Hepatic IFNL4 Gene Activation in Hepatocellular Carcinoma Patients with Regard to Etiology
Abstract
:1. Introduction
2. Results
2.1. The TCGA HCC Patient Sample with Regard to Etiology
2.2. IFN Gene Signature in Nonmalignant and in Malignant Liver Tissues
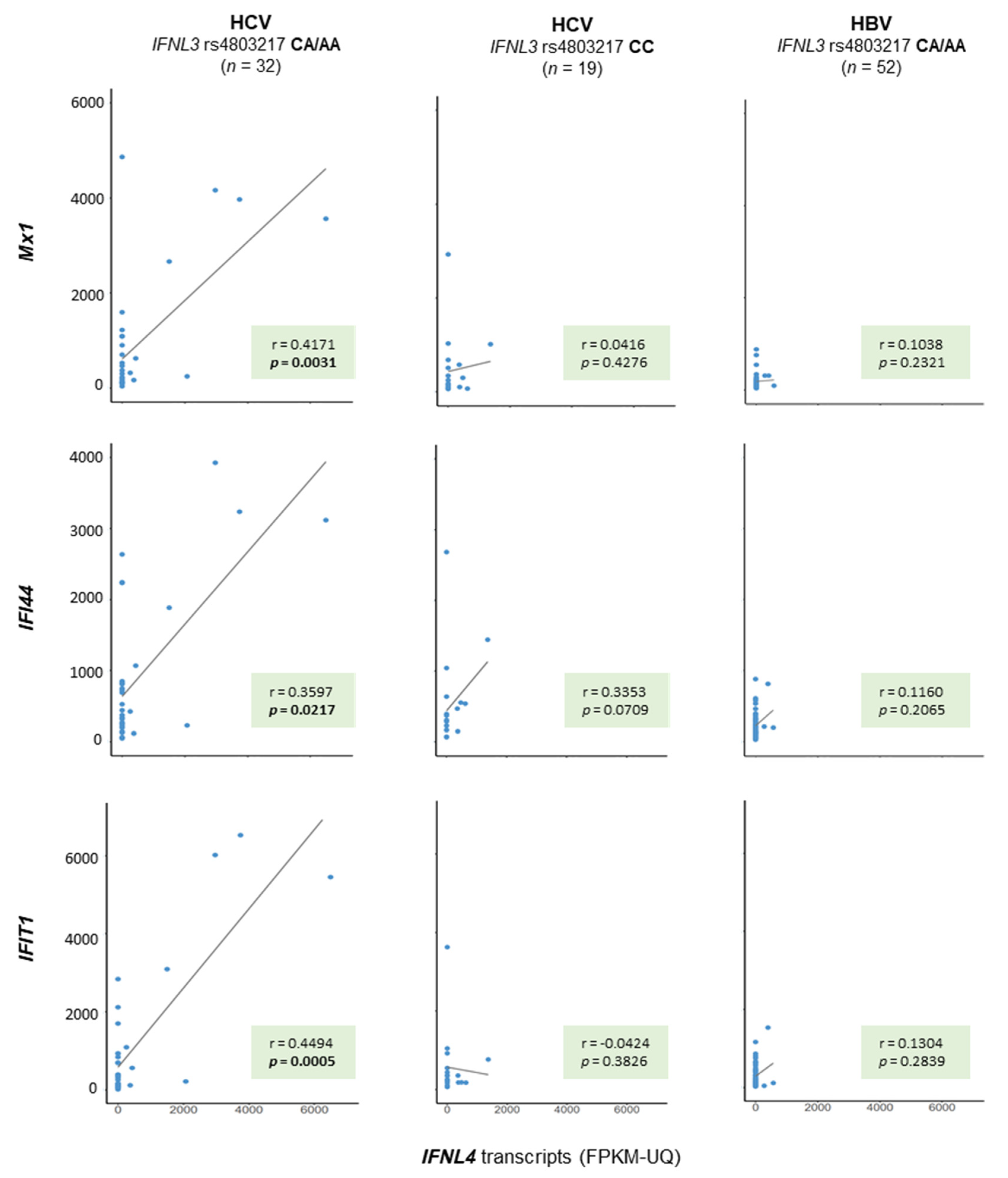
2.3. ISG Gene Activation with Regard to IFNL Genotypes
3. Discussion
4. Materials and Methods
4.1. TCGA Data
4.2. Reading Out IFNL Genotypes
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Burden of Disease Cancer Collaboration; Fitzmaurice, C.; Akinyemiju, T.F.; Al Lami, F.H.; Alam, T.; Alizadeh-Navaei, R.; Allen, C.; Alsharif, U.; Alvis-Guzman, N.; Amini, E.; et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: A systematic analysis for the global burden of disease study. JAMA Oncol. 2018, 4, 1553–1568. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Liu, Z.; Jiang, Y.; Yuan, H.; Fang, Q.; Cai, N.; Suo, C.; Jin, L.; Zhang, T.; Chen, X. The trends in incidence of primary liver cancer caused by specific etiologies: Results from the Global Burden of Disease Study 2016 and implications for liver cancer prevention. J. Hepatol. 2019, 70, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S. The mutational landscape of hepatocellular carcinoma. Clin. Mol. Hepatol. 2015, 21, 220–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulze, K.; Imbeaud, S.; Letouze, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Inada, Y.; Mizukoshi, E.; Seike, T.; Tamai, T.; Iida, N.; Kitahara, M.; Yamashita, T.; Arai, K.; Terashima, T.; Fushimi, K.; et al. Characteristics of immune response to tumor-associated antigens and immune cell profile in patients with hepatocellular carcinoma. Hepatology 2019, 69, 653–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfister, D.; Nunez, N.G.; Pinyol, R.; Govaere, O.; Pinter, M.; Szydlowska, M.; Gupta, R.; Qiu, M.; Deczkowska, A.; Weiner, A.; et al. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature 2021, 592, 450–456. [Google Scholar] [CrossRef]
- Mihm, S.; Frese, M.; Meier, V.; Wietzke-Braun, P.; Scharf, J.G.; Bartenschlager, R.; Ramadori, G. Interferon type I gene expression in chronic hepatitis C. Lab. Investig. 2004, 84, 1148–1159. [Google Scholar] [CrossRef]
- Suslov, A.; Boldanova, T.; Wang, X.; Wieland, S.; Heim, M.H. Hepatitis B virus does not interfere with innate immune responses in the human liver. Gastroenterology 2018, 154, 1778–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patzwahl, R.; Meier, V.; Ramadori, G.; Mihm, S. Enhanced expression of interferon-regulated genes in the liver of patients with chronic hepatitis C virus infection: Detection by suppression-subtractive hybridization. J. Virol. 2001, 75, 1332–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mihm, S. Activation of type I and type III interferons in chronic hepatitis C. J. Innate Immun. 2015, 7, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Prokunina-Olsson, L.; Muchmore, B.; Tang, W.; Pfeiffer, R.M.; Park, H.; Dickensheets, H.; Hergott, D.; Porter-Gill, P.; Mumy, A.; Kohaar, I.; et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013, 45, 164–171. [Google Scholar] [CrossRef]
- Key, F.M.; Peter, B.; Dennis, M.Y.; Huerta-Sanchez, E.; Tang, W.; Prokunina-Olsson, L.; Nielsen, R.; Andres, A.M. Selection on a variant associated with improved viral clearance drives local, adaptive pseudogenization of interferon lambda 4 (IFNL4). PLoS Genet. 2014, 10, e1004681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amanzada, A.; Kopp, W.; Spengler, U.; Ramadori, G.; Mihm, S. Interferon-lambda4 (IFNL4) transcript expression in human liver tissue samples. PLoS ONE 2013, 8, e84026. [Google Scholar] [CrossRef] [PubMed]
- Murakawa, M.; Asahina, Y.; Kawai-Kitahata, F.; Nakagawa, M.; Nitta, S.; Otani, S.; Nagata, H.; Kaneko, S.; Asano, Y.; Tsunoda, T.; et al. Hepatic IFNL4 expression is associated with non-response to interferon-based therapy through the regulation of basal interferon-stimulated gene expression in chronic hepatitis C patients. J. Med. Virol. 2017, 89, 1241–1247. [Google Scholar] [CrossRef]
- Noureddin, M.; Rotman, Y.; Zhang, F.; Park, H.; Rehermann, B.; Thomas, E.; Liang, T.J. Hepatic expression levels of interferons and interferon-stimulated genes in patients with chronic hepatitis C: A phenotype-genotype correlation study. Genes Immun. 2015, 16, 321–329. [Google Scholar] [CrossRef]
- Terczynska-Dyla, E.; Bibert, S.; Duong, F.H.; Krol, I.; Jorgensen, S.; Collinet, E.; Kutalik, Z.; Aubert, V.; Cerny, A.; Kaiser, L.; et al. Reduced IFNlambda4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nat. Commun. 2014, 5, 5699. [Google Scholar] [CrossRef]
- O’Brien, T.R.; Jackson, S.S. What have we learned from studies of IFN-lambda variants and hepatitis C virus infection? J. Interferon Cytokine Res. 2019, 39, 618–626. [Google Scholar] [CrossRef]
- Yan, Y.; Zheng, L.; Du, Q.; Cui, X.; Dong, K.; Guo, Y.; Geller, D.A. Interferon regulatory factor 1 (IRF-1) downregulates checkpoint kinase 1 (CHK1) through miR-195 to upregulate apoptosis and PD-L1 expression in hepatocellular carcinoma (HCC) cells. Br. J. Cancer 2021. [Google Scholar] [CrossRef]
- Yan, Y.; Zheng, L.; Du, Q.; Yan, B.; Geller, D.A. Interferon regulatory factor 1 (IRF-1) and IRF-2 regulate PD-L1 expression in hepatocellular carcinoma (HCC) cells. Cancer Immunol. Immunother. 2020, 69, 1891–1903. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zheng, L.; Du, Q.; Yazdani, H.; Dong, K.; Guo, Y.; Geller, D.A. Interferon regulatory factor 1(IRF-1) activates anti-tumor immunity via CXCL10/CXCR3 axis in hepatocellular carcinoma (HCC). Cancer Lett. 2021, 506, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Huschka, H.; Mihm, S. Interferon-lambda (IFNL) germline variations and their significance for HCC and PDAC progression: An analysis of The Cancer Genome Atlas (TCGA) data. BMC Cancer 2020, 20, 1131. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. Available online: https://wwwR-projectorg/ (accessed on 14 July 2021).


| Total | Unknown Risk | Nonviral | HBV | HCV | HBV + HCV | p-Value | |
|---|---|---|---|---|---|---|---|
| (n = 377) | (n = 54) | (n = 43) | (n = 144) | (n = 48) | (n = 88) | ||
| Age, mean ± SD (years) | 59.5 ± 13.5 | 61.1 ± 17.1 | 67.0 ± 9.5 | 57.5 ± 13.0 | 60.2 ± 8.6 | 57.7 ± 14.4 | <0.0001 |
| Gender, m/f (n) | 255/122 | 20/34 | 31/12 | 104/40 | 38/10 | 62/26 | <0.0001 1 |
| 0.7385 2 | |||||||
| 3 Ethnicity (n (%)) | |||||||
| White American | 187 (51.0) | 38 (74.5) | 35 (83.3) | 42 (29.6) | 31 (66.0) | 41 (48.2) | <0.0001 |
| Asian American | 161 (43.9) | 11 (21.6) | 5 (11.9) | 97 (68.3) | 7 (14.9) | 41 (48.2) | |
| Black/African American | 17 (4.6) | 1 (2.0) | 2 (4.8) | 3 (2.1) | 8 (17.0) | 3 (3.5) | |
| Natives | 2 (0.5) | 1 (2.0) | 0 | 0 | 1 (2.1) | 0 | |
| 4 Tumor grade (n (%)) | |||||||
| G1 | 55 (14.8) | 8 (15.4) | 6 (14.0) | 11 (7.6) | 10 (21.7) | 20 (23.0) | |
| G2 | 180 (48.4) | 30 (57.7) | 22 (51.2) | 55 (38.2) | 26 (56.5) | 47 (54.0) | <0.0001 |
| G3 | 124 (33.3) | 13 (25.0) | 15 (34.9) | 67 (46.5) | 10 (21.7) | 19 (21.8) | |
| G4 | 13 (3.5) | 1 (1.9) | 0 | 11 (7.6) | 0 | 1 (1.2) | |
| 4 AJCC tumor stage (n (%)) | <0.0001 1 | ||||||
| I | 175 (49.6) | 24 (50.0) | 15 (44.1) | 84 (59.6) | 26 (59.1) | 26 (30.2) | |
| II | 87 (24.7) | 8 (16.7) | 11 (32.4) | 35 (24.8) | 12 (27.3) | 21 (24.4) | 0.2660 5 |
| III | 86 (24.4) | 15 (31.3) | 7 (20.6) | 19 (13.5) | 6 (13.6) | 39 (45.4) | |
| IV | 5 (1.4) | 1 (2.1) | 1 (2.9) | 3 (2.1) | 0 | 0 | |
| IFNL3 rs4803217 | |||||||
| CC:CA:AA | 227:123:26 | 29:21:4 | 25:16:2 | 100:35:8 | 19:19:10 | 54:52:2 | 0.0005 |
| MAF | 0.232 | 0.269 | 0.232 | 0.178 | 0.406 | 0.259 | 0.0003 |
| SNP | Alleles | MAF | D’ (Red) and r2 (Yellow) | ||||
|---|---|---|---|---|---|---|---|
| AS | EUR | rs4803217 | rs117648444 | rs12979860 | rs368234815 | ||
| IFNL3 rs4803217 | C/A | 0.153 | 0.308 | 1.0 | 0.990 | 0.980 | 0.978 |
| IFNL4 rs117648444 | G/A | 0.024 | 0.118 | 0.119 | 1.0 | 1.000 | 1.000 |
| IFNL4 rs12979860 | C/T | 0.156 | 0.309 | 0.944 | 0.120 | 1.0 | 0.997 |
| IFNL4 rs368234815 | TT/ΔG | 0.159 | 0.311 | 0.892 | 0.114 | 0.943 | 1.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huschka, H.; Mihm, S. Hepatic IFNL4 Gene Activation in Hepatocellular Carcinoma Patients with Regard to Etiology. Int. J. Mol. Sci. 2021, 22, 7803. https://doi.org/10.3390/ijms22157803
Huschka H, Mihm S. Hepatic IFNL4 Gene Activation in Hepatocellular Carcinoma Patients with Regard to Etiology. International Journal of Molecular Sciences. 2021; 22(15):7803. https://doi.org/10.3390/ijms22157803
Chicago/Turabian StyleHuschka, Henriette, and Sabine Mihm. 2021. "Hepatic IFNL4 Gene Activation in Hepatocellular Carcinoma Patients with Regard to Etiology" International Journal of Molecular Sciences 22, no. 15: 7803. https://doi.org/10.3390/ijms22157803
APA StyleHuschka, H., & Mihm, S. (2021). Hepatic IFNL4 Gene Activation in Hepatocellular Carcinoma Patients with Regard to Etiology. International Journal of Molecular Sciences, 22(15), 7803. https://doi.org/10.3390/ijms22157803






