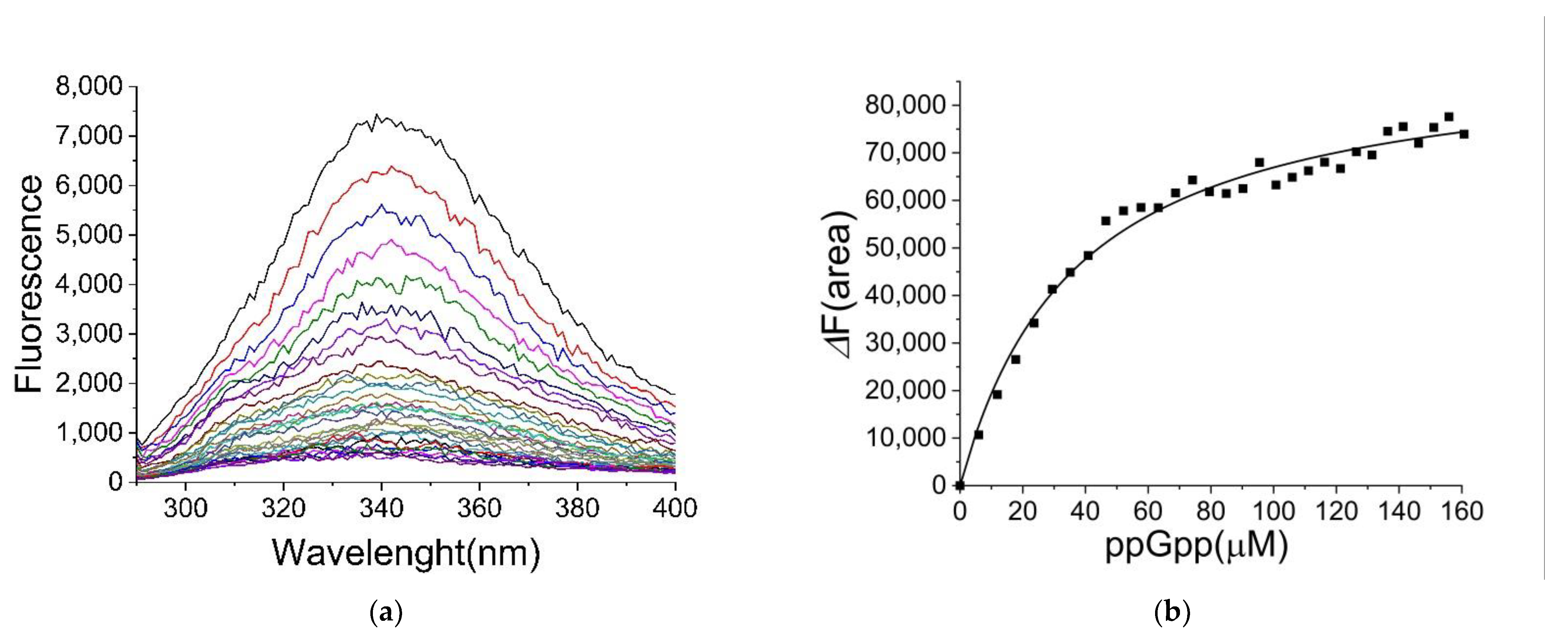
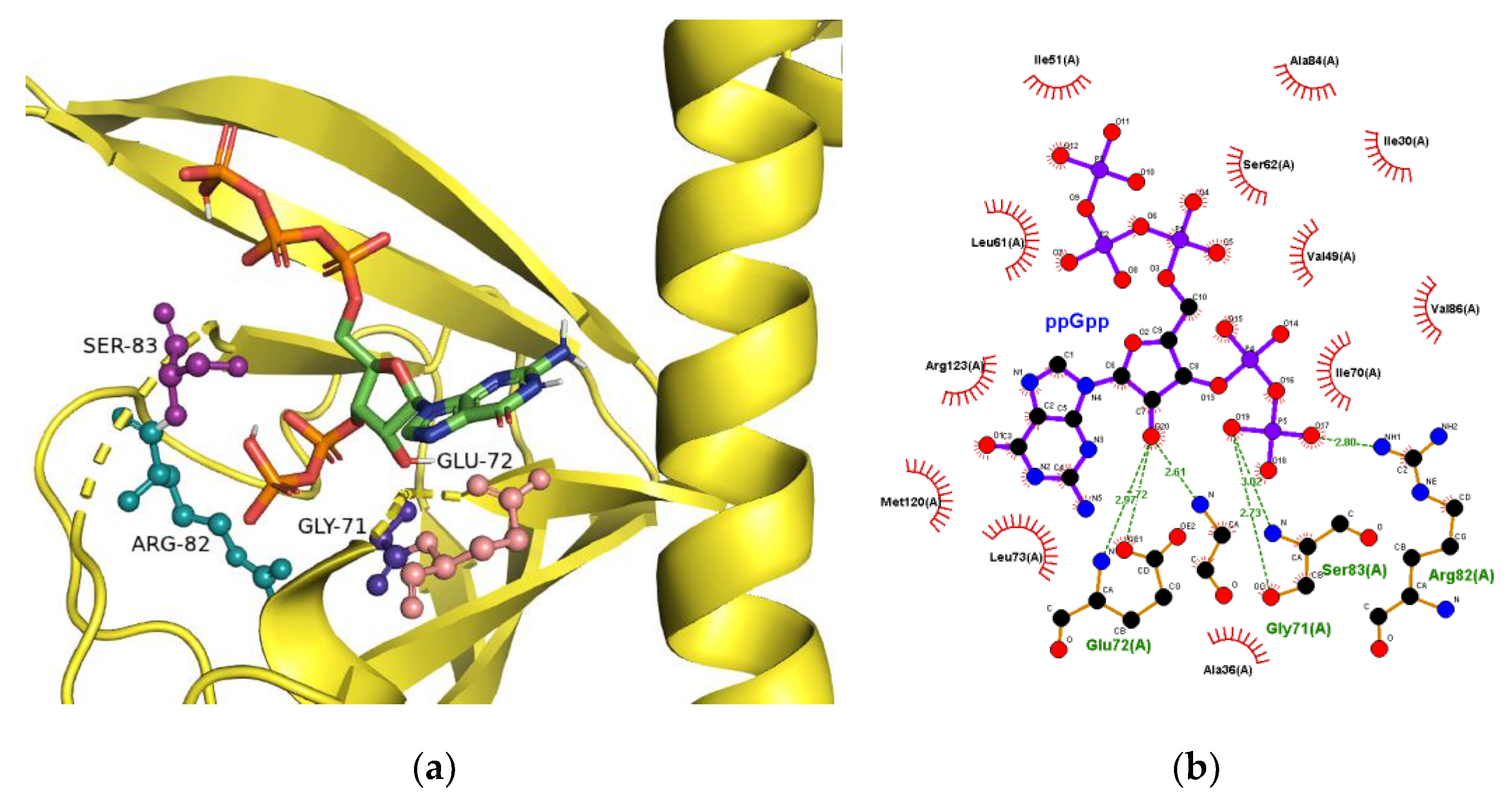
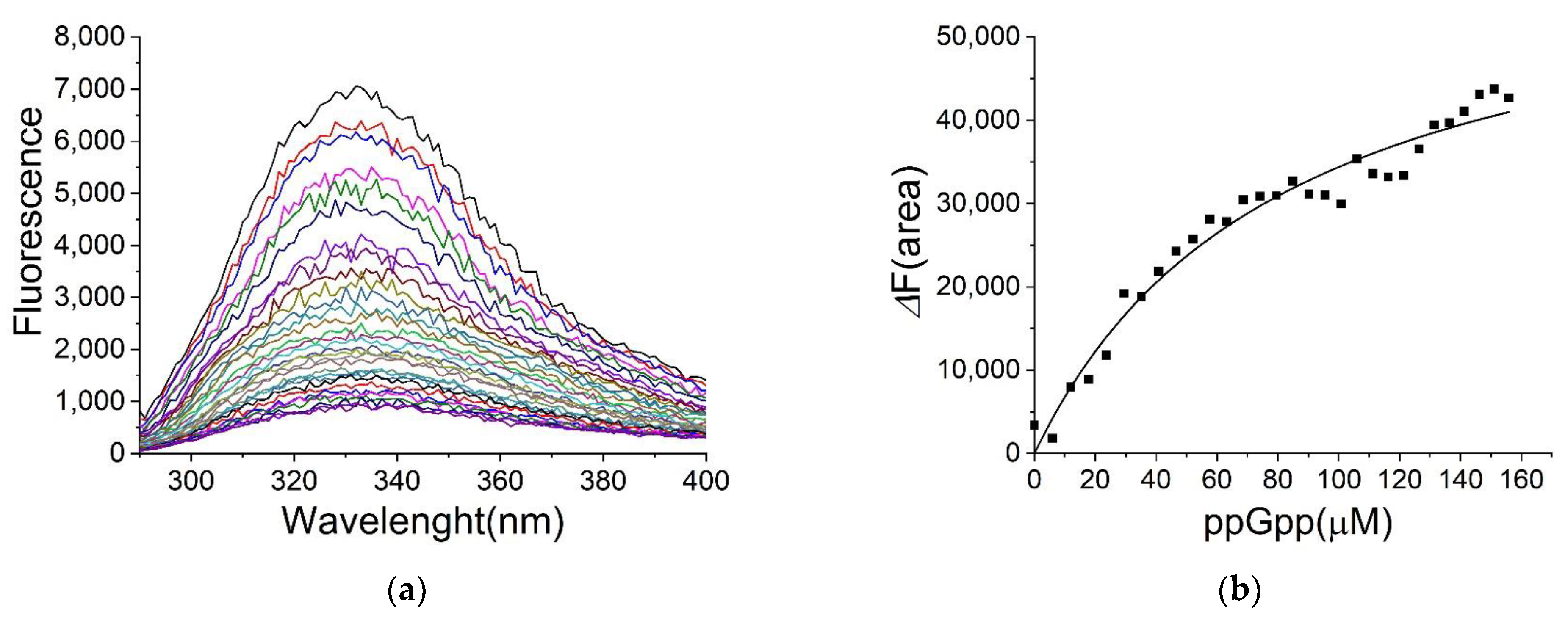
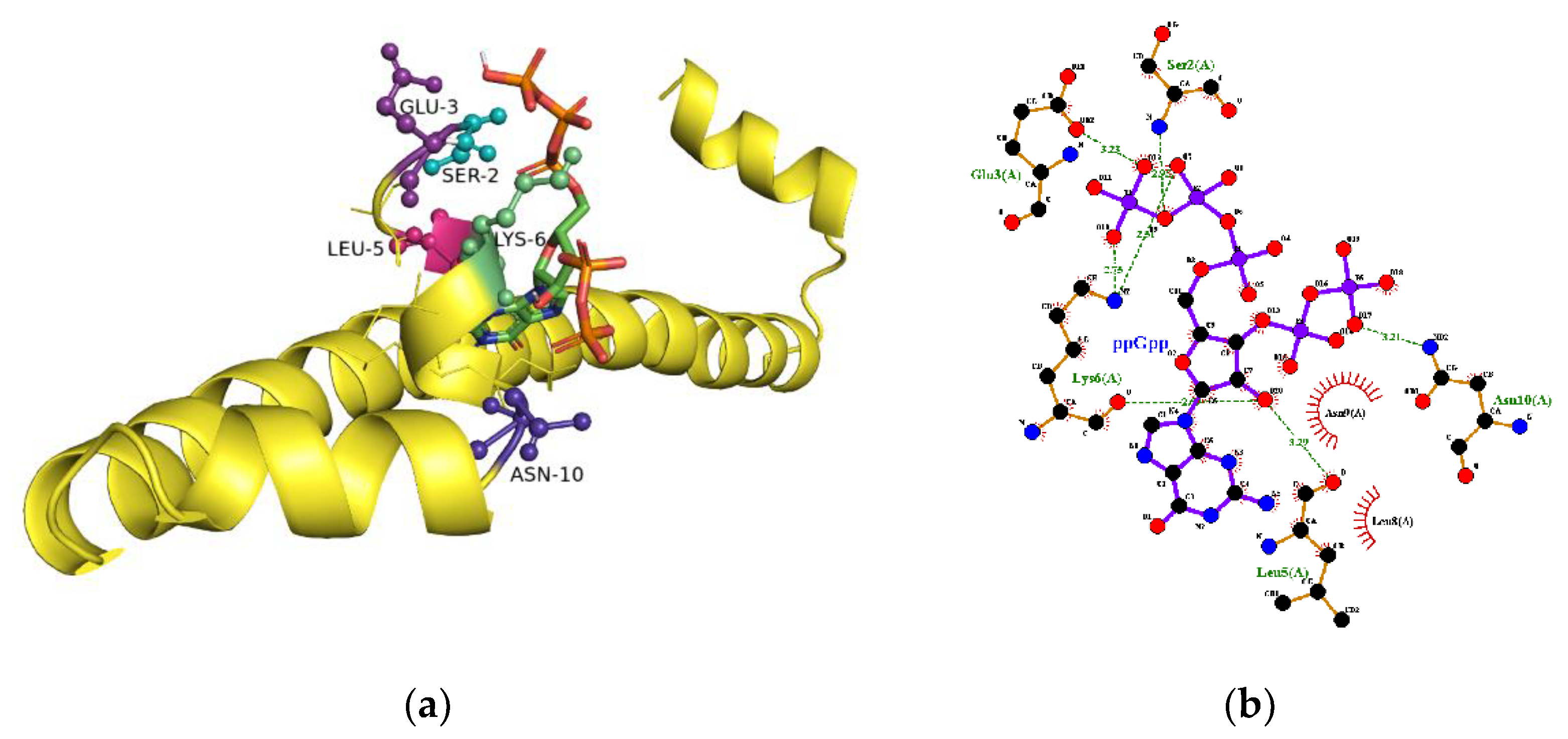
| Ligand | KD (µM) |
|---|---|
| ppGpp | 35 ± 3 |
| GDP | 62 ± 4 |
| GTP | 35 ± 3 |
| cGMP | 17 ± 1 |
| cAMP | 22 ± 3 |
| ATP | 33 ± 4 |
| cAMP a | 25 and 11 a |
| Binding Pocket Residue | 1g6n (cAMP Form) | 1n9i (cGMP Form) |
|---|---|---|
| Q80 | −9.90 | −10.53 |
| E81 | −7.07 | −10.43 |
| R82 | −9.42 | −13.48 |
| S83 | −10.05 | −14.01 |
| A84 | −10.33 | −12.60 |
| Ligand | KD (µM) |
|---|---|
| ppGpp | 80 ± 11 |
| GDP | 72 ± 6 |
| GTP | 120 ± 25 |
| cAMP | 270 ± 40 |
| ATP | NS 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duysak, T.; Tran, T.T.; Afzal, A.R.; Jung, C.-H. Fluorescence Spectroscopic Analysis of ppGpp Binding to cAMP Receptor Protein and Histone-Like Nucleoid Structuring Protein. Int. J. Mol. Sci. 2021, 22, 7871. https://doi.org/10.3390/ijms22157871
Duysak T, Tran TT, Afzal AR, Jung C-H. Fluorescence Spectroscopic Analysis of ppGpp Binding to cAMP Receptor Protein and Histone-Like Nucleoid Structuring Protein. International Journal of Molecular Sciences. 2021; 22(15):7871. https://doi.org/10.3390/ijms22157871
Chicago/Turabian StyleDuysak, Taner, Thanh Tuyen Tran, Aqeel Rana Afzal, and Che-Hun Jung. 2021. "Fluorescence Spectroscopic Analysis of ppGpp Binding to cAMP Receptor Protein and Histone-Like Nucleoid Structuring Protein" International Journal of Molecular Sciences 22, no. 15: 7871. https://doi.org/10.3390/ijms22157871
APA StyleDuysak, T., Tran, T. T., Afzal, A. R., & Jung, C.-H. (2021). Fluorescence Spectroscopic Analysis of ppGpp Binding to cAMP Receptor Protein and Histone-Like Nucleoid Structuring Protein. International Journal of Molecular Sciences, 22(15), 7871. https://doi.org/10.3390/ijms22157871






