Exosomes as New Biomarkers and Drug Delivery Tools for the Prevention and Treatment of Various Diseases: Current Perspectives
Abstract
1. Introduction
2. Exosomes Biology
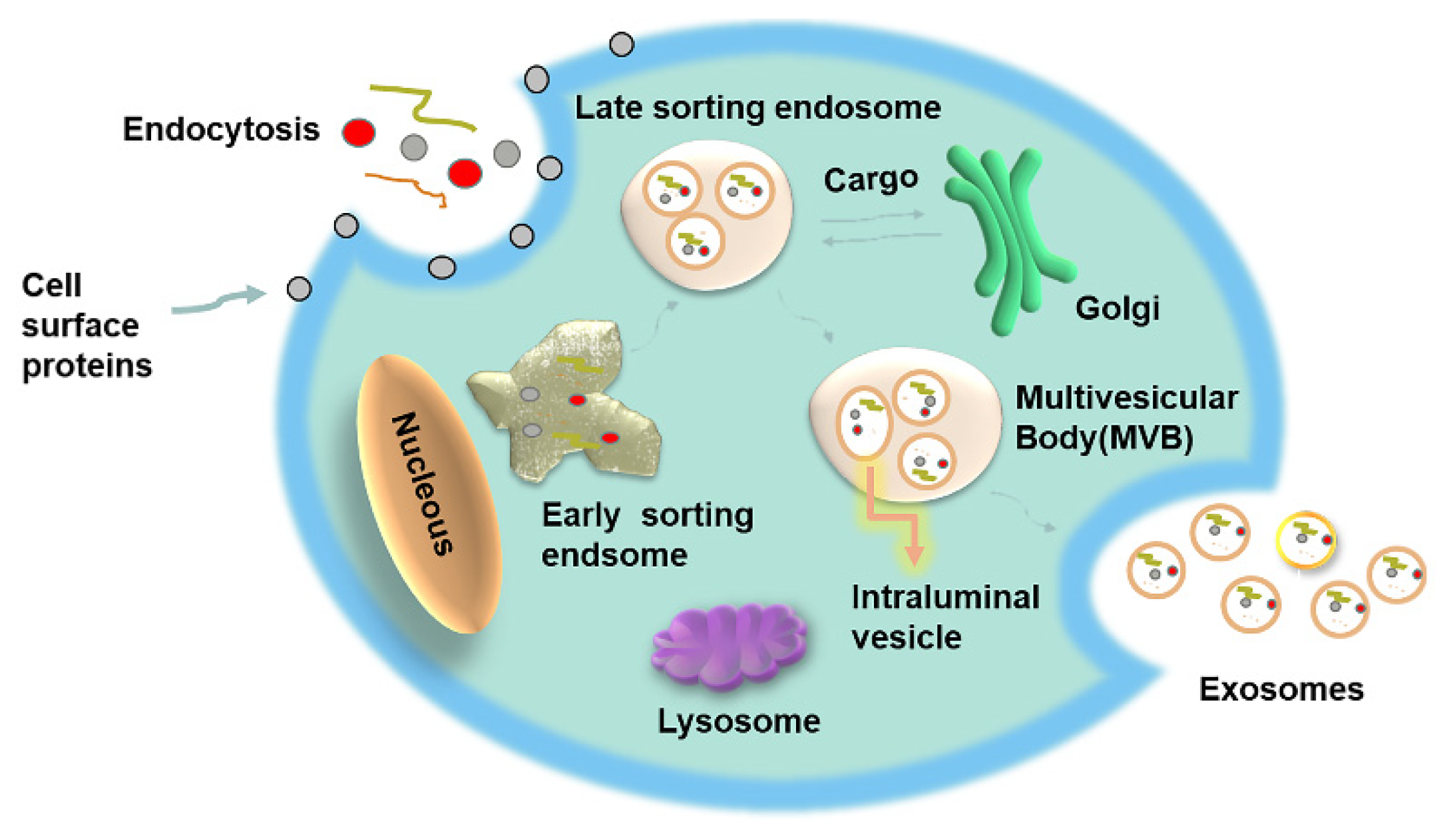
2.1. Formation, Structure and Circulation
2.2. Exosomes: Biogenesis and Cellular Absorption
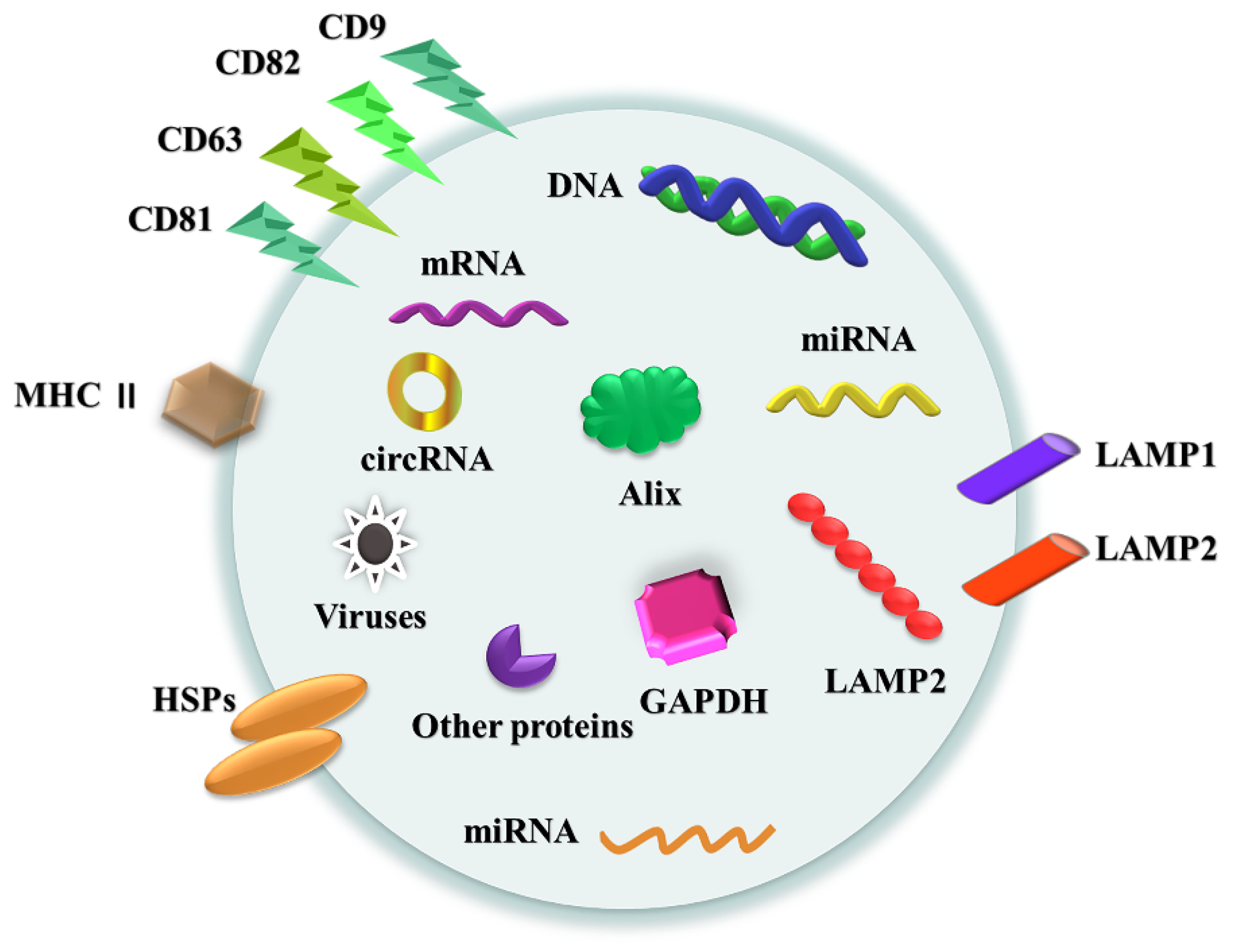
2.3. Exosomes Classification
3. Involvement of Exosomes in Disease Immunopathology
3.1. Exosomes and Tumor Environment
3.2. Exosomes and Digestive Environment
3.3. Exosomes and Cardiovascular Diseases
3.4. Exosomes and Glioblastoma
3.5. Exosomes, the Endocrine System and Cancer
3.6. Exosomes and the Urinary System
3.7. Exosomes in Metabolic Diseases
3.8. Exosomes in Viral Pathogenesis
3.9. Exosomes in Transplantation
3.10. Anti-Inflammatory and Antimicrobial Vesicles
4. Exosomes as a Means of Drug Delivery
4.1. Exosomes and Chemical Synthesis
4.2. Exosomes and Peptidomes
4.2.1. Protein
4.2.2. Nucleic Acids
4.2.3. Lipids
4.3. Exosomes and Natural Medicine Monomers
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deatherage, B.L.; Cookson, B.T. Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012, 80, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Kalra, H.; Mathivanan, S. ExoCarta as a resource for exosomal research. J. Extracell. Vesicles 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Caby, M.P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef]
- Pisitkun, T.; Shen, R.F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef]
- Frühbeis, C.; Fröhlich, D.; Kuo, W.P.; Amphornrat, J.; Thilemann, S.; Saab, A.S.; Kirchhoff, F.; Möbius, W.; Goebbels, S.; Nave, K.A.; et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013, 11, e1001604. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Korkut, C.; Ataman, B.; Ramachandran, P.; Ashley, J.; Barria, R.; Gherbesi, N.; Budnik, V. Trans-synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell 2009, 139, 393–404. [Google Scholar] [CrossRef]
- Machtinger, R.; Laurent, L.C.; Baccarelli, A.A. Extracellular vesicles: Roles in gamete maturation, fertilization and embryo implantation. Hum. Reprod. Update 2016, 22, 182–193. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Ailawadi, S.; Wang, X.; Gu, H.; Fan, G.C. Pathologic function and therapeutic potential of exosomes in cardiovascular disease. Biochim. Biophys. Acta 2015, 1852, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kulshreshtha, A.; Ahmad, T.; Agrawal, A.; Ghosh, B. Proinflammatory role of epithelial cell-derived exosomes in allergic airway inflammation. J. Allergy Clin. Immunol. 2013, 131, 1194–1203, 1203.e1–e14. [Google Scholar] [CrossRef]
- Kadiu, I.; Narayanasamy, P.; Dash, P.K.; Zhang, W.; Gendelman, H.E. Biochemical and biologic characterization of exosomes and microvesicles as facilitators of HIV-1 infection in macrophages. J. Immunol. 2012, 189, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Vella, L.J.; Sharples, R.A.; Lawson, V.A.; Masters, C.L.; Cappai, R.; Hill, A.F. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J. Pathol. 2007, 211, 582–590. [Google Scholar] [CrossRef]
- Kibria, G.; Ramos, E.K.; Wan, Y.; Gius, D.R.; Liu, H. Exosomes as a Drug Delivery System in Cancer Therapy: Potential and Challenges. Mol. Pharm. 2018, 15, 3625–3633. [Google Scholar] [CrossRef] [PubMed]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef]
- Davies, R.T.; Kim, J.; Jang, S.C.; Choi, E.J.; Gho, Y.S.; Park, J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip 2012, 12, 5202–5210. [Google Scholar] [CrossRef]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.; Hole, P.; Carr, B.; Redman, C.W.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 780–788. [Google Scholar] [CrossRef]
- Pospichalova, V.; Svoboda, J.; Dave, Z.; Kotrbova, A.; Kaiser, K.; Klemova, D.; Ilkovics, L.; Hampl, A.; Crha, I.; Jandakova, E.; et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J. Extracell. Vesicles 2015, 4, 25530. [Google Scholar] [CrossRef]
- Fu, Y.; Xiong, S. Tagged extracellular vesicles with the RBD of the viral spike protein for delivery of antiviral agents against SARS-COV-2 infection. J. Control Release Off. J. Control Release Soc. 2021, 335, 584–595. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Hauser, P.; Wang, S.; Didenko, V.V. Apoptotic Bodies: Selective Detection in Extracellular Vesicles. Methods Mol. Biol. 2017, 1554, 193–200. [Google Scholar] [PubMed]
- Leventis, P.A.; Grinstein, S. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 2010, 39, 407–427. [Google Scholar] [CrossRef] [PubMed]
- Cloos, A.S.; Ghodsi, M.; Stommen, A.; Vanderroost, J.; Dauguet, N.; Pollet, H.; D’Auria, L.; Mignolet, E.; Larondelle, Y.; Terrasi, R.; et al. Interplay Between Plasma Membrane Lipid Alteration, Oxidative Stress and Calcium-Based Mechanism for Extracellular Vesicle Biogenesis From Erythrocytes During Blood Storage. Front. Physiol. 2020, 11, 712. [Google Scholar] [CrossRef]
- Meldolesi, J. Exosomes and Ectosomes in Intercellular Communication. Curr. Biol. CB 2018, 28, R435–R444. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Desai, N.N.; Qureshi, M.Z.; Librelotto, D.R.N.; Gasparri, M.L.; Bishayee, A.; Nabavi, S.M.; Curti, V.; Daglia, M. Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol. Adv. 2018, 36, 328–334. [Google Scholar] [CrossRef]
- Pluchino, S.; Smith, J.A. Explicating Exosomes: Reclassifying the Rising Stars of Intercellular Communication. Cell 2019, 177, 225–227. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Raposo, G. Exosomes—Vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.; Sadik, M.; Alaarg, A.; Smith, C.I.; Lehtiö, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef]
- Whiteside, T.L. The potential of tumor-derived exosomes for noninvasive cancer monitoring: An update. Expert Rev. Mol. Diagn. 2018, 18, 1029–1040. [Google Scholar] [CrossRef]
- Whiteside, T.L. The emerging role of plasma exosomes in diagnosis, prognosis and therapies of patients with cancer. Contemp. Oncol. 2018, 22, 38–40. [Google Scholar] [CrossRef]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef] [PubMed]
- McAndrews, K.M.; Kalluri, R. Mechanisms associated with biogenesis of exosomes in cancer. Mol. Cancer 2019, 18, 52. [Google Scholar] [CrossRef]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126 Pt 24, 5553–5565. [Google Scholar] [CrossRef]
- van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Buschow, S.I.; Liefhebber, J.M.; Wubbolts, R.; Stoorvogel, W. Exosomes contain ubiquitinated proteins. Blood Cells Mol. Dis. 2005, 35, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yáñez-Mó, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 288, 11649–11661. [Google Scholar] [CrossRef] [PubMed]
- Nolte-’t Hoen, E.N.; Buermans, H.P.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.; t Hoen, P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef]
- Cai, H.; Reinisch, K.; Ferro-Novick, S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 2007, 12, 671–682. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; You, S.; Spinelli, C.; Morley, S.; Zandian, M.; Aspuria, P.J.; Cavallini, L.; Ciardiello, C.; Reis Sobreiro, M.; Morello, M.; et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget 2015, 6, 11327–11341. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Didiot, M.C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; DiFiglia, M.; Kiebish, M.A.; et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 2016, 5, 32570. [Google Scholar] [CrossRef]
- Xu, R.; Greening, D.W.; Rai, A.; Ji, H.; Simpson, R.J. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods 2015, 87, 11–25. [Google Scholar] [CrossRef]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Mathivanan, S.; Ji, H.; Simpson, R.J. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell. Proteom. MCP 2013, 12, 587–598. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Guescini, M.; Genedani, S.; Stocchi, V.; Agnati, L.F. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J. Neural Transm. 2010, 117, 1–4. [Google Scholar] [CrossRef]
- Ferguson, S.W.; Nguyen, J. Exosomes as therapeutics: The implications of molecular composition and exosomal heterogeneity. J. Control Release Off. J. Control Release Soc. 2016, 228, 179–190. [Google Scholar] [CrossRef]
- Khan, S.; Jutzy, J.M.; Valenzuela, M.M.; Turay, D.; Aspe, J.R.; Ashok, A.; Mirshahidi, S.; Mercola, D.; Lilly, M.B.; Wall, N.R. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS ONE 2012, 7, e46737. [Google Scholar] [CrossRef] [PubMed]
- Lässer, C.; Alikhani, V.S.; Ekström, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjöstrand, M.; Gabrielsson, S.; Lötvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef]
- Street, J.M.; Barran, P.E.; Mackay, C.L.; Weidt, S.; Balmforth, C.; Walsh, T.S.; Chalmers, R.T.; Webb, D.J.; Dear, J.W. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J. Transl. Med. 2012, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Vojtech, L.; Woo, S.; Hughes, S.; Levy, C.; Ballweber, L.; Sauteraud, R.P.; Strobl, J.; Westerberg, K.; Gottardo, R.; Tewari, M.; et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014, 42, 7290–7304. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, K.; Wu, S.; Cui, M.; Xu, T. Focus on Mesenchymal Stem Cell-Derived Exosomes: Opportunities and Challenges in Cell-Free Therapy. Stem Cells Int. 2017, 2017, 6305295. [Google Scholar] [CrossRef]
- He, C.; Hua, W.; Liu, J.; Fan, L.; Wang, H.; Sun, G. Exosomes derived from endoplasmic reticulum-stressed liver cancer cells enhance the expression of cytokines in macrophages via the STAT3 signaling pathway. Oncol. Lett. 2020, 20, 589–600. [Google Scholar] [CrossRef]
- Li, D.; Wang, Y.; Jin, X.; Hu, D.; Xia, C.; Xu, H.; Hu, J. NK cell-derived exosomes carry miR-207 and alleviate depression-like symptoms in mice. J. Neuroinflamm. 2020, 17, 126. [Google Scholar] [CrossRef]
- Zhao, D.; Yu, Z.; Li, Y.; Wang, Y.; Li, Q.; Han, D. GelMA combined with sustained release of HUVECs derived exosomes for promoting cutaneous wound healing and facilitating skin regeneration. J. Mol. Histol. 2020, 51, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Poggio, M.; Hu, T.; Pai, C.C.; Chu, B.; Belair, C.D.; Chang, A.; Montabana, E.; Lang, U.E.; Fu, Q.; Fong, L.; et al. Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory. Cell 2019, 177, 414–427.e13. [Google Scholar] [CrossRef]
- Sanderson, R.D.; Bandari, S.K.; Vlodavsky, I. Proteases and glycosidases on the surface of exosomes: Newly discovered mechanisms for extracellular remodeling. Matrix Biol. J. Int. Soc. Matrix Biol. 2019, 75–76, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, S.; Foroni, C.; Zanaglio, C.; Re, F.; Polverelli, N.; Turra, A.; Morello, E.; Farina, M.; Cattina, F.; Gandolfi, L.; et al. Feasibility of tumor-derived exosome enrichment in the onco-hematology leukemic model of chronic myeloid leukemia. Int. J. Mol. Med. 2019, 44, 2133–2144. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Gougelet, A. Exosomal microRNAs as a potential therapeutic strategy in hepatocellular carcinoma. World J. Hepatol. 2018, 10, 785–789. [Google Scholar] [CrossRef]
- Li, I.; Nabet, B.Y. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol. Cancer 2019, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, S.; Yang, L.; Li, Y. The role of PD-1 and PD-L1 in T-cell immune suppression in patients with hematological malignancies. J. Hematol. Oncol. 2013, 6, 74. [Google Scholar] [CrossRef]
- Francisco, L.M.; Salinas, V.H.; Brown, K.E.; Vanguri, V.K.; Freeman, G.J.; Kuchroo, V.K.; Sharpe, A.H. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J. Exp. Med. 2009, 206, 3015–3029. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, X.; Zeng, K.; Xu, M.; He, B.; Pan, Y.; Sun, H.; Pan, B.; Xu, X.; Xu, T.; et al. DNA-methylation-mediated silencing of miR-486-5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/β-catenin signal pathways. Cell Death Dis. 2018, 9, 1037. [Google Scholar] [CrossRef] [PubMed]
- Theodoraki, M.N.; Yerneni, S.S.; Hoffmann, T.K.; Gooding, W.E.; Whiteside, T.L. Clinical Significance of PD-L1(+) Exosomes in Plasma of Head and Neck Cancer Patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 896–905. [Google Scholar] [CrossRef]
- Skokos, D.; Botros, H.G.; Demeure, C.; Morin, J.; Peronet, R.; Birkenmeier, G.; Boudaly, S.; Mécheri, S. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J. Immunol. 2003, 170, 3037–3045. [Google Scholar] [CrossRef]
- Bhatnagar, S.; Shinagawa, K.; Castellino, F.J.; Schorey, J.S. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood 2007, 110, 3234–3244. [Google Scholar] [CrossRef]
- Kim, S.H.; Bianco, N.R.; Shufesky, W.J.; Morelli, A.E.; Robbins, P.D. MHC class II+ exosomes in plasma suppress inflammation in an antigen-specific and Fas ligand/Fas-dependent manner. J. Immunol. 2007, 179, 2235–2241. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Zhou, A.; Li, X.; Zhu, S.; Wang, Y.; Zhang, S.; Li, P. The Significance of Exosomal RNAs in the Development, Diagnosis, and Treatment of Gastric Cancer. Genes 2021, 12, 73. [Google Scholar] [CrossRef]
- Ogorevc, E.; Kralj-Iglic, V.; Veranic, P. The role of extracellular vesicles in phenotypic cancer transformation. Radiol. Oncol. 2013, 47, 197–205. [Google Scholar] [CrossRef]
- Hannafon, B.N.; Ding, W.Q. Intercellular communication by exosome-derived microRNAs in cancer. Int. J. Mol. Sci. 2013, 14, 14240–14269. [Google Scholar] [CrossRef] [PubMed]
- Che, Y.; Geng, B.; Xu, Y.; Miao, X.; Chen, L.; Mu, X.; Pan, J.; Zhang, C.; Zhao, T.; Wang, C.; et al. Helicobacter pylori-induced exosomal MET educates tumour-associated macrophages to promote gastric cancer progression. J. Cell. Mol. Med. 2018, 22, 5708–5719. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Zhang, L.; Chi, J.; Li, H.; Liu, X.; Hu, T.; Li, R.; Guo, Y.; Zhang, X.; Wang, H.; et al. Helicobacter pylori Infection Impairs Endothelial Function through an Exosome-Mediated Mechanism. J. Am. Heart Assoc. 2020, 9, e014120. [Google Scholar] [CrossRef] [PubMed]
- Gidlöf, O.; Andersson, P.; van der Pals, J.; Götberg, M.; Erlinge, D. Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology 2011, 118, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Sakata, Y.; Suna, S.; Nakatani, D.; Usami, M.; Hara, M.; Kitamura, T.; Hamasaki, T.; Nanto, S.; Kawahara, Y.; et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ. Res. 2013, 113, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.K.; Zhu, J.Q.; Zhang, J.T.; Li, Q.; Li, Y.; He, J.; Qin, Y.W.; Jing, Q. Circulating microRNA: A novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J. 2010, 31, 659–666. [Google Scholar] [CrossRef]
- Lee, C.; Mitsialis, S.A.; Aslam, M.; Vitali, S.H.; Vergadi, E.; Konstantinou, G.; Sdrimas, K.; Fernandez-Gonzalez, A.; Kourembanas, S. Exosomes mediate the cytoprotective action of mesenchymal stromal cells on hypoxia-induced pulmonary hypertension. Circulation 2012, 126, 2601–2611. [Google Scholar] [CrossRef]
- Ahir, B.K.; Ozer, H.; Engelhard, H.H.; Lakka, S.S. MicroRNAs in glioblastoma pathogenesis and therapy: A comprehensive review. Crit. Rev. Oncol. Hematol. 2017, 120, 22–33. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Shi, L.; Wan, Y.; Sun, G.; Gu, X.; Qian, C.; Yan, W.; Zhang, S.; Pan, T.; Wang, Z.; You, Y. Functional differences of miR-125b on the invasion of primary glioblastoma CD133-negative cells and CD133-positive cells. Neuromol. Med. 2012, 14, 303–316. [Google Scholar] [CrossRef]
- Ciafrè, S.A.; Galardi, S.; Mangiola, A.; Ferracin, M.; Liu, C.G.; Sabatino, G.; Negrini, M.; Maira, G.; Croce, C.M.; Farace, M.G. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005, 334, 1351–1358. [Google Scholar] [CrossRef]
- Sun, Y.C.; Wang, J.; Guo, C.C.; Sai, K.; Wang, J.; Chen, F.R.; Yang, Q.Y.; Chen, Y.S.; Wang, J.; To, T.S.; et al. MiR-181b sensitizes glioma cells to teniposide by targeting MDM2. BMC Cancer 2014, 14, 611. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Ma, R.; Zhang, L.; Li, H.; Tang, Y.; Du, G.; Niu, D.; Yin, D. The Role of Exosomes in Thyroid Cancer and Their Potential Clinical Application. Front. Oncol. 2020, 10, 596132. [Google Scholar] [CrossRef]
- Lee, J.C.; Zhao, J.T.; Gundara, J.; Serpell, J.; Bach, L.A.; Sidhu, S. Papillary thyroid cancer-derived exosomes contain miRNA-146b and miRNA-222. J. Surg. Res. 2015, 196, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Li, G.; Chen, W.; Song, L.; Wei, T.; Li, Z.; Gong, R.; Lei, J.; Shi, H.; Zhu, J. Plasma Exosomal miR-146b-5p and miR-222-3p are Potential Biomarkers for Lymph Node Metastasis in Papillary Thyroid Carcinomas. OncoTargets Ther. 2020, 13, 1311–1319. [Google Scholar] [CrossRef] [PubMed]
- Khatami, F.; Tavangar, S.M. Liquid Biopsy in Thyroid Cancer: New Insight. Int. J. Hematol. Oncol. Stem Cell Res. 2018, 12, 235–248. [Google Scholar] [PubMed]
- Samsonov, R.; Burdakov, V.; Shtam, T.; Radzhabova, Z.; Vasilyev, D.; Tsyrlina, E.; Titov, S.; Ivanov, M.; Berstein, L.; Filatov, M.; et al. Plasma exosomal miR-21 and miR-181a differentiates follicular from papillary thyroid cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 12011–12021. [Google Scholar] [CrossRef]
- Wang, Z.; Lv, J.; Zou, X.; Huang, Z.; Zhang, H.; Liu, Q.; Jiang, L.; Zhou, X.; Zhu, W. A three plasma microRNA signature for papillary thyroid carcinoma diagnosis in Chinese patients. Gene 2019, 693, 37–45. [Google Scholar] [CrossRef]
- Dai, D.; Tan, Y.; Guo, L.; Tang, A.; Zhao, Y. Identification of exosomal miRNA biomarkers for diagnosis of papillary thyroid cancer by small RNA sequencing. Eur. J. Endocrinol. 2020, 182, 111–121. [Google Scholar] [CrossRef]
- Liang, M.; Yu, S.; Tang, S.; Bai, L.; Cheng, J.; Gu, Y.; Li, S.; Zheng, X.; Duan, L.; Wang, L.; et al. A Panel of Plasma Exosomal miRNAs as Potential Biomarkers for Differential Diagnosis of Thyroid Nodules. Front. Genet. 2020, 11, 449. [Google Scholar] [CrossRef]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef]
- Borges, F.T.; Melo, S.A.; Özdemir, B.C.; Kato, N.; Revuelta, I.; Miller, C.A.; Gattone, V.H., 2nd; LeBleu, V.S.; Kalluri, R. TGF-β1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J. Am. Soc. Nephrol. JASN 2013, 24, 385–392. [Google Scholar] [CrossRef]
- Okada, H. A new look at tubulointerstitial communication with exosomes. J. Am. Soc. Nephrol. JASN 2013, 24, 330–332. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Daryabor, G.; Kabelitz, D.; Kalantar, K. An update on immune dysregulation in obesity-related insulin resistance. Scand. J. Immunol. 2019, 89, e12747. [Google Scholar] [CrossRef] [PubMed]
- Huang-Doran, I.; Zhang, C.Y.; Vidal-Puig, A. Extracellular Vesicles: Novel Mediators of Cell Communication in Metabolic Disease. Trends Endocrinol. Metab. TEM 2017, 28, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.D.; Sheng, H.; Dias, P.; Jubayer Rahman, M.; Bashratyan, R.; Regn, D.; Marquardt, K. Autoimmune Responses to Exosomes and Candidate Antigens Contribute to Type 1 Diabetes in Non-Obese Diabetic Mice. Curr. Diabetes Rep. 2017, 17, 130. [Google Scholar] [CrossRef] [PubMed]
- Guay, C.; Menoud, V.; Rome, S.; Regazzi, R. Horizontal transfer of exosomal microRNAs transduce apoptotic signals between pancreatic beta-cells. Cell Commun. Signal. CCS 2015, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.Z.; Liu, Y.M.; Niu, X.; Yin, J.Y.; Hu, B.; Guo, S.C.; Fan, Y.; Wang, Y.; Wang, N.S. Exosomes secreted by human urine-derived stem cells could prevent kidney complications from type I diabetes in rats. Stem Cell Res. Ther. 2016, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Wu, H.; Liu, Y.; Zhao, M.; Li, D.; Lu, Q. Recent advances of exosomes in immune modulation and autoimmune diseases. Autoimmunity 2016, 49, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Shang, Q.; Pan, Z.; Bai, Y.; Li, Z.; Zhang, H.; Zhang, Q.; Guo, C.; Zhang, L.; Wang, Q. Exosomes from Adipose-Derived Stem Cells Attenuate Adipose Inflammation and Obesity through Polarizing M2 Macrophages and Beiging in White Adipose Tissue. Diabetes 2018, 67, 235–247. [Google Scholar] [CrossRef]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef] [PubMed]
- Samuelson, I.; Vidal-Puig, A.J. Fed-EXosome: Extracellular vesicles and cell-cell communication in metabolic regulation. Essays Biochem. 2018, 62, 165–175. [Google Scholar] [PubMed]
- Karolina, D.S.; Tavintharan, S.; Armugam, A.; Sepramaniam, S.; Pek, S.L.; Wong, M.T.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. Circulating miRNA profiles in patients with metabolic syndrome. J. Clin. Endocrinol. Metab. 2012, 97, E2271–E2276. [Google Scholar] [CrossRef]
- Dini, L.; Tacconi, S.; Carata, E.; Tata, A.M.; Vergallo, C.; Panzarini, E. Microvesicles and exosomes in metabolic diseases and inflammation. Cytokine Growth Factor Rev. 2020, 51, 27–39. [Google Scholar] [CrossRef]
- Szczepanski, M.J.; Szajnik, M.; Welsh, A.; Whiteside, T.L.; Boyiadzis, M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica 2011, 96, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Gilligan, K.E.; Dwyer, R.M. Engineering Exosomes for Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 1122. [Google Scholar] [CrossRef]
- Binenbaum, Y.; Fridman, E.; Yaari, Z.; Milman, N.; Schroeder, A.; Ben David, G.; Shlomi, T.; Gil, Z. Transfer of miRNA in Macrophage-Derived Exosomes Induces Drug Resistance in Pancreatic Adenocarcinoma. Cancer Res. 2018, 78, 5287–5299. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Hu, C. Exosomal transfer of miR-214 mediates gefitinib resistance in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018, 507, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Liu, Y.; Fang, X.; Liu, Y.; Fang, L.; Lin, L.; Liu, X.; Wang, N. Tumor-derived microRNA-494 promotes angiogenesis in non-small cell lung cancer. Angiogenesis 2015, 18, 373–382. [Google Scholar] [CrossRef]
- Hsu, H.H.; Kuo, W.W.; Shih, H.N.; Cheng, S.F.; Yang, C.K.; Chen, M.C.; Tu, C.C.; Viswanadha, V.P.; Liao, P.H.; Huang, C.Y. FOXC1 Regulation of miR-31-5p Confers Oxaliplatin Resistance by Targeting LATS2 in Colorectal Cancer. Cancers 2019, 11, 1576. [Google Scholar] [CrossRef] [PubMed]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef]
- Yang, Y.C.; Liu, G.J.; Yuan, D.F.; Li, C.Q.; Xue, M.; Chen, L.J. Influence of exosome-derived miR-21on chemotherapy resistance of esophageal cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1513–1519. [Google Scholar] [PubMed]
- Qin, X.; Guo, H.; Wang, X.; Zhu, X.; Yan, M.; Wang, X.; Xu, Q.; Shi, J.; Lu, E.; Chen, W.; et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019, 20, 12. [Google Scholar] [CrossRef]
- Fang, Y.; Zhou, W.T.; Rong, Y.F.; Kuang, T.T.; Xu, X.F.; Wu, W.C.; Wang, D.S.; Lou, W.H. Exosomal miRNA-106b from cancer-associated fibroblast promotes gemcitabine resistance in pancreatic cancer. Exp. Cell Res. 2019, 383, 111543. [Google Scholar] [CrossRef]
- Crenshaw, B.J.; Gu, L.; Sims, B.; Matthews, Q.L. Exosome Biogenesis and Biological Function in Response to Viral Infections. Open Virol J. 2018, 12, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.R.; Kashanchi, F.; Jacobson, S. Exosomes in Viral Disease. Neurotherapeutics 2016, 13, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Alenquer, M.; Amorim, M.J. Exosome Biogenesis, Regulation, and Function in Viral Infection. Viruses 2015, 7, 5066–5083. [Google Scholar] [CrossRef]
- Chugh, P.E.; Sin, S.H.; Ozgur, S.; Henry, D.H.; Menezes, P.; Griffith, J.; Eron, J.J.; Damania, B.; Dittmer, D.P. Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog. 2013, 9, e1003484. [Google Scholar] [CrossRef]
- Sanche, S.; Lin, Y.T.; Xu, C.; Romero-Severson, E.; Hengartner, N.; Ke, R. High Contagiousness and Rapid Spread of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1470–1477. [Google Scholar] [CrossRef]
- Kumar, A.; Faiq, M.A.; Pareek, V.; Raza, K.; Narayan, R.K.; Prasoon, P.; Kumar, P.; Kulandhasamy, M.; Kumari, C.; Kant, K.; et al. Relevance of SARS-CoV-2 related factors ACE2 and TMPRSS2 expressions in gastrointestinal tissue with pathogenesis of digestive symptoms, diabetes-associated mortality, and disease recurrence in COVID-19 patients. Med. Hypotheses 2020, 144, 110271. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef]
- Zhang, C.; Shi, L.; Wang, F.S. Liver injury in COVID-19: Management and challenges. Lancet. Gastroenterol. Hepatol. 2020, 5, 428–430. [Google Scholar] [CrossRef]
- Lenassi, M.; Cagney, G.; Liao, M.; Vaupotic, T.; Bartholomeeusen, K.; Cheng, Y.; Krogan, N.J.; Plemenitas, A.; Peterlin, B.M. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 2010, 11, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Dukers, D.F.; Meij, P.; Vervoort, M.B.; Vos, W.; Scheper, R.J.; Meijer, C.J.; Bloemena, E.; Middeldorp, J.M. Direct immunosuppressive effects of EBV-encoded latent membrane protein 1. J. Immunol. 2000, 165, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.; Singh, S.; Henderson, J.; Krishnamurthy, P. Mechanisms of COVID-19-induced cardiovascular disease: Is sepsis or exosome the missing link? J. Cell. Physiol. 2020, 236, 3366–3382. [Google Scholar] [CrossRef] [PubMed]
- Arenaccio, C.; Anticoli, S.; Manfredi, F.; Chiozzini, C.; Olivetta, E.; Federico, M. Latent HIV-1 is activated by exosomes from cells infected with either replication-competent or defective HIV-1. Retrovirology 2015, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Jabbari, N.; Karimipour, M.; Khaksar, M.; Akbariazar, E.; Heidarzadeh, M.; Mojarad, B.; Aftab, H.; Rahbarghazi, R.; Rezaie, J. Tumor-derived extracellular vesicles: Insights into bystander effects of exosomes after irradiation. Lasers Med. Sci. 2020, 35, 531–545. [Google Scholar] [CrossRef]
- Huang, H.I.; Lin, J.Y.; Chiang, H.C.; Huang, P.N.; Lin, Q.D.; Shih, S.R. Exosomes Facilitate Transmission of Enterovirus A71 From Human Intestinal Epithelial Cells. J. Infect. Dis. 2020, 222, 456–469. [Google Scholar] [CrossRef]
- Marino, J.; Babiker-Mohamed, M.H.; Crosby-Bertorini, P.; Paster, J.T.; LeGuern, C.; Germana, S.; Abdi, R.; Uehara, M.; Kim, J.I.; Markmann, J.F.; et al. Donor exosomes rather than passenger leukocytes initiate alloreactive T cell responses after transplantation. Sci. Immunol. 2016, 1, aaf8759. [Google Scholar] [CrossRef]
- Chen, L.; Huang, H.; Zhang, W.; Ding, F.; Fan, Z.; Zeng, Z. Exosomes Derived From T Regulatory Cells Suppress CD8+ Cytotoxic T Lymphocyte Proliferation and Prolong Liver Allograft Survival. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 4877–4884. [Google Scholar] [CrossRef]
- Liu, Y.; Holmes, C. Tissue Regeneration Capacity of Extracellular Vesicles Isolated from Bone Marrow-Derived and Adipose-Derived Mesenchymal Stromal/Stem Cells. Front. Cell Dev. Biol. 2021, 9, 648098. [Google Scholar] [CrossRef]
- Ramirez-Bajo, M.J.; Rovira, J.; Lazo-Rodriguez, M.; Banon-Maneus, E.; Tubita, V.; Moya-Rull, D.; Hierro-Garcia, N.; Ventura-Aguiar, P.; Oppenheimer, F.; Campistol, J.M.; et al. Impact of Mesenchymal Stromal Cells and Their Extracellular Vesicles in a Rat Model of Kidney Rejection. Front. Cell Dev. Biol. 2020, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- van Rhijn-Brouwer, F.C.C.; van Balkom, B.W.M.; Papazova, D.A.; Hazenbrink, D.H.M.; Meijer, A.J.; Brete, I.; Briceno, V.; van Zuilen, A.D.; Toorop, R.J.; Fledderus, J.O.; et al. Paracrine Proangiogenic Function of Human Bone Marrow-Derived Mesenchymal Stem Cells Is Not Affected by Chronic Kidney Disease. Stem Cells Int. 2019, 2019, 1232810. [Google Scholar] [CrossRef]
- Hosseiniyan Khatibi, S.M.; Kheyrolahzadeh, K.; Barzegari, A.; Rahbar Saadat, Y.; Zununi Vahed, S. Medicinal signaling cells: A potential antimicrobial drug store. J. Cell. Physiol. 2020, 235, 7731–7746. [Google Scholar] [CrossRef]
- Harrell, C.R.; Jovicic, N.; Djonov, V.; Arsenijevic, N.; Volarevic, V. Mesenchymal Stem Cell-Derived Exosomes and Other Extracellular Vesicles as New Remedies in the Therapy of Inflammatory Diseases. Cells 2019, 8, 1605. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Francos, S.; Eiro, N.; Costa, L.A.; Escudero-Cernuda, S.; Fernández-Sánchez, M.L.; Vizoso, F.J. Mesenchymal Stem Cells as a Cornerstone in a Galaxy of Intercellular Signals: Basis for a New Era of Medicine. Int. J. Mol. Sci. 2021, 22, 3576. [Google Scholar] [CrossRef] [PubMed]
- Pierce, L.M.; Kurata, W.E. Priming with Toll-Like Receptor 3 Agonist Poly(I:C) Enhances Content of Innate Immune Defense Proteins but Not MicroRNAs in Human Mesenchymal Stem Cell-Derived Extracellular Vesicles. Front. Cell Dev. Biol. 2021, 9, 676356. [Google Scholar] [CrossRef]
- Marrazzo, P.; Pizzuti, V.; Zia, S.; Sargenti, A.; Gazzola, D.; Roda, B.; Bonsi, L.; Alviano, F. Microfluidic Tools for Enhanced Characterization of Therapeutic Stem Cells and Prediction of Their Potential Antimicrobial Secretome. Antibiotics 2021, 10, 750. [Google Scholar] [CrossRef] [PubMed]
- Pugholm, L.H.; Bæk, R.; Søndergaard, E.K.; Revenfeld, A.L.; Jørgensen, M.M.; Varming, K. Phenotyping of Leukocytes and Leukocyte-Derived Extracellular Vesicles. J. Immunol. Res. 2016, 2016, 6391264. [Google Scholar] [CrossRef]
- Youn, Y.J.; Shrestha, S.; Lee, Y.B.; Kim, J.K.; Lee, J.H.; Hur, K.; Mali, N.M.; Nam, S.W.; Kim, S.H.; Lee, S.; et al. Neutrophil-derived trail is a proinflammatory subtype of neutrophil-derived extracellular vesicles. Theranostics 2021, 11, 2770–2787. [Google Scholar] [CrossRef]
- Hong, C.W. Extracellular Vesicles of Neutrophils. Immune Netw. 2018, 18, e43. [Google Scholar] [CrossRef]
- Wang, C.; Börger, V.; Sardari, M.; Murke, F.; Skuljec, J.; Pul, R.; Hagemann, N.; Dzyubenko, E.; Dittrich, R.; Gregorius, J.; et al. Mesenchymal Stromal Cell-Derived Small Extracellular Vesicles Induce Ischemic Neuroprotection by Modulating Leukocytes and Specifically Neutrophils. Stroke 2020, 51, 1825–1834. [Google Scholar] [CrossRef]
- Wang, J.; Xia, J.; Huang, R.; Hu, Y.; Fan, J.; Shu, Q.; Xu, J. Mesenchymal stem cell-derived extracellular vesicles alter disease outcomes via endorsement of macrophage polarization. Stem Cell Res. Ther. 2020, 11, 424. [Google Scholar] [CrossRef] [PubMed]
- Akbar, N.; Paget, D.; Choudhury, R.P. Extracellular Vesicles in Innate Immune Cell Programming. Biomedicines 2021, 9, 713. [Google Scholar] [CrossRef] [PubMed]
- Mirzakhani, M.; Mohammadnia-Afrouzi, M.; Shahbazi, M.; Mirhosseini, S.A.; Hosseini, H.M.; Amani, J. The exosome as a novel predictive/diagnostic biomarker of rejection in the field of transplantation. Clin. Immunol. 2019, 203, 134–141. [Google Scholar] [CrossRef]
- Pascucci, L.; Coccè, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Viganò, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control Release Off. J. Control Release Soc. 2014, 192, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.C.; Kim, O.Y.; Yoon, C.M.; Choi, D.S.; Roh, T.Y.; Park, J.; Nilsson, J.; Lötvall, J.; Kim, Y.K.; Gho, Y.S. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano 2013, 7, 7698–7710. [Google Scholar] [CrossRef]
- Busatto, S.; Pham, A.; Suh, A.; Shapiro, S.; Wolfram, J. Organotropic drug delivery: Synthetic nanoparticles and extracellular vesicles. Biomed. Microdevices 2019, 21, 46. [Google Scholar] [CrossRef]
- Pasut, G.; Paolino, D.; Celia, C.; Mero, A.; Joseph, A.S.; Wolfram, J.; Cosco, D.; Schiavon, O.; Shen, H.; Fresta, M. Polyethylene glycol (PEG)-dendron phospholipids as innovative constructs for the preparation of super stealth liposomes for anticancer therapy. J. Control Release Off. J. Control Release Soc. 2015, 199, 106–113. [Google Scholar] [CrossRef]
- Lathwal, S.; Yerneni, S.S.; Boye, S.; Muza, U.L.; Takahashi, S.; Sugimoto, N.; Lederer, A.; Das, S.R.; Campbell, P.G.; Matyjaszewski, K. Engineering exosome polymer hybrids by atom transfer radical polymerization. Proc. Natl. Acad. Sci. USA 2021, 118, e2020241118. [Google Scholar] [CrossRef]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef]
- Kim, H.S.; Choi, D.Y.; Yun, S.J.; Choi, S.M.; Kang, J.W.; Jung, J.W.; Hwang, D.; Kim, K.P.; Kim, D.W. Proteomic analysis of microvesicles derived from human mesenchymal stem cells. J. Proteome Res. 2012, 11, 839–849. [Google Scholar] [CrossRef]
- Simpson, R.J.; Jensen, S.S.; Lim, J.W. Proteomic profiling of exosomes: Current perspectives. Proteomics 2008, 8, 4083–4099. [Google Scholar] [CrossRef]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef]
- Brosseau, C.; Colas, L.; Magnan, A.; Brouard, S. CD9 Tetraspanin: A New Pathway for the Regulation of Inflammation? Front. Immunol. 2018, 9, 2316. [Google Scholar] [CrossRef]
- Wang, H.X.; Li, Q.; Sharma, C.; Knoblich, K.; Hemler, M.E. Tetraspanin protein contributions to cancer. Biochem. Soc. Trans. 2011, 39, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Simons, M. Exosomes: Vesicular carriers for intercellular communication in neurodegenerative disorders. Cell Tissue Res. 2013, 352, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Mizrak, A.; Bolukbasi, M.F.; Ozdener, G.B.; Brenner, G.J.; Madlener, S.; Erkan, E.P.; Ströbel, T.; Breakefield, X.O.; Saydam, O. Genetically engineered microvesicles carrying suicide mRNA/protein inhibit schwannoma tumor growth. Mol. Ther. J. Am. Soc. Gene Ther. 2013, 21, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control Release Off. J. Control Release Soc. 2015, 207, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Sterzenbach, U.; Putz, U.; Low, L.H.; Silke, J.; Tan, S.S.; Howitt, J. Engineered Exosomes as Vehicles for Biologically Active Proteins. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 1269–1278. [Google Scholar] [CrossRef]
- Yuan, D.; Zhao, Y.; Banks, W.A.; Bullock, K.M.; Haney, M.; Batrakova, E.; Kabanov, A.V. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials 2017, 142, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, H.; Sheokand, N.; Kumar, S.; Chauhan, A.S.; Kumar, M.; Jakhar, P.; Boradia, V.M.; Raje, C.I.; Raje, M. Exosomes: Tunable Nano Vehicles for Macromolecular Delivery of Transferrin and Lactoferrin to Specific Intracellular Compartment. J. Biomed. Nanotechnol. 2016, 12, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Batagov, A.O.; Kurochkin, I.V. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3’-untranslated regions. Biol. Direct 2013, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, L.; Giurato, G.; Cicchini, C.; Montaldo, C.; Mancone, C.; Tarallo, R.; Battistelli, C.; Alonzi, T.; Weisz, A.; Tripodi, M. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep. 2016, 17, 799–808. [Google Scholar] [CrossRef]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Donoso-Quezada, J.; Ayala-Mar, S.; González-Valdez, J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic 2021, 22, 204–220. [Google Scholar] [CrossRef]
- Hough, K.P.; Wilson, L.S.; Trevor, J.L.; Strenkowski, J.G.; Maina, N.; Kim, Y.I.; Spell, M.L.; Wang, Y.; Chanda, D.; Dager, J.R.; et al. Unique Lipid Signatures of Extracellular Vesicles from the Airways of Asthmatics. Sci. Rep. 2018, 8, 10340. [Google Scholar] [CrossRef]
- Aswad, H.; Forterre, A.; Wiklander, O.P.; Vial, G.; Danty-Berger, E.; Jalabert, A.; Lamazière, A.; Meugnier, E.; Pesenti, S.; Ott, C.; et al. Exosomes participate in the alteration of muscle homeostasis during lipid-induced insulin resistance in mice. Diabetologia 2014, 57, 2155–2164. [Google Scholar] [CrossRef]
- Beloribi-Djefaflia, S.; Siret, C.; Lombardo, D. Exosomal lipids induce human pancreatic tumoral MiaPaCa-2 cells resistance through the CXCR4-SDF-1α signaling axis. Oncoscience 2015, 2, 15–30. [Google Scholar] [CrossRef] [PubMed]
- EL Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Zhong, C.; Wall, N.R.; Zu, Y.; Sui, G. Therapeutic Application of Natural Medicine Monomers in Cancer Treatment. Curr. Med. Chem. 2017, 24, 3681–3697. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Melzig, M.F. The Stability of Medicinal Plant microRNAs in the Herb Preparation Process. Molecules 2018, 23, 919. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.R.; Fong, M.Y.; Somlo, G.; Wu, J.; Swiderski, P.; Wu, X.; Wang, S.E. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016, 26, 217–228. [Google Scholar] [CrossRef]
- Huang, F.; Du, J.; Liang, Z.; Xu, Z.; Xu, J.; Zhao, Y.; Lin, Y.; Mei, S.; He, Q.; Zhu, J.; et al. Large-scale analysis of small RNAs derived from traditional Chinese herbs in human tissues. Sci. China Life Sci. 2019, 62, 321–332. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef]
- Sluijter, J.P.G.; Davidson, S.M.; Boulanger, C.M.; Buzás, E.I.; de Kleijn, D.P.V.; Engel, F.B.; Giricz, Z.; Hausenloy, D.J.; Kishore, R.; Lecour, S.; et al. Extracellular vesicles in diagnostics and therapy of the ischaemic heart: Position Paper from the Working Group on Cellular Biology of the Heart of the European Society of Cardiology. Cardiovasc. Res. 2018, 114, 19–34. [Google Scholar] [CrossRef]
- Iaccino, E.; Mimmi, S.; Dattilo, V.; Marino, F.; Candeloro, P.; Di Loria, A.; Marimpietri, D.; Pisano, A.; Albano, F.; Vecchio, E.; et al. Monitoring multiple myeloma by idiotype-specific peptide binders of tumor-derived exosomes. Mol. Cancer 2017, 16, 159. [Google Scholar] [CrossRef]
- Yao, J.; Yang, M.; Duan, Y. Chemistry, biology, and medicine of fluorescent nanomaterials and related systems: New insights into biosensing, bioimaging, genomics, diagnostics, and therapy. Chem. Rev. 2014, 114, 6130–6178. [Google Scholar] [CrossRef]
- Zhang, M.; Zang, X.; Wang, M.; Li, Z.; Qiao, M.; Hu, H.; Chen, D. Exosome-based nanocarriers as bio-inspired and versatile vehicles for drug delivery: Recent advances and challenges. J. Mater. Chem. B 2019, 7, 2421–2433. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Weng, A.; Melzig, M.F. MicroRNAs as New Bioactive Components in Medicinal Plants. Planta Med. 2016, 82, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
| Type of Tumor | Source | Function | References |
|---|---|---|---|
| Colorectal cancer (CRC) | Serum and plasma | MiR-486-5p promotes the proliferation and migration of CRC cells by activating the signal pathways of pleomorphic adenomatoid gene 2 (PLAGL2), insulin-like growth factor 2 (IGF2) and β-catenin in vivo and in vitro. | [74] |
| Head and neck squamous cell carcinomas | Plasma | Downregulate CD69 expression on effector T cells to inhibit antitumor response | [75] |
| Prostate cancer | Tumor tissue | Suppress the function of T cells in the draining lymph node and block anti-PD-L1 antibodies | [66] |
| Melanoma | Plasma | Suppress the function of CD8 + T cells and cause failure of anti-PD-1 therapy | [69] |
| Disease | Exosomal miRNAs | Target or Pathway | References |
|---|---|---|---|
| Acute myeloid leukemia | Exosomes with MICA/B (MHC I chain-related proteins A and B) | By downregulating NKG2D receptor expression | [116] |
| Brain cancer | Brain endothelial cells | Rhodamine 123, PTX, DOX | [117] |
| Breast cancer | MiR-365 in macrophage-derived exosomes | The triphospho-nucleotide pool, the enzyme cytidine deaminase | [118] |
| Leukemia | MiR-210 | CD107a | [119] |
| Lung cancer | MiR-494 | Suppresses PTEN (PTEN (phosphatase and tensin homolog deleted on chromosome ten), it is located at 10q23.3 and the transcriptional product is 515 kb mRNA). | [120] |
| Colorectal cancer | MiR-31-5p in (tumor-derived exosomes) TDEs | LATS2 | [121] |
| Nasopharyngeal cancer | MiR-24-3p | ND | [122] |
| Esophageal cancer | MiR-21 in TDEs | PDCD4 | [123] |
| Head and neck cancer | MiR-196a in cancer associate fibroblasts (CAF)- derived exosomes | CDKN1B and ING5 | [124] |
| Pancreatic cancer | MiR-106b in CAFs-derived exosomes | TP53INP1 | [125] |
| Virus | Source | Function | References |
|---|---|---|---|
| Avian influenza (H5N1) | miR-483-3P | Increased production of proinflammatory cytokines in vascular endothelial cells | [136] |
| HIV | Nef | Susceptibility to infection and apoptosis of CD4 cells | [134,137] |
| KSHV | miRNA and others | IL6 production and cellular metabolism | [129] |
| Coronavirus | CD9 | Proviral | [138] |
| EV-A71 | Viral protein and nucleic acid | Virus spread | [139] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Li, S.; Dupuy, A.; Mai, H.l.; Sailliet, N.; Logé, C.; Robert, J.-M.H.; Brouard, S. Exosomes as New Biomarkers and Drug Delivery Tools for the Prevention and Treatment of Various Diseases: Current Perspectives. Int. J. Mol. Sci. 2021, 22, 7763. https://doi.org/10.3390/ijms22157763
Liu Q, Li S, Dupuy A, Mai Hl, Sailliet N, Logé C, Robert J-MH, Brouard S. Exosomes as New Biomarkers and Drug Delivery Tools for the Prevention and Treatment of Various Diseases: Current Perspectives. International Journal of Molecular Sciences. 2021; 22(15):7763. https://doi.org/10.3390/ijms22157763
Chicago/Turabian StyleLiu, Qi, Shiying Li, Amandine Dupuy, Hoa le Mai, Nicolas Sailliet, Cédric Logé, J.-Michel H. Robert, and Sophie Brouard. 2021. "Exosomes as New Biomarkers and Drug Delivery Tools for the Prevention and Treatment of Various Diseases: Current Perspectives" International Journal of Molecular Sciences 22, no. 15: 7763. https://doi.org/10.3390/ijms22157763
APA StyleLiu, Q., Li, S., Dupuy, A., Mai, H. l., Sailliet, N., Logé, C., Robert, J.-M. H., & Brouard, S. (2021). Exosomes as New Biomarkers and Drug Delivery Tools for the Prevention and Treatment of Various Diseases: Current Perspectives. International Journal of Molecular Sciences, 22(15), 7763. https://doi.org/10.3390/ijms22157763







