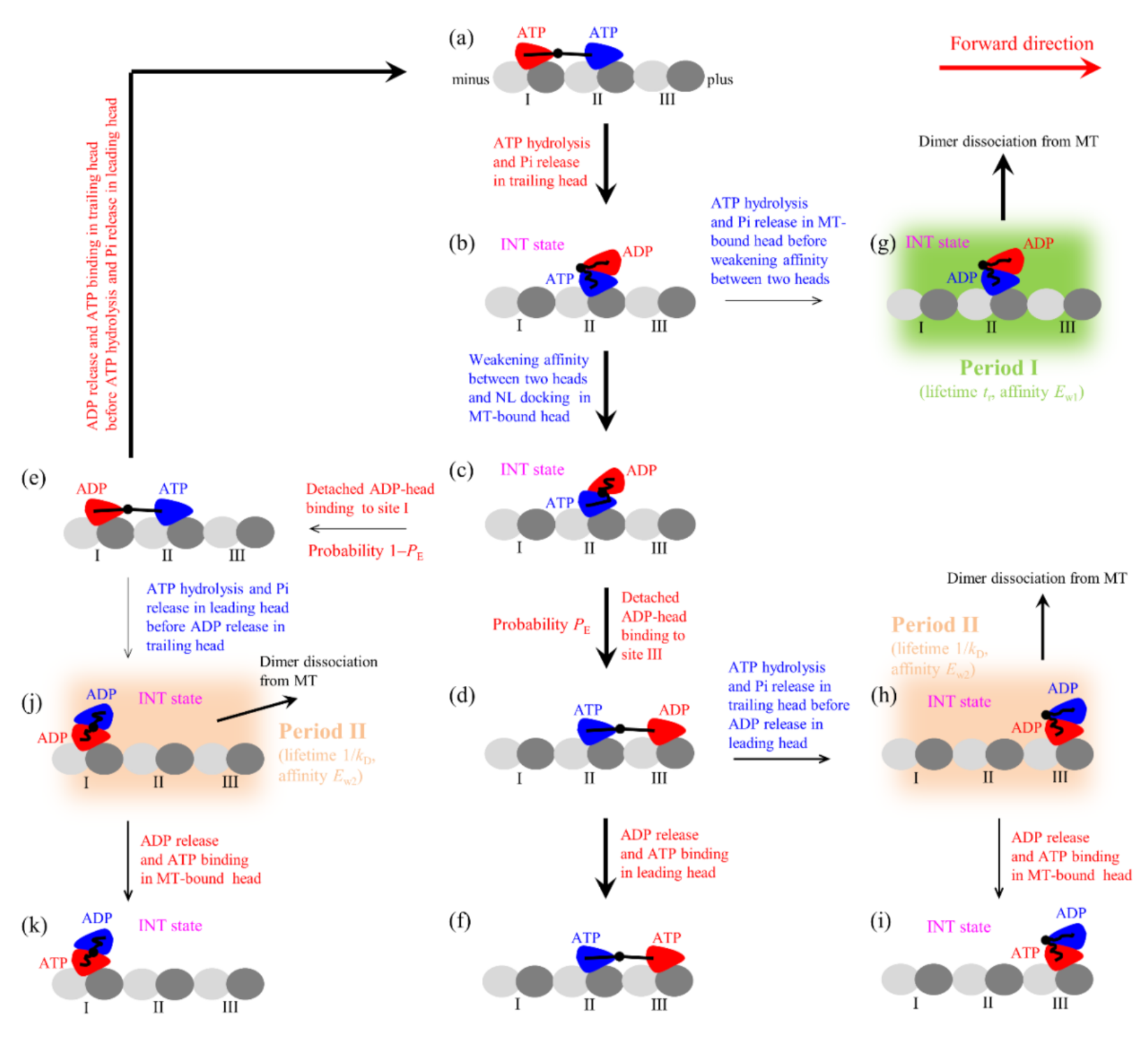
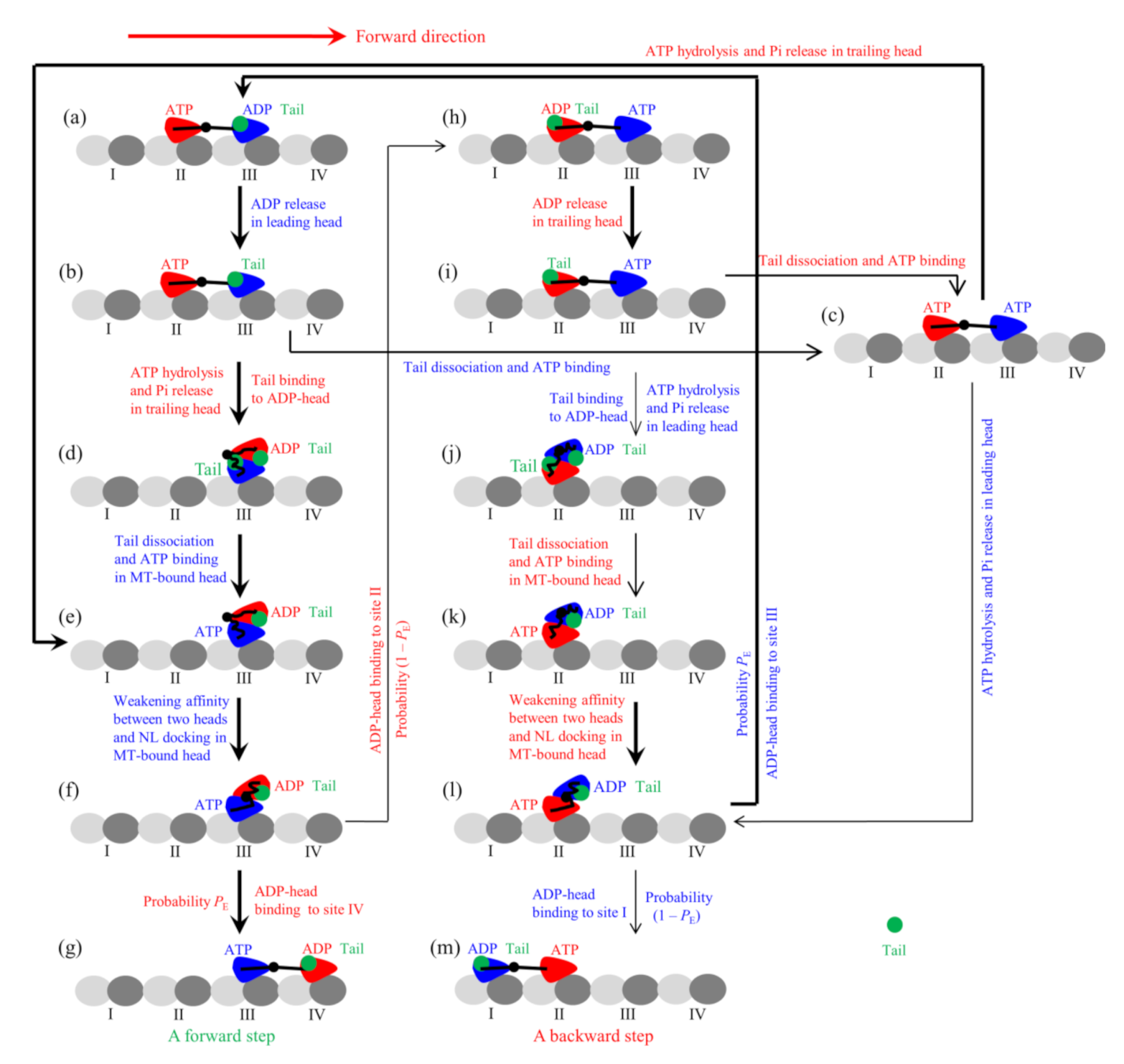
2.1. Dynamics of Single Eg5-ΔTail Moving on Single MT
In this section, on the basis of the model for the chemomechanical coupling of Eg5-ΔTail (
Figure 1, see Methods for detailed descriptions), we study the dynamics of the single Eg5-ΔTail motor moving on the single MT. Consider a load
F on the stalk of the motor, with
F < 0 (
F > 0) being the backward (forward) load. As derived before [
16], the load dependence of effective probability
PE (defined in
Figure 1) has the form
where
ED is the energy change associated with both the NL docking and the conformational change of the head induced by ATP binding,
d(+) is characteristic distance for the movement of the detached head from INT position to either the forward or backward binding site on MT, and
being the thermal energy. Note that in the pathway for FL-Eg5 (
Figure 2, see Methods for detailed descriptions), which is modified from that for Eg5-ΔTail,
PE has the same form.
Before presenting equations for velocity and run length, let us define rate constants of the ATPase activity and NL docking. We denote by
k(+) the rate of ATP transition to ADP in the head with the forward NL orientation (e.g., the trailing head) and by
k(−) the rate of ATP transition to ADP in the head without the forward NL orientation (e.g., the leading head). Rates
k(+) and
k(−) are independent of the force on the NLs, which are consistent with the available experimental data (see, e.g., [
17,
18,
19] for detailed discussion). We denote by
kD the rate of ADP release from the head bound to MT, which for simplicity is treated here to be independent of NL direction and force on NL. Since both the rate of weakening the affinity of the MT-bound ATP-head to the detached ADP-head and the rate of NL docking of the MT-bound ATP-head are determined by the rate of the large conformational change of the ATP-head, the three rates have approximately the same values. Thus, we use
kNL to represent both the rate of NL docking and that of weakening between the two heads.
Considering that
kNL >>
k(+) (see later) and the weak MT-binding periods (Period I and Period II) occur occasionally and when it occurs the motor has a large probability to dissociate from MT, the overall ATPase rates of trailing and leading heads can be approximately calculated with [
16]
The total ATPase rate of the motor is
kT +
kL. The overall forward stepping rate of the motor is
PEkT and the overall backward stepping rate is (1 −
PE)
kL. The velocity of the motor can then be written as
where
d = 8.2 nm is the step size.
The occurrence probability of Period I,
PI, can be calculated with [
16,
20]
where it is considered that under no or a backward load (
F 0) on the stalk the NL of the MT-bound head in INT state before NL docking is not in the forward orientation, with the rate of ATP transition to ADP equal to
k(−), while under a forward load larger than or equal to 4 pN (
F 4 pN) the NL of the MT-bound head in INT state before NL docking is driven to be in the forward orientation, with the rate of ATP transition to ADP equal to
k(+). Under the forward load smaller than 4 pN, the following phenomenological form can be adopted for the rate of ATP transition to ADP in the MT-bound head [
20]
where
k =
k(−) when
F = 0 and
k =
k(+) when
F = 4 pN. The occurrence probability of Period I can then be calculated with
In Period I, since Ew1 is very small the motor can dissociate from MT with a nearly 100% probability even under no load, giving the dissociation probability in Period I PdI 1 under any load.
The occurrence probability of Period II,
PII, can be calculated with [
16]
The dissociation probability,
PdII, in Period II can be calculated with
where
kdII is the dissociation rate during Period II, with
kw0 being the dissociation rate under no load and
being the distance parameter for dissociation. To be consistent with the Debye length of about 1 nm in solution, we take
= 1 nm.
For approximation, in this work we neglect the dissociation of kinesin in the strong MT-binding state. Thus, the dissociation rate can be calculated with [
16,
20]
where
kT is calculated by Equation (2),
kL is calculated by Equation (3),
PI is calculated by Equations (5)–(8),
PdI = 1,
PII is calculated by Equation (9), and
PdII is calculated by Equations (10) and (11). The run length can then be calculated with
where
v is calculated with Equation (4) and
is calculated with Equation (12).
Before presenting our theoretical results using the above equations, we firstly discuss the choice of the parameter values. The biochemical data for kinesin-1 head showed that without NL the ATPase rate of the head is reduced largely while the ADP release rate is not affected [
21], implying that the interaction of NL in the forward (or docked) direction enhances the rate of ATP transition to ADP. Thus, we take the rate constant of ATP transition to ADP in Eg5 head with the NL in the forward direction being also larger than that in the head without in the forward direction, with
k(−) =
k(+)/15. The velocity at the large forward load is determined mainly by
k(+). To be consistent with the single-molecule data of Valentine et al. [
11] at the large forward load, we take
k(+) = 12.8
. Note that the above value of
k(+) is close to that determined biochemically [
22]. Since the curve form of velocity versus load is determined mainly by
ED and
d(+), to be consistent with the single-molecule data of Valentine et al. [
11], we adjust values of
ED and
d(+), with
ED = 2.5
kBT and
d(+) = 2.2 nm. Note that this value of
ED = 2.5
kBT is consistent with the experimentally measured free energy change associated with NL docking for kinesin-1 head to be smaller than 1
kBT [
23] and the atomistic MD simulated free energy change associated with the large conformational change to be only about 1.7
kBT for kinesin-1 head [
24]. Since the run length for Eg5 under no load is determined mainly by
kNL,
kD and
kw0 besides the unloaded velocity, to be consistent with the single-molecule data of Valentine et al. [
12] for the unloaded run length, we adjust values of
kNL,
kD and
kw0, with
kNL = 200
,
kD = 50
and
kw0 = 20
. The above value of
kD is similar to that measured biochemically for Eg5 [
22]. The above value of
kNL for Eg5 (200
) is much smaller than that for kinesin-1 (1500
) determined before [
16,
20], which is also consistent with the biochemical results for Eg5 and kinesin-1 [
22,
25]. For clarity, the parameter values are listed in
Table 1.
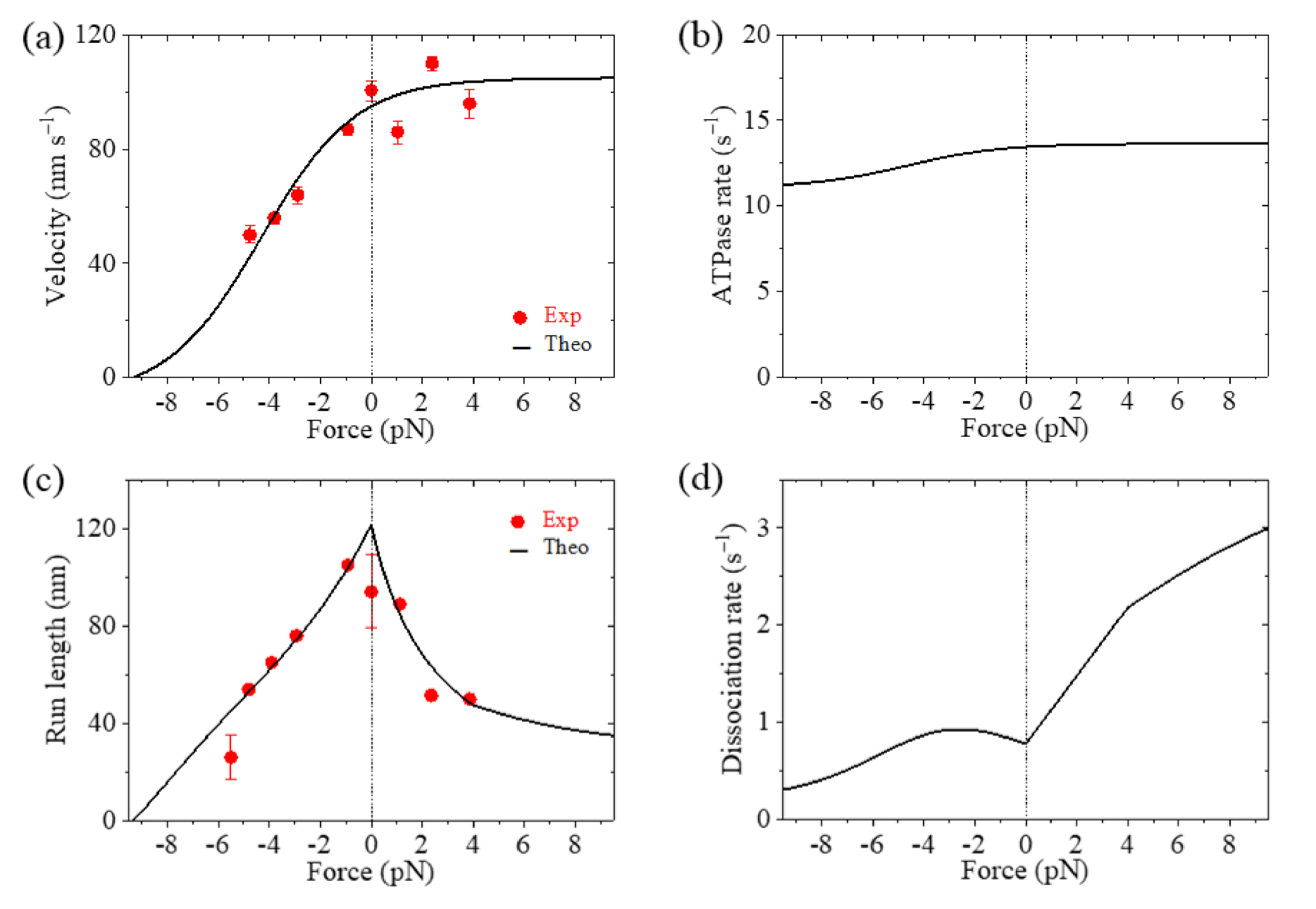
The theoretical results of velocity, ATPase rate, run length and dissociation rate versus load for Eg5-ΔTail are shown in
Figure 3a–d, respectively, where the available single-molecule data [
11,
12] are also shown. It is seen that the theoretical results for the velocity and run length versus load are consistent with the experimental data (
Figure 3a,c). The results show that the stall force, at which the velocity and run length are equal to zero, is about 9.3 pN (
Figure 3a,c). From
Figure 3b, it is seen that the ATPase rate of the motor changes only slightly with the increase in the magnitude of backward load and is kept almost unchanged with the forward load. This feature is consistent with the experimental data for kinesin-1 dimer showing that the ATPase rate is insensitive to the variation of the NL length (equivalent to the variation of the internal force) [
26]. From
Figure 3d, it is seen that the dissociation rate increases rapidly with the increase of the forward load, while the dissociation rate increases slightly with the increase in the magnitude of the backward load smaller than 2.6 pN and then decreases slightly with the further increase in the magnitude of the backward load.
2.2. Dynamics of Single FL-Eg5 Moving on Single MT
In this section, on the basis of the chemomechanical coupling pathway for FL-Eg5 (
Figure 2, see Methods for detailed descriptions), which is modified from that for Eg5-ΔTail by considering the nucleotide-dependent interaction between the tail and head, we study the dynamics of the single FL-Eg5 motor moving on the single MT. As discussed in the model for FL-Eg5 (see Methods), since the tail has a near-zero rate to release from the ADP-head, its release from the ADP-head can be neglected. The tail has a low rate to release from ϕ-head, which is denoted by
kr. Only after the release of the tail from ϕ-head can ATP bind to the ϕ-head, implying that at saturating ATP after the release of the tail from ϕ-head ATP can bind immediately. In addition, we argue here that the strong interaction between the tail and ADP-head induces the conformational change of the head, increasing the weak affinity
Ew2 of the ADP-head to MT and affecting the interaction potential of the ADP-head with MT. Thus, the binding of the tail to ADP-head could also affect
d(+).
From the pathway (
Figure 2), it is noted that both the rate for the fraction of transitions from
Figure 2a to b to c to e and that for the fraction of transitions from
Figure 2a to b to d are equal to
k(+). The proportion of the fraction of transitions from
Figure 2a to b to d (i.e., the proportion of the fraction of the transitions where the tail has not been released in
Figure 2d) in the total transitions from
Figure 2a to e can be approximately written as
. The rate of transition from
Figure 2d to e is
kr. Thus, considering
kNL >>
k(+) the average rate of the total transitions from
Figure 2a to e to f can be approximately calculated with
. Considering
kNL >>
k(+) the rate for the transition from
Figure 2h to i to c to e to f can be approximately written as
. Since during the processive motion the state of the motor with the trailing head in ATP state and the leading head in ADP state (
Figure 2a) occurs with probability
PE while the state of the motor with the trailing head in ADP state and the leading head in ATP state (
Figure 2h) occurs with probability 1 −
PE, the overall ATPase rate of the trailing head can then be approximately written as
where
(see above).
Similarly, considering
kNL >>
k(−) the overall ATPase rates of the leading head can then be approximately written as
where
(see above).
From the pathway (
Figure 2), it is noted that the velocity can still be written in the form of Equation (4), where
PE is still calculated by Equation (1) and
kT and
kL are calculated by Equations (14) and (15), respectively. The occurrence probability of Period I,
PI, can also be calculated by Equations (5)–(8). The dissociation probability in Period I is still
PdI 1, for upon Pi release the time of Period I is so short (in the order of 10 μs) that the tail cannot bind to the ADP-head during the period. The occurrence probability of Period II,
PII, can still be calculated by Equation (9), and the dissociation probability
PdII in Period II can also be calculated by Equations (10) and (11). For simplicity, we neglect the very small occurrence probability of state of
Figure 2j, from which Period I can also occur occasionally. The dissociation rate can then still be expressed in the form of Equation (12) and the run length can still be written in the form of Equation (13).
As discussed above, for FL-Eg5, except for parameters
kw0,
d(+) and
kr other parameters have the same values as those for Eg5-ΔTail. Here, we take
kw0 = 5
for FL-Eg5 (
Table 1), which is 4-fold smaller than that for Eg5-ΔTail. For FL-Eg5, besides taking
d(+) = 2.2 nm, which is the same as that for Eg5-ΔTail, we also take
d(+) = 8.2 nm, which is equal to
d. To be consistent with the experimental data of the velocity under near-zero load measured by Shimamoto et al. [
13], we adjust value of
kr, with
kr = 4.6
(
Table 1).
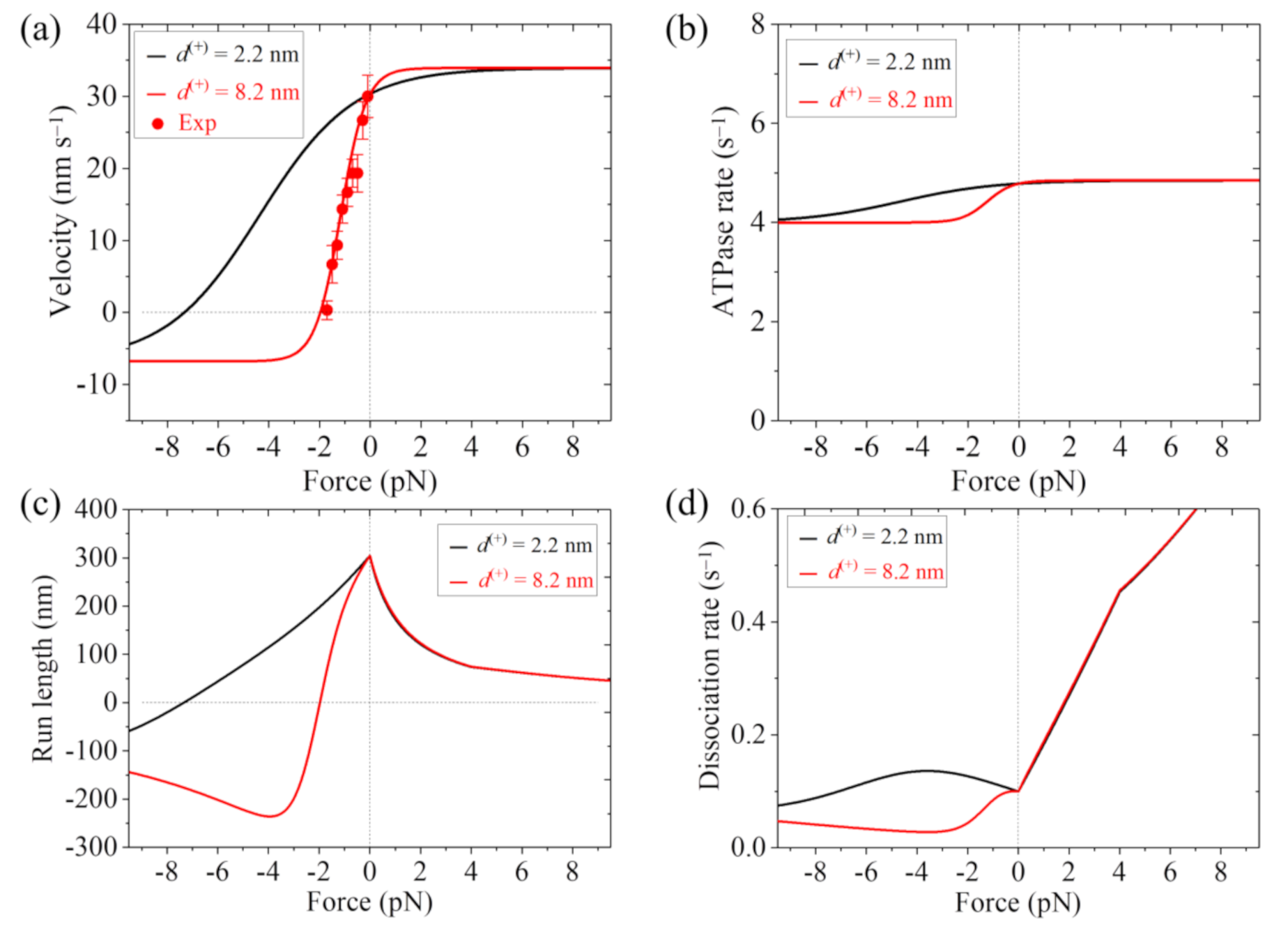
In
Figure 4a–d, we show the theoretical results of velocity, ATPase rate, run length and dissociation rate versus load, respectively, for FL-Eg5, where the experimental data of Shimamoto et al. [
13] are also shown. It is seen that the theoretical results of the velocity versus load with
d(+) = 8.2 nm are in good agreement with the experimental data (
Figure 4a). The results show that with
d(+) = 2.2 nm the stall force for FL-Eg5 is about 7.3 pN (
Figure 4a,c), which is smaller than 9.3 pN for Eg5-ΔTail with the same
d(+) = 2.2 nm (
Figure 3a,c). With
d(+) = 8.2 nm the stall force for FL-Eg5 is only about 2 pN (
Figure 4a,c). Comparing
Figure 3b and
Figure 4b it is seen that with the tail the ATPase rate of Eg5 is reduced by more than 2-fold, which is consistent with the experimental data of Bodrug et al. [
14]. Comparing
Figure 3c and
Figure 4c it is seen that for FL-Eg5 the run length under no load is more than 2-fold larger than that for Eg5-ΔTail, although the velocity for the former is smaller than that for the latter (
Figure 3a and
Figure 4a), which is consistent with the experimental data of Bodrug et al. [
14]. From
Figure 4d, it is seen that under the forward load the dissociation rate versus load for FL-Eg5 shows the similar feature to that for Eg5-ΔTail. Under the backward load, for FL-Eg5 with
d(+) = 2.2 nm the dissociation rate increases slightly with the increase in the magnitude of the load smaller than 3.6 pN and then decreases slightly with the further increase in the magnitude of the backward load, which is similar to that for Eg5-ΔTail. For FL-Eg5 with
d(+) = 2.2 nm the dissociation rate under the backward load around the stall force is about 3.3-fold smaller than that for Eg5-ΔTail, similar to the value of
kw0 for FL-Eg5 relative to that for Eg5-ΔTail. Interestingly, from
Figure 4d, it is seen that for FL-Eg5 with
d(+) = 8.2 nm the dissociation rate decreases monotonically and largely with the increase in the magnitude of the backward load (behaving as catch-bond characteristic) before the stall force of about 2 pN, which is in sharp contrast to that for Eg5-ΔTail and that for FL-Eg5 with
d(+) = 2.2 nm. More interestingly, by comparing
Figure 3d with
Figure 4d, it is seen that for FL-Eg5 with
d(+) = 8.2 nm the dissociation rate under the backward load around the stall force is much (about 10-fold) smaller than that for Eg5-ΔTail, although
kw0 for FL-Eg5 is only 4-fold smaller than that for Eg5-ΔTail. Since the low dissociation rate under the backward load around the stall force is critical for exerting long-time force to slide apart two antiparallel MTs by the motor whereas the unloaded velocity is insignificant for exerting force, the above results give an explanation of why Eg5 motor with the tail has the smaller velocity and smaller stall force or larger
d(+) than without the tail.
2.3. Dynamics of Sliding Apart Two Antiparallel MTs by Multiple Motors
In this section we study numerically the force generated by multiple kinesin-5 motors to slide apart two antiparallel MTs. The numerical simulation procedure is described as follows (refer to
Figure S2 in Supplementary Materials).
To calculate the force, we fix the two antiparallel MT filaments. A pair of kinesin-5 heads at one end of the stalk binds to one of the two antiparallel MT filaments with rate
ka, where
ka is proportional to the concentration of kinesin-5 motors in solution and the MT overlapping length. Then, the other pair of heads at the opposite end of the stalk can bind to the antiparallel MT filament with rate
. Note that when only one pair of heads is bound to MT, the other pair of heads at the opposite ends of the stalk is considered to be in the same position along the
x direction (parallel to the MT filament). After two pairs of heads binding to MTs, the two pairs move independently on the MTs. Each pair of heads takes a forward (plus-end) step with rate
kF =
PEkT, takes a backward (minus-end) step with rate
kB = (1 −
PE)
kL, and detaches from MT with rate
. After one pair of heads detaching from MT, the detached pair of heads moves immediately to the position of the other MT-bound pair of heads along the
x direction. The MT-bound pair of heads can detach from the MT still with rate
and the detached pair of heads can rebind to the former MT with rate
. (Since when only one pair of heads is bound to MT no force can be generated on MTs, the movement of the pair of heads on MT is not needed to consider). If the detachment of the MT-bound pairs of heads occurs before the rebinding of the detached pair of heads to MT, the tetramer is dissociated into solution. When one kinesin-5 tetramer (termed as, e.g., the
ith motor) is bound to MTs in the overlap, denoting by
the center-of-mass position of one pair of heads in the MT with the plus end in the
x direction and by
the center-of-mass position of the other pair of heads in the MT with the plus end in the −
x direction, the force generated by the
ith motor to slide apart MTs can be calculated with
, where
K5 is the stretching elastic coefficient of kinesin-5 stalk. When
N kinesin-5 motors are bound to the overlapping MTs, the total force generated can be calculated by
. For simplicity of analysis, here we do not consider the interaction between any two kinesin-5 motors. The above procedure is simulated with Monte-Carlo algorithm (see
Section S1 in Supplementary Materials), as done before [
27,
28].
The forward stepping rate kF = PEkT, backward stepping rate kB = (1 − PE)kL and dissociation rate for one pair of heads of Eg5-ΔTail and those of FL-Eg5 are determined above. We take both Eg5-ΔTail and FL-Eg5 having the same binding rate ka, which is taken as a variable parameter in this work. The rebinding rate and the elastic coefficient of the stalk for Eg5-ΔTail and those for FL-Eg5 are determined as follows.
Since in the system of cargo transported by multiple kinesin-1 motors the rebinding rate of one detached kinesin-1 motor to MT (denoted by
) is available in the literature (see below), the rebinding rate
can be approximately determined from
, as described as follows. Consider that a Brownian particle is connected to a fixed point via a stalk of length
l and the stalk can rotate in one-dimensional potential
, where
is the rotation angle and
k is constant. Thus, the position of the Brownian particle is
. The probability of the stalk with rotation angle
has the form
The probability of the position of the Brownian particle can then be written as
As it is known, in solution when the distance between a particle and its partner is larger than the Debye length
a = 1 nm, nearly no interaction between them exists. Thus, the binding rate of the Brownian particle to its partner that is kept in the position of distance
l away from the point to which the stalk of the Brownian particle is connected can be approximately calculated with
, where
C is a constant that is inversely proportional to the dissociation rate of the particle from its partner. Let
l =
l1 and
l =
l5 be the stalk lengths of kinesin-1 and kinesin-5 motors, respectively. Then, the ratio of
to
can be calculated by
where
represents
kw0 for kinesin-5 head in ADP state and
represents
kw0 for kinesin-1 head in ADP state. As determined above,
= 20
for Eg5-ΔTail and
= 5
for FL-Eg5 (see
Table 1). As determined before,
= 5
[
16]. The available data gave
l5 = 61.3 nm [
4]. As done before [
29], we take
l1 = 35 nm. We have checked that varying values of
l5 and
l1 has nearly no effect on the results presented in this work. As shown experimentally before [
30] and used in a widespread manner [
31,
32], we take
= 5
. With above parameter values and from Equation (18) we obtain
1.25
for Eg5-ΔTail and
5
for FL-Eg5, which are nearly independent of the value of
k. Thus, in our numerical simulations we take
= 1.25
and 5
for Eg5-ΔTail and FL-Eg5, respectively.
As done before in [
27,
28,
29], it is considered that the kinesin stalk can be stretched elastically. Since both Eg5-ΔTail and FL-Eg5 having the same length, they have the same stretching elastic coefficient, with the value being determined as follows. The available experimental data showed that the elastic coefficient for kinesin-1 stalk is
K1 = 0.3 pN/nm [
33]. Thus, the elastic coefficient for kinesin-5 stalk can be estimated as
= 0.17 pN/nm. We take this value of
K5 in our simulations. However, we have checked that varying value of
K5 has nearly no effect on the results presented in this work.
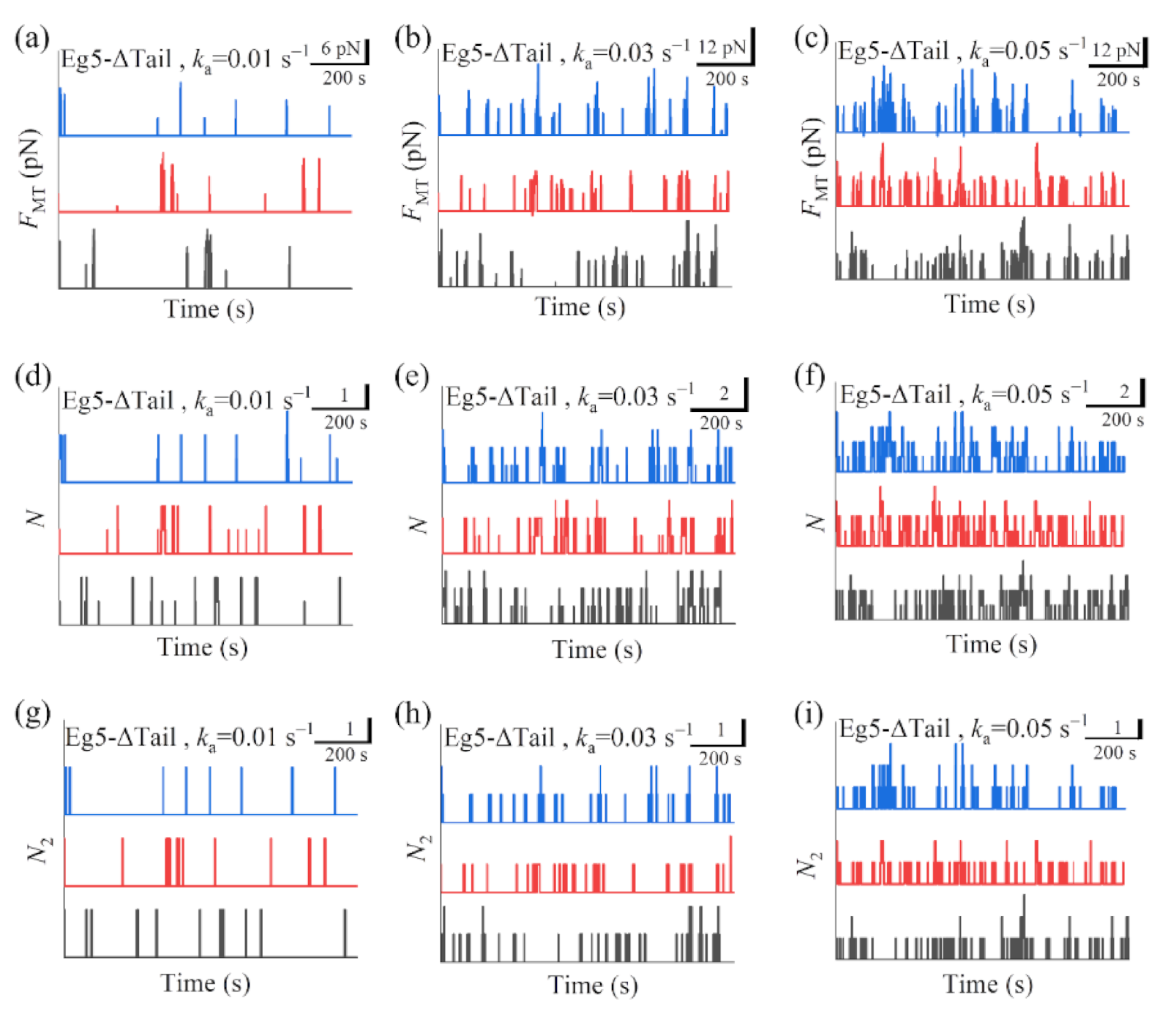
In
Figure 5a–c we show some simulated results for the temporal evolution of the force
FMT generated by multiple Eg5-ΔTail motors under different values of motor-binding rate
ka, where
t = 0 corresponds to the moment when only one motor binds to the overlapping MTs. The corresponding results for the total number of Eg5-ΔTail motors,
N, in the MT overlap are shown in
Figure 5d–f. The corresponding results for the number of Eg5-ΔTail motors with two pairs of heads bound to the MTs,
N2, are shown in
Figure 5g–i (noting that
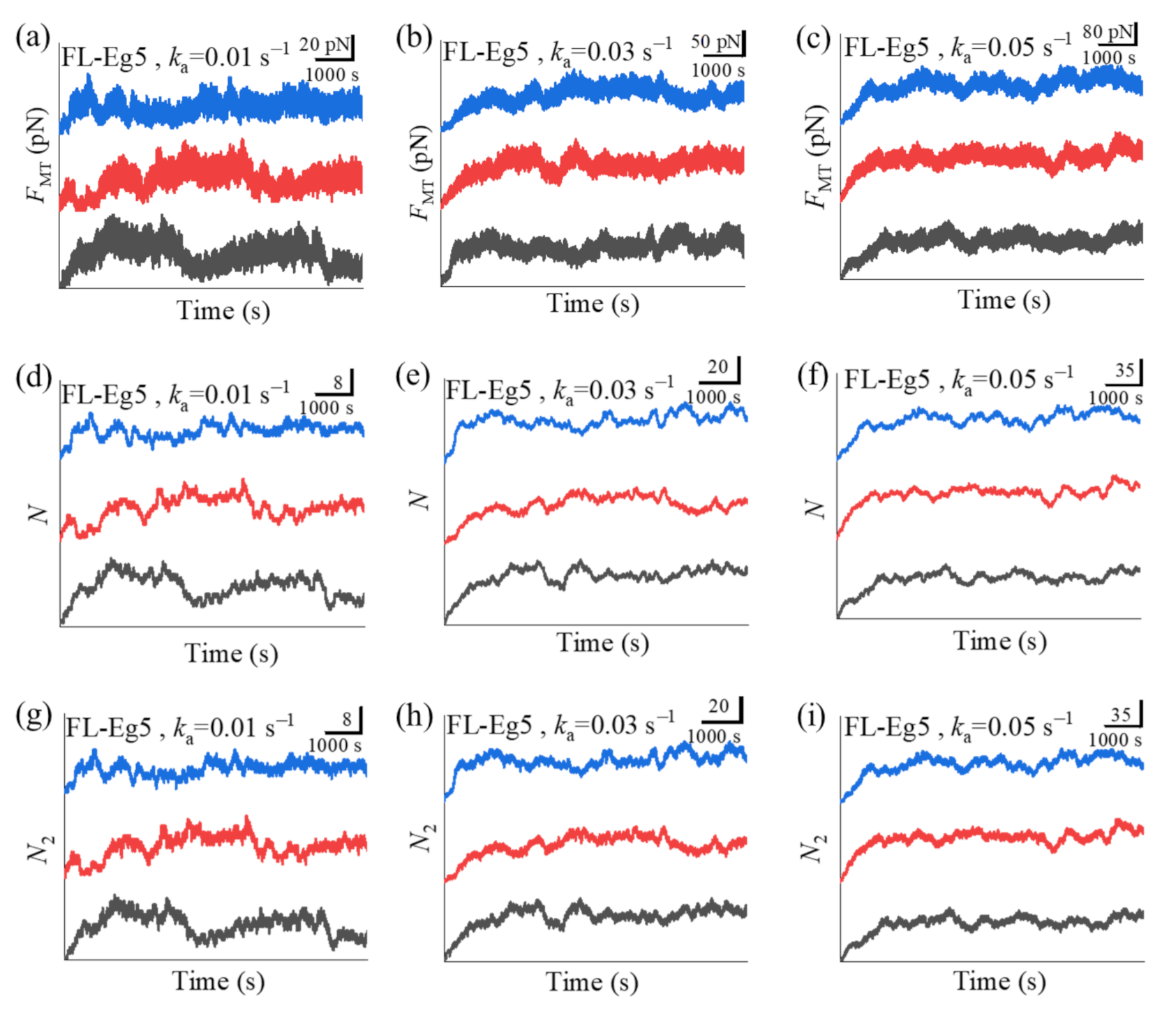
N2 corresponds to the effective number of the motors that can generate the force). For comparison between the case of Eg5-ΔTail motors and that of FL-Eg5 motors, in
Figure 6 we show some simulated results for the temporal evolution of
FMT,
N and
N2 for the case of FL-Eg5 motors (with
d(+) = 8.2 nm) under the same values of
ka as those in
Figure 5.
Our results show that for the case of Eg5-ΔTail motors no steady force
FMT can be generated in the range of
ka from 0.005
to 0.06
(
Figure 5a–c). This is because no steady number of Eg5-ΔTail motors bound to MTs in the overlap can be reached (
Figure 5d–i). By contrast, our results show that for the case of FL-Eg5 motors the force
FMT increases gradually until the maximum steady value is reached at any given
ka in the range from 0.005
to 0.06
(
Figure 6a–c), which is due to the gradual increase in the number of FL-Eg5 motors bound to the MTs in the overlap until the maximum steady number is reached (
Figure 6d–i). The rather different feature for the force
FMT generated by FL-Eg5 motors from that by Eg5-ΔTail is due mainly to the dissociation rate for FL-Eg5 before stall force having catch-bond characteristic (see
Figure 4d), the dissociation rate around the stall force for the FL-Eg5 being much (about 10-fold) smaller than that for the Eg5-ΔTail (see
Figure 3d and
Figure 4d) and the rebinding rate for the FL-Eg5 being 4-fold larger than that for the Eg5-ΔTail (see above). By comparing
Figure 6d–f with
Figure 6g–i it is noted that nearly all FL-Eg5 motors in the overlap are those with two pairs of heads bound to MTs (with
N only slightly larger than
N2). It is interestingly noted that the simulated curves of
FMT versus time for FL-Eg5 motors (
Figure 6a–c) resemble the experimental data measured by Shimamoto et al. [
13] and by Bodrug et al. [
14], with both the simulated and experimental results showing that
FMT increases over time and becomes leveled off to a large steady value. For Eg5-ΔTail motors, the simulated results (
Figure 5a–c) are also similar to the experimental results showing that
FMT only fluctuates around a small value [
14]. Moreover, from
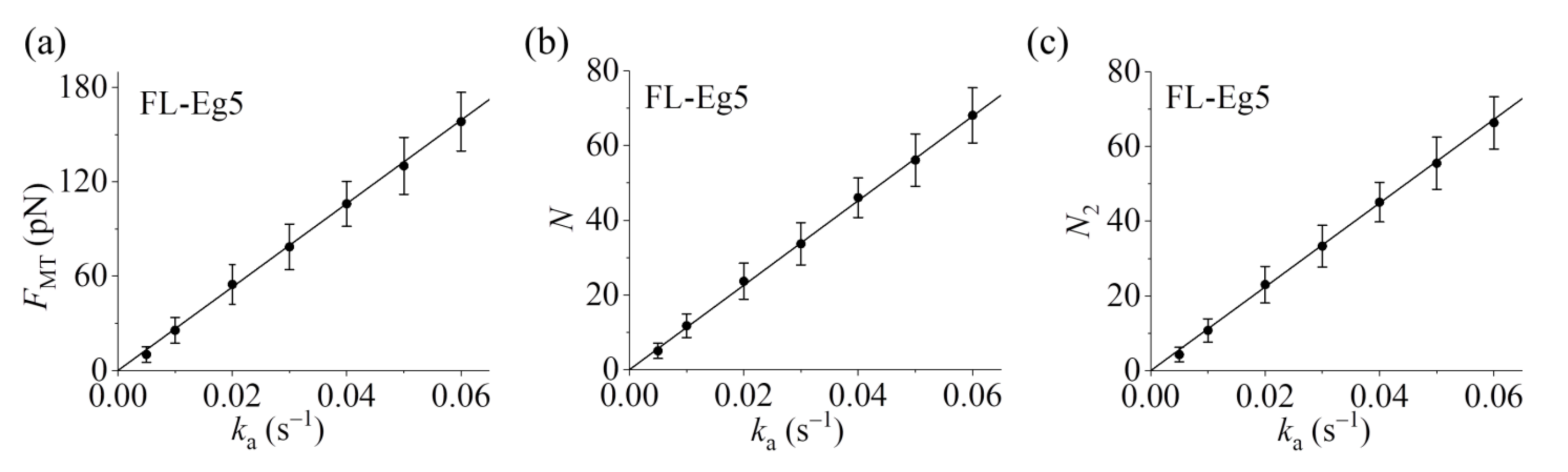
Figure 6a–c it is seen that the maximum steady force
FMT generated by FL-Eg5 motors increases linearly with the increase of
ka, which can be also seen clearly from
Figure 7 where the maximum steady
FMT,
N and
N2 versus
ka are shown. Since
ka is proportional to the concentration of kinesin-5 tetramers in solution and the MT overlapping length, the results of
Figure 7 imply that for a given concentration of kinesin-5 tetramers the maximum steady
FMT,
N and
N2 increase linearly with the increase of the MT overlapping length. These results are also consistent with the experimental data of Bodrug et al. [
14]. Similarly, for a given MT overlapping length the maximum steady
FMT,
N and
N2 increases linearly with the increase in the concentration of kinesin-5 tetramers, which is consistent with the experimental data of Shimamoto et al. [
13].
It is noted that the experimental data of Bodrug et al. [
14] indicated that at high concentration of KCl (100 mM) mainly two FL-Eg5 motors can form a cluster while at 25 mM KCl no cluster can be formed. Here, for simplicity, we have not considered the formation of FL-Eg5 clusters in our simulations, which is applicable to the case of low concentration of KCl (closer to physiological conditions). At the high concentration of KCl (100 mM), the formation of clusters could further reduce the dissociation rate of the motors, as proposed before [
14], and thus further facilitate the generation of the steady MT-sliding force by FL-Eg5 motors, which will be studied theoretically and numerically in the future.
In addition, we also study numerically the dynamics of Eg5 motors moving within the overlapping MTs (see
Section S2 in Supplementary Materials), where one MT is fixed and the other antiparallel MT is free. The numerical results showed that while Eg5-ΔTail shows bi-directional movement with frequent directional reversals FL-Eg5 shows unidirectional movement with infrequent directional reversals (see
Section S2 and Figure S3 in Supplementary Materials). These results resemble well the experimental data of Bodrug et al. [
14]. The movement of the free MT by multiple FL-Eg5 motors is also simulated, with the velocity being nearly independent of
ka (see
Section S2 and Figure S4 in Supplementary Materials).
Taken together, the results presented in this section show that in the same range of ka from 0.005 to 0.06 , Eg5-ΔTail motors cannot generate steady force to slide apart two antiparallel MTs whereas FL-Eg5 motors can generate the steady force. Moreover, the steady force generated by FL-Eg5 motors increases linearly with the increase of ka, namely the steady force increases linearly with the increase of the MT overlapping length for a given concentration of FL-Eg5 motors and increases linearly with the increase of FL-Eg5 concentrations for a given MT overlapping length. These results explain why FL-Eg5 rather than Eg5-ΔTail motors are used to slide apart two antiparallel MTs in cells.