Selective Chemical Activation of Piezo1 in Leukemia Cell Membrane: Single Channel Analysis
Abstract
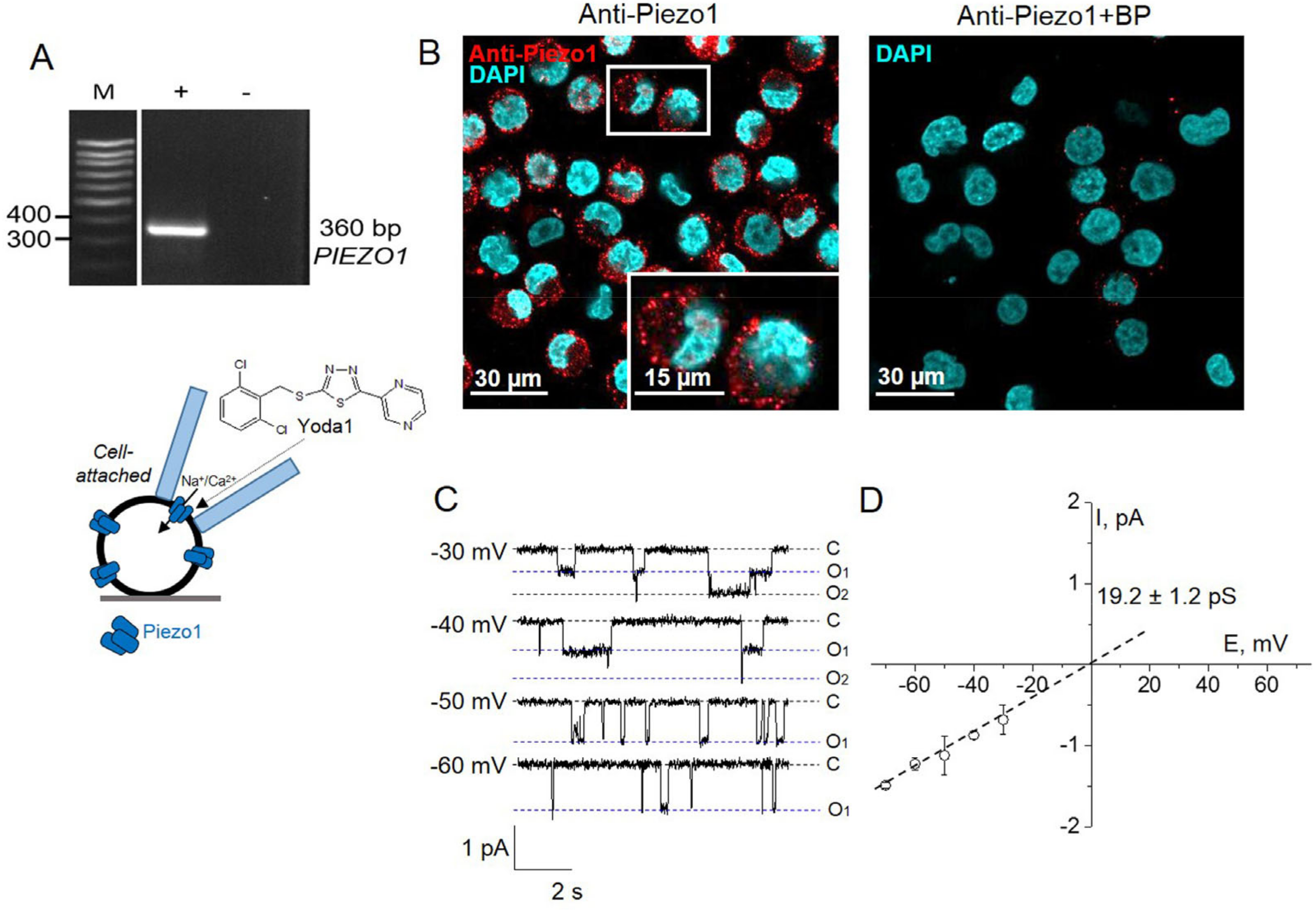
1. Introduction
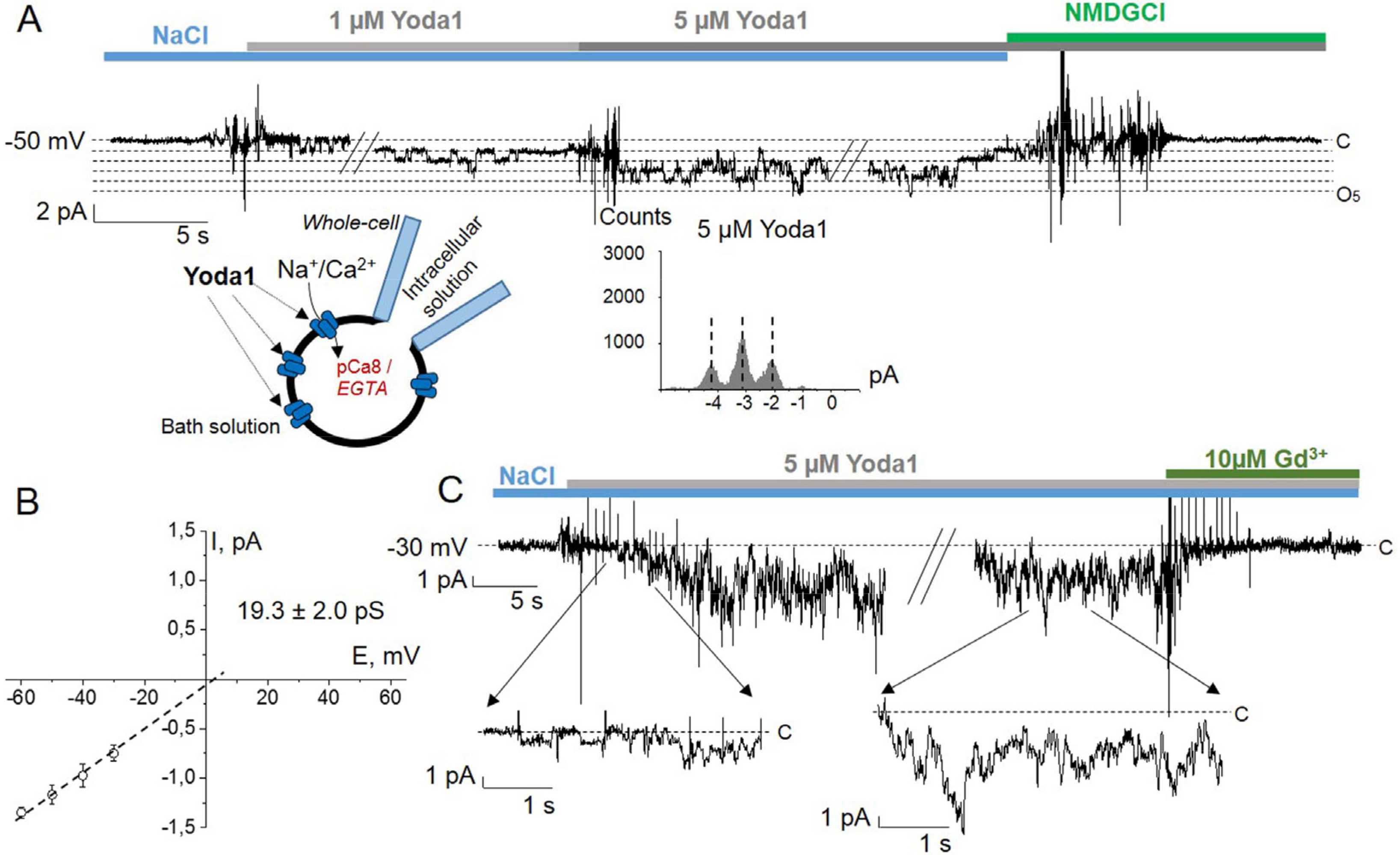
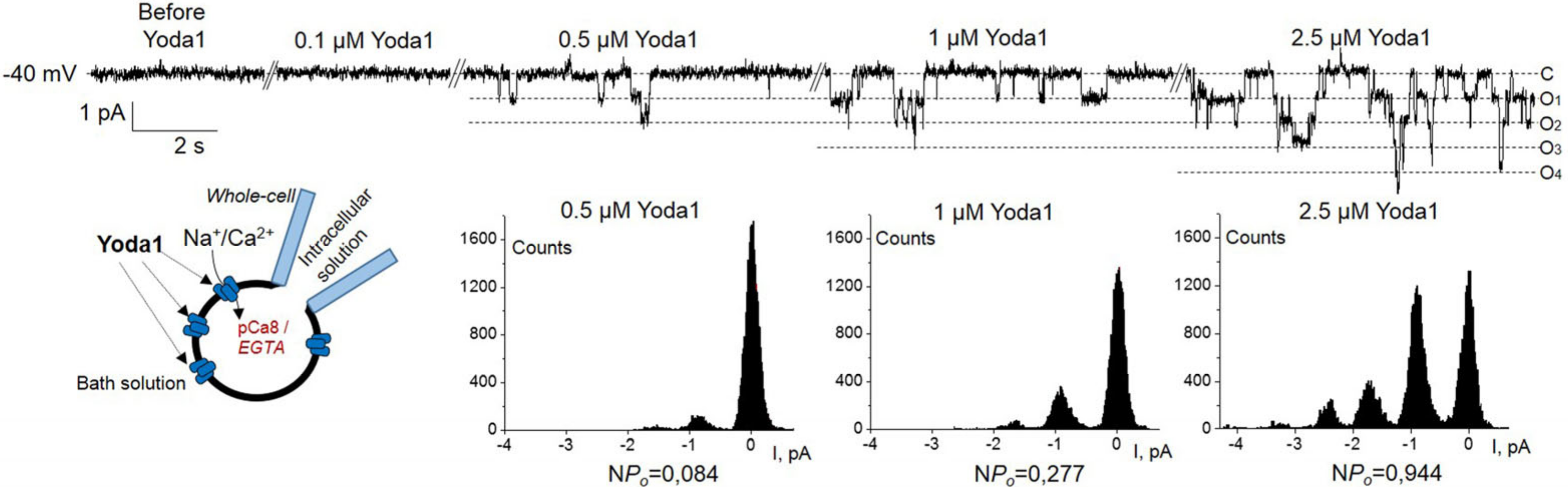
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. RT-PCR
4.3. Immunofluorescence
4.4. Electrophysiology
4.4.1. Patch-Clamp Setup
4.4.2. Solutions
4.4.3. Data Analysis and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, H.; Kamm, R.D.; Lee, R.T. Cell mechanics and mechanotransduction: Pathways, probes, and physiology. Am. J. Physiol. Cell Physiol. 2004, 287, C1–C11. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Xu, X.Z. Mechanosensitive channels: In touch with Piezo. Curr. Biol. 2010, 20, R936–R938. [Google Scholar] [CrossRef] [PubMed]
- Nikolaev, Y.A.; Cox, C.D.; Ridone, P.; Rohde, P.R.; Cordero-Morales, J.F.; Vásquez, V.; Laver, D.R.; Martinac, B. Mammalian TRP ion channels are insensitive to membrane stretch. J. Cell Sci. 2019, 132, jcs238360. [Google Scholar] [CrossRef]
- Startek, J.B.; Boonen, B.; Talavera, K.; Meseguer, V. TRP Channels as Sensors of Chemically-Induced Changes in Cell Membrane Mechanical Properties. Int. J. Mol. Sci. 2019, 20, 371. [Google Scholar] [CrossRef]
- Coste, B.; Mathur, J.; Schmidt, M.; Earley, T.J.; Ranade, S.; Petrus, M.J.; Dubin, A.E.; Patapoutian, A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 2010, 330, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Taberner, F.J.; Prato, V.; Schaefer, I.; Schrenk-Siemens, K.; Heppenstall, P.A.; Lechner, S.G. Structure-guided examination of the mechanogating mechanism of PIEZO2. Proc. Natl. Acad. Sci. USA 2019, 116, 14260–14269. [Google Scholar] [CrossRef] [PubMed]
- De Felice, D.; Alaimo, A. Mechanosensitive Piezo channels in cancer: Focus on altered calcium signaling in cancer cells and in tumor progression. Cancers 2020, 12, 1780. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, M.; Liu, G.; Zhang, X.; Zhi, L.; Zhao, J.; Wang, G. The function of Piezo1 in colon cancer metastasis and its potential regulatory mechanism. J. Cancer Res. Clin. Oncol. 2020, 146, 1139–1152. [Google Scholar] [CrossRef]
- Syeda, R.; Xu, J.; Dubin, A.E.; Coste, B.; Mathur, J.; Huynh, T.; Matzen, J.; Lao, J.; Tully, D.C.; Engels, I.H.; et al. Chemical activation of the mechanotransduction channel Piezo1. Elife 2015, 4, e07369. [Google Scholar] [CrossRef]
- Botello-Smith, W.M.; Jiang, W.; Zhang, H.; Ozkan, A.D.; Lin, Y.C.; Pham, C.N.; Lacroix, J.J.; Luo, Y. A mechanism for the activation of the mechanosensitive Piezo1 channel by the small molecule Yoda1. Nat. Commun. 2019, 10, 4503. [Google Scholar] [CrossRef]
- Staruschenko, A.V.; Vedernikova, E.A. Mechanosensitive cation channels in human leukaemia cells: Calcium permeation and blocking effect. J. Physiol. 2002, 541, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Staruschenko, A.; Negulyaev, Y.A.; Morachevskaya, E.A. Actin cytoskeleton disassembly affects conductive properties of stretch-activated cation channels in leukaemia cells. Biochim. Biophys. Acta 2005, 1669, 53–60. [Google Scholar] [CrossRef][Green Version]
- Chubinskiy-Nadezhdin, V.I.; Negulyaev, Y.A.; Morachevskaya, E.A. Cholesterol depletion-induced inhibition of stretch-activated channels is mediated via actin rearrangement. Biochem. Biophys. Res. Commun. 2011, 412, 80–85. [Google Scholar] [CrossRef]
- Wu, C.; Orozco, C.; Boyer, J.; Leglise, M.; Goodale, J.; Batalov, S.; Hodge, C.L.; Haase, J.; Janes, J.; Huss, J.W., 3rd; et al. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome. Biol. 2009, 10, R130. [Google Scholar] [CrossRef]
- Wang, Y.; Chi, S.; Guo, H.; Li, G.; Wang, L.; Zhao, Q.; Rao, Y.; Zu, L.; He, W.; Xiao, B. A lever-like transduction pathway for long-distance chemical- and mechano-gating of the mechanosensitive Piezo1 channel. Nat. Commun. 2018, 9, 1300. [Google Scholar] [CrossRef] [PubMed]
- Bae, C.; Sachs, F.; Gottlieb, P.A. The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry 2011, 50, 6295–6300. [Google Scholar] [CrossRef]
- Coste, B.; Xiao, B.; Santos, J.S.; Syeda, R.; Grandl, J.; Spencer, K.S.; Kim, S.E.; Schmidt, M.; Mathur, J.; Dubin, A.E.; et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 2012, 483, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Rezania, S.; Kammerer, S.; Sokolowski, A.; Devaney, T.; Gorischek, A.; Jahn, S.; Hackl, H.; Groschner, K.; Windpassinger, C.; et al. Piezo1 forms mechanosensitive ion channels in the human MCF-7 breast cancer cell line. Sci. Rep. 2015, 5, 8364. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.C.; Park, H.J.; Kim, J.G.; Lee, I.H.; Cho, H.; Park, C.; Sung, T.S.; Koh, S.D.; Park, S.W.; Bae, Y.M. The Piezo2 ion channel is mechanically activated by low-threshold positive pressure. Sci. Rep. 2019, 9, 6446. [Google Scholar] [CrossRef] [PubMed]
- Nosyreva, E.D.; Thompson, D.; Syeda, R. Identification and functional characterization of the Piezo1 channel pore domain. J. Biol. Chem. 2021, 296, 100225. [Google Scholar] [CrossRef] [PubMed]
- Chubinskiy-Nadezhdin, V.I.; Negulyaev, Y.A.; Morachevskaya, E.A. Functional coupling of ion channels in the process of mechano-dependent activation in the membrane of K562 cells. Cell Tiss. Biol. 2019, 13, 470–477. [Google Scholar] [CrossRef]
- Morachevskaya, E.; Sudarikova, A.; Negulyaev, Y. Mechanosensitive channel activity and F-actin organization in cholesterol-depleted human leukaemia cells. Cell Biol. Int. 2007, 31, 374–381. [Google Scholar] [CrossRef]
- Cahalan, S.M.; Lukacs, V.; Ranade, S.S.; Chien, S.; Bandell, M.; Patapoutian, A. Piezo1 links mechanical forces to red blood cell volume. Elife 2015, 4, e07370. [Google Scholar] [CrossRef] [PubMed]
- Alper, S.L. Genetic Diseases of PIEZO1 and PIEZO2 Dysfunction. Curr. Top. Membr. 2017, 79, 97–134. [Google Scholar] [CrossRef]
- Bianchi, N.; Ongaro, F.; Chiarabelli, C.; Gualandi, L.; Mischiati, C.; Bergamini, P.; Gambari, R. Induction of erythroid differentiation of human K562 cells by cisplatin analogs. Biochem. Pharmacol. 2000, 60, 31–40. [Google Scholar] [CrossRef]
- Sudarikova, A.V.; Vasileva, V.Y.; Vassilieva, I.O.; Negulyaev, Y.A.; Morachevskaya, E.A.; Chubinskiy-Nadezhdin, V.I. Extracellular protease trypsin activates amiloride-insensitive sodium channels in human leukemia cells. J. Cell Biochem. 2019, 120, 461–469. [Google Scholar] [CrossRef]
- Ermakov, Y.A.; Kamaraju, K.; Sengupta, K.; Sukharev, S. Gadolinium ions block mechanosensitive channels by altering the packing and lateral pressure of anionic lipids. Biophys. J. 2010, 98, 1018–1027. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, S.H. Guide to data Acquisition and Analysis. In Single-Channel Recording, 2nd ed.; Sakmann, B., Neher, E., Eds.; Springer: Boston, MA, USA, 1995; p. 80. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasileva, V.; Morachevskaya, E.; Sudarikova, A.; Negulyaev, Y.; Chubinskiy-Nadezhdin, V. Selective Chemical Activation of Piezo1 in Leukemia Cell Membrane: Single Channel Analysis. Int. J. Mol. Sci. 2021, 22, 7839. https://doi.org/10.3390/ijms22157839
Vasileva V, Morachevskaya E, Sudarikova A, Negulyaev Y, Chubinskiy-Nadezhdin V. Selective Chemical Activation of Piezo1 in Leukemia Cell Membrane: Single Channel Analysis. International Journal of Molecular Sciences. 2021; 22(15):7839. https://doi.org/10.3390/ijms22157839
Chicago/Turabian StyleVasileva, Valeria, Elena Morachevskaya, Anastasia Sudarikova, Yuri Negulyaev, and Vladislav Chubinskiy-Nadezhdin. 2021. "Selective Chemical Activation of Piezo1 in Leukemia Cell Membrane: Single Channel Analysis" International Journal of Molecular Sciences 22, no. 15: 7839. https://doi.org/10.3390/ijms22157839
APA StyleVasileva, V., Morachevskaya, E., Sudarikova, A., Negulyaev, Y., & Chubinskiy-Nadezhdin, V. (2021). Selective Chemical Activation of Piezo1 in Leukemia Cell Membrane: Single Channel Analysis. International Journal of Molecular Sciences, 22(15), 7839. https://doi.org/10.3390/ijms22157839





