Tuning Beforehand: A Foresight on RNA Interference (RNAi) and In Vitro-Derived dsRNAs to Enhance Crop Resilience to Biotic and Abiotic Stresses
Abstract
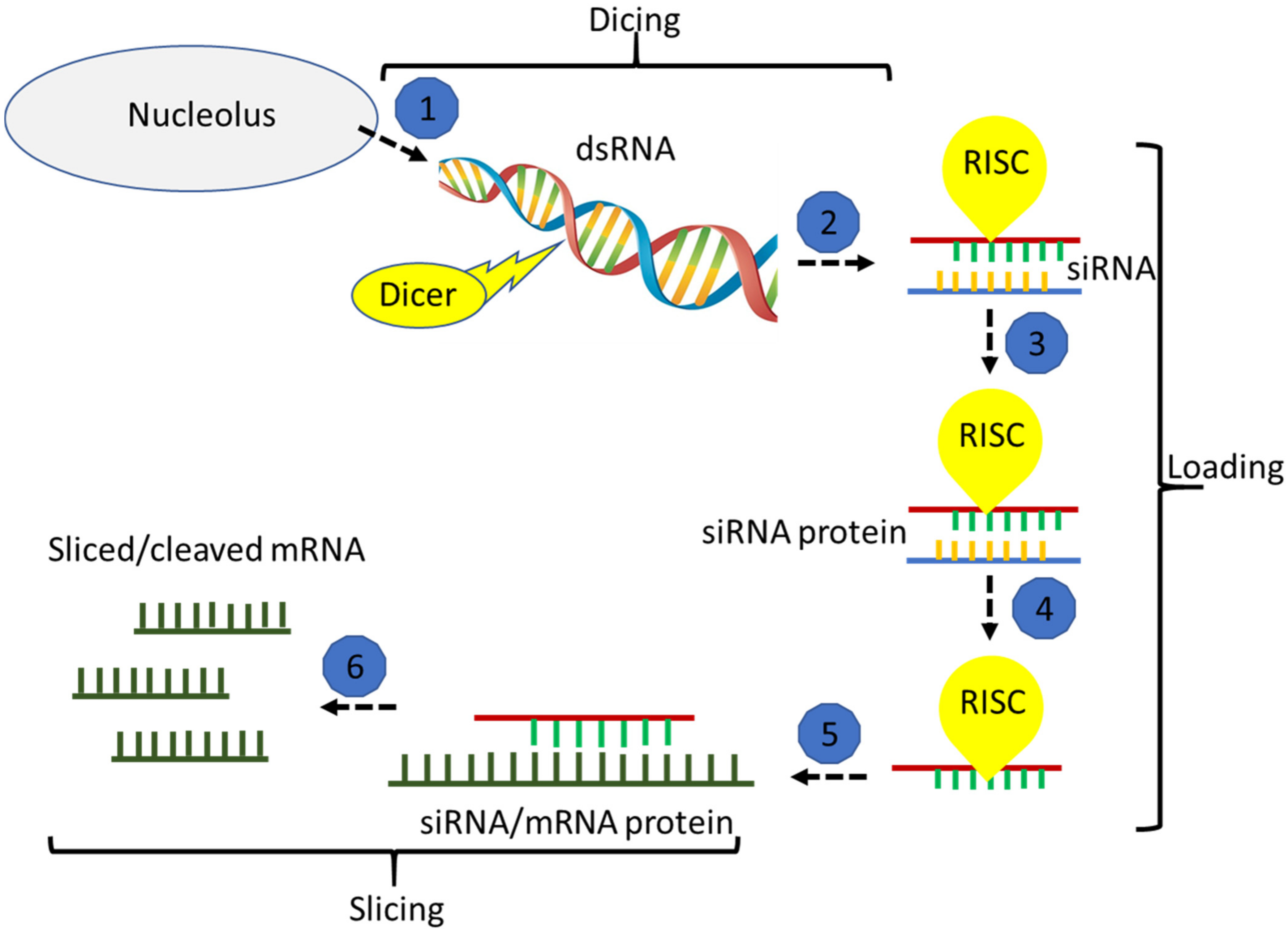
:1. Introduction
2. Vision One: Biotic Stress
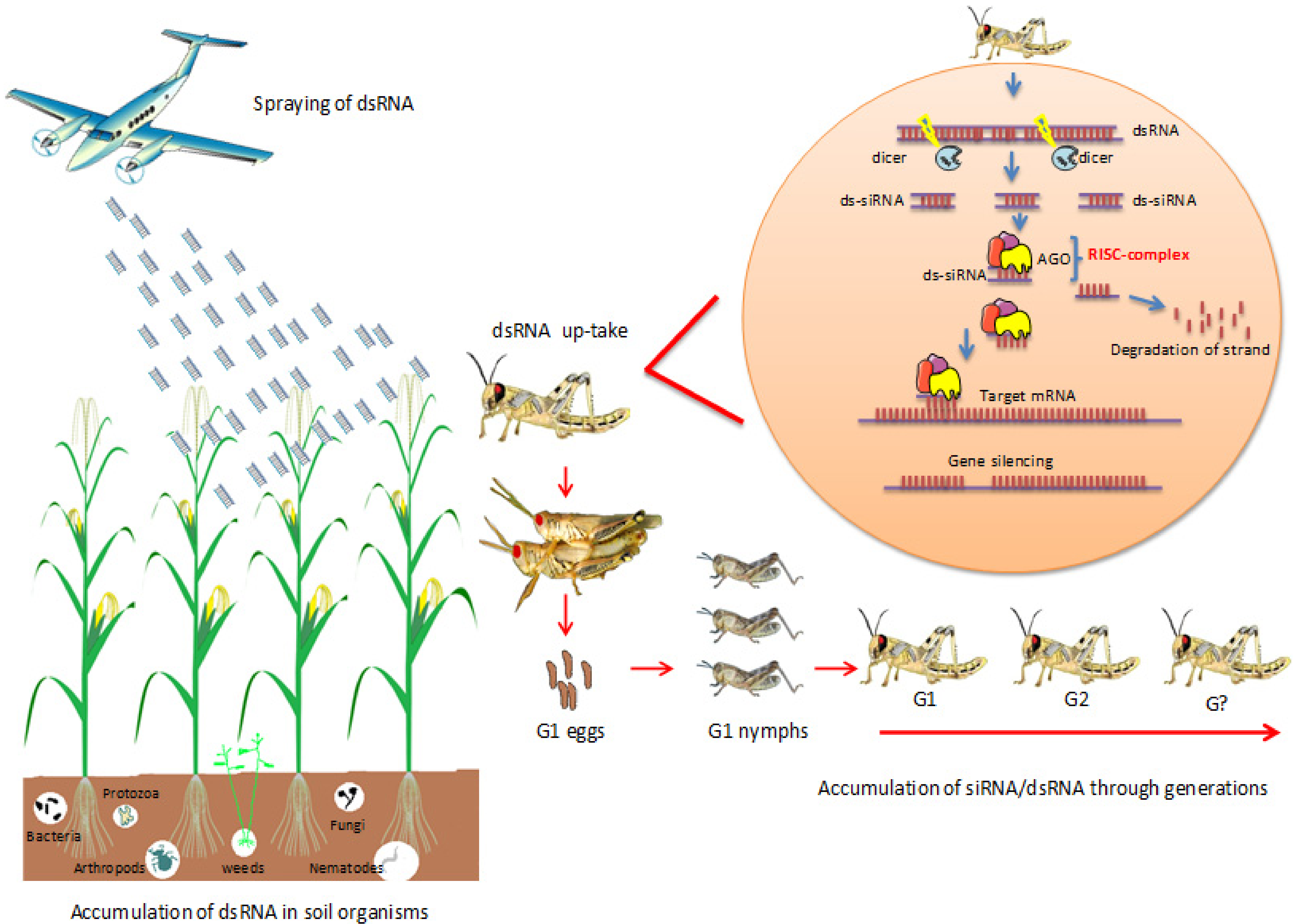
2.1. Fate of DsRNAs in Soil and Soil-Living Organisms
2.2. Long-Lasting Gene Silencing in Insects
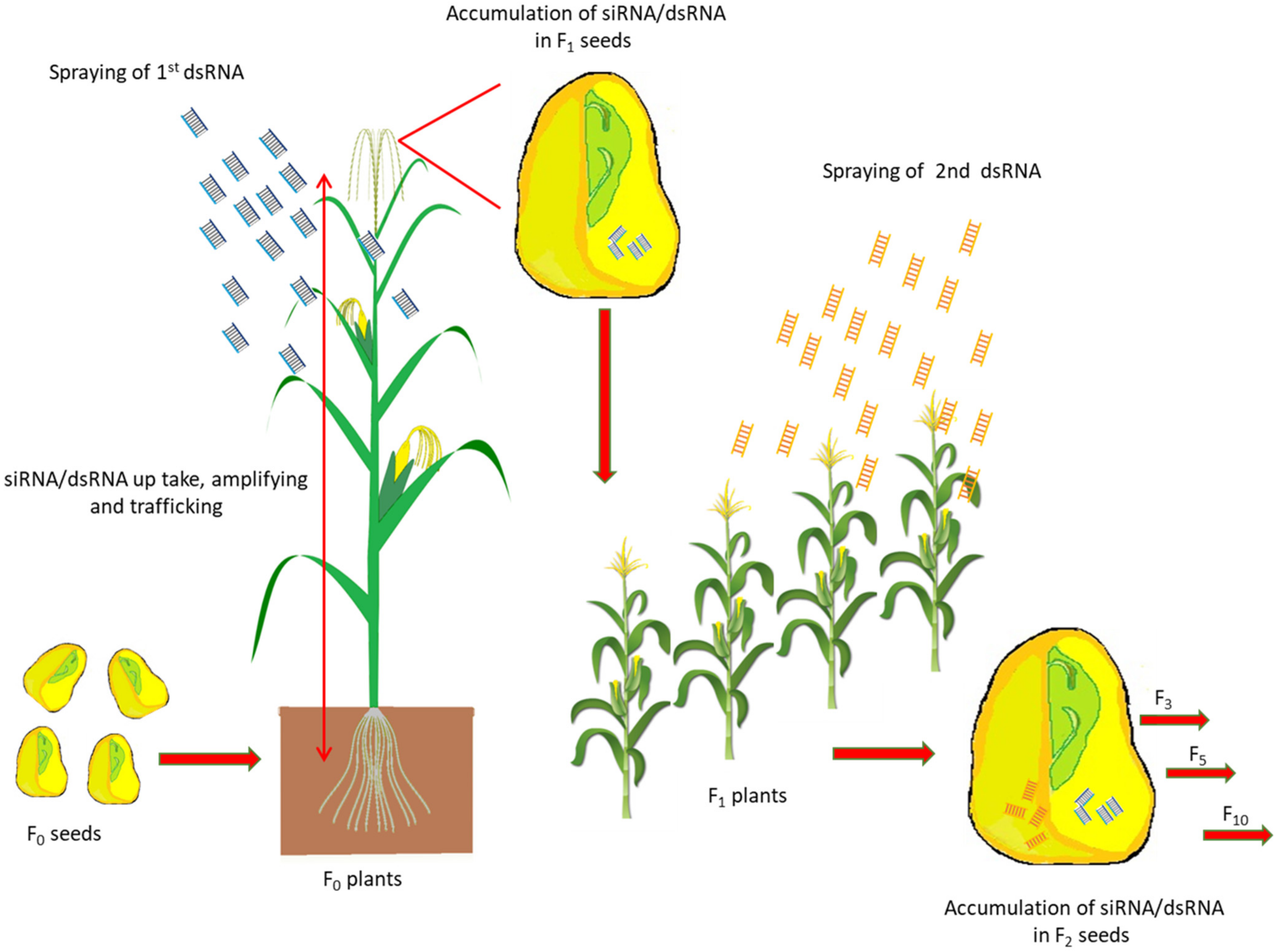
2.3. Accumulation of SiRNA/DsRNA
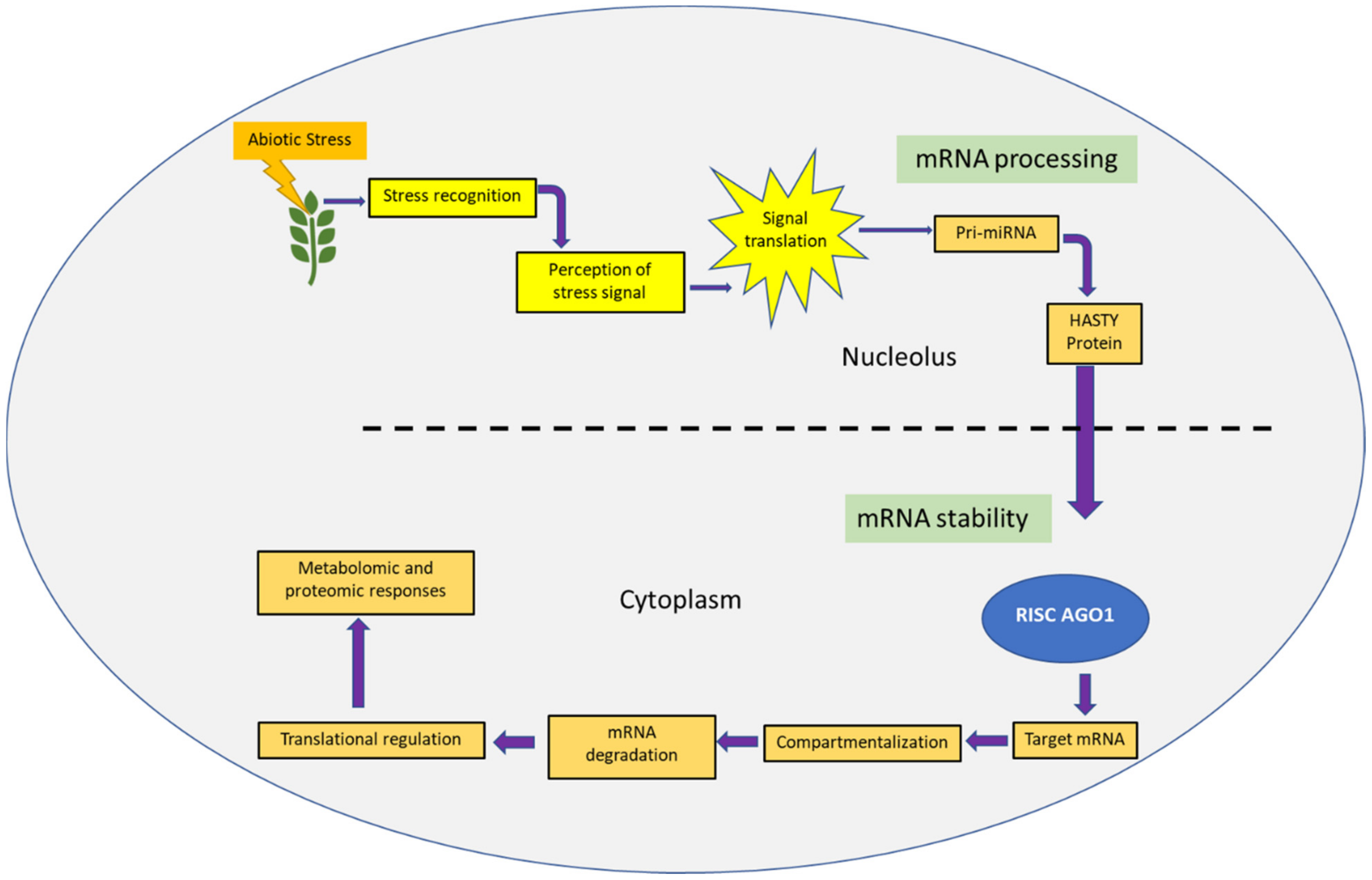
3. Vision Two: Abiotic Stress
3.1. RNAi for Abiotic Stress Tolerance
3.2. Drought Stress Tolerance
| Target | Plant | Reference | |
|---|---|---|---|
| Tobamovirus, potyvirus, and alfamovirus | Tobacco | [80] | |
| Sugarcane mosaic virus SCMV | Maize | [81] | |
| Seed-borne mosaic virus (PSBMV) | Pea | [82] | |
| Cymbidium mosaic virus (CymMV) | Orchid | [83] | |
| Tobacco Mosaic Virus p126 replicase (TMV) | Tobacco | [22] | |
| Zucchini yellow mosaic virus (ZYMV) | Cucurbits | [84] | |
| Papaya ringspot virus CP | Papaya tree | [85] | |
| Fungi Fusarium graminearum | Barley | [26] | |
| Colorado potato beetle | Potato | [20] | |
| Plutella xylostella | Brassica | [86] | |
| Fungi Phakopsora pachyrhizi | Soybean | [87] | |
| Botrytis cinerea | Tomato, strawberry, iceberg lettuce, onion, and rose | [19] | |
| miRNA | Plant | Examples in Abiotic | Reference |
| miR156 | Arabidopsis | Heat | [88] |
| miR156, miR160, and miR164 | Wheat | Drought | [89] |
| miR156, miR159, and miR160 | Wheat | Heat | [89] |
| miR156, miR159 and miR319 | Maize | Drought | [90] |
| miR157 | Maize | Salinity | [91] |
| miR159 | Wheat | Heat | [76] |
| Rice | Heat/Cold | [92] | |
| Durum wheat | Drought | [93] | |
| Arabidopsis | Heat | [57] | |
| Rice | Cold | [57] | |
| miR160 | Rice | Heat | [94] |
| Rice | Drought | [95] | |
| Rice | Salinity/Drought | [96] | |
| Wheat | Salinity | [97,98] | |
| miR164, and miR1029 | Wheat | Cold/Drought | [99] |
| miR166 | Wheat | Drought | [93] |
| miR168, and miR474 | Maize | Drought | [59,100] |
| miR169 | Rice | Drought | [55] |
| Arabidopsis | Drought | [101] | |
| Wheat | Heat/Cold | [102,103] | |
| Wheat | Heat | [104] | |
| miR319 | Rice | Cold | [105] |
| miR393, miR394, and miR164 | Arabidopsis | Cold | [57] |
| miR393, miR166, and miR172 | Wheat | Heat | [57,76] |
| miR396, and miR394 | Rice | Drought/Heat | [92] |
| miR393 | Arabidopsis | Salinity | [31] |
| Rice | Salinity/Drought | [54,60] | |
| miR394 | Wheat | Drought | [106] |
| Rice | Heat | [107] | |
| miR408 | Rice | Drought | [79] |
| Wheat | Heat | [97] | |
| miR529 | Rice | Drought/Salinity | [92] |
| miR444 | wheat | Salinity | [98] |
| Barley | Salinity | [108,109] | |
| miR827 | Maize | Drought/Salinity | [100] |
| miR855 | Wheat | Heat/Cold/Drought | [99] |
| miR5049 | Durum wheat | Drought/Salinity | [93] |
| Wheat | Salinity | [98] | |
| miR5064 | Barley | Drought | [110] |
| Emmer wheat | Drought | [111] | |
| miR1030 | Barley | Drought | [112] |
| Rice | Drought/Heat | [55,64,94] | |
| Barley | Cold/Salinity | [110] | |
| Wheat | Salinity | [112] | |
3.3. Salt Stress Tolerance
3.4. Tolerance to Stress Induced by Heat and Cold
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Guo, J.; Peng, H.; Liu, P.; Kang, Z.; Guo, J. Host-induced gene silencing: A powerful strategy to control diseases of wheat and barley. Int. J. Mol. Sci. 2019, 20, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubrovina, A.S.; Aleynova, O.A.; Alexander, V.K.; Suprun Andrey, R.; Ogneva, Z.V.; Kiselev, K.V. Induction of Transgene Suppression in Plants via External Application of Synthetic dsRNA. Int. J. Mol. Sci. 2019, 20, 1585. [Google Scholar] [CrossRef] [Green Version]
- Dalakouras, A.; Wassenegger, M.; Dadami, E.; Ganopoulos, I.; Pappas, M.L.; Papadopoulou, K. Genetically modified organism-free RNA interference: Exogenous application of RNA molecules in plants. Plant. Physiol. 2020, 182, 38–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abhary, M.; Rezk, A. RNAi Technology: APotential Tool in Plant Breeding. In Advances in Plant Breeding Strategies: Biotechnology and Molecular Tools; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
- Pradhan, B.; Naqvi, A.R.; Saraf, S.; Mukherjee, S.K.; Dey, N. Prediction and characterization of Tomato leaf curl New Delhi virus (ToLCNDV) responsive novel microRNAs in Solanum lycopersicum. Virus Res. 2015, 195, 183–195. [Google Scholar] [CrossRef]
- Homem, R.A.; Davies, T.G.E. An overview of functional genomic tools in deciphering insecticide resistance. Curr. Opin. Insect Sci. 2018, 27, 103–110. [Google Scholar] [CrossRef]
- Belles, X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu. Rev. Entomol. 2010, 5, 111–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, A.; Perrimon, N.A. functional RNAi screen for regulators of receptor tyrosine kinase and ERK signaling. Nature 2006, 444, 230–234. [Google Scholar] [CrossRef]
- Frizzi, A.; Huang, S. Tapping RNA silencing pathways for plant biotechnology. Plant Biotechnol. J. 2010, 8, 655–677. [Google Scholar] [CrossRef] [PubMed]
- Huvenne, H.; Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 2010, 56, 227–235. [Google Scholar] [CrossRef]
- Bhatia, V.; Bhattacharya, R.; Uniyal, P.; Singh, R.; Niranjan, R. Host generated siRNAs attenuate expression of serine protease gene in Myzus persicae. PLoS ONE 2012, 7, e46343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA 2006, 103, 14302–14306. [Google Scholar] [CrossRef] [Green Version]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Sindhu, A.S.; Maier, T.R.; Mitchum, M.G.; Hussey, R.S.; Davis, E.L. Effective and specific in planta RNAi in cyst nematodes: Expression interference of four parasitism genes reduces parasitic success. J. Exp. Botany 2009, 60, 315–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 2010, 22, 3130–3141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, A.; Kumar, N.; Weber, L.; Keller, H.; Imani, J.; Kogel, K.-H. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase–encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. USA 2013, 110, 19324–19329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdellatef, E.; Will, T.; Koch, A.; Imani, J.; Vilcinskas, A.; Kogel, K.H. Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae. Plant Biotechnol. J. 2015, 13, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weiberg, A.; Lin, F.-M.; Thomma, B.P.H.J.; Huang, H.D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef]
- San Miguel, K.; Scott, J.G. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manag. Sci. 2016, 72, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, R.; Rajasekaran, K.; Cary, J.W. RNA Interference (RNAi) as a Potential Tool for Control of Mycotoxin Contamination in Crop Plants: Concepts and Considerations. Front. Plant Sci. 2017, 8, 200. [Google Scholar] [CrossRef] [Green Version]
- Konakalla, N.C.; Kaldis, A.; Berbati, M.; Masarapu, H.; Voloudakis, A.E. Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta 2016, 244, 961–969. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D.A. plant miRNA contributes to antibacterial resistance by repressing auxin signalling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, N.; Datta, K.S.; Datta, K. RNA Interference in Designing Transgenic Crops. Landes Biosci. 2010, 1, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Khraiwesh, B.; Zhu, J.-K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 137–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.S.; Wani, S.H. MicroRNAs as potential targets for abiotic stress tolerance in plants. Front. Plant Sci. 2016, 7, 8173389–8173817. [Google Scholar] [CrossRef]
- Kumar, V.; Khare, T.; Shriram, V.; Wani, S.H. Plant small RNAs: The essential epigenetic regulators of gene expression for salinity stress responses and tolerance. Plant Cell Rep. 2017, 37, 61–75. [Google Scholar] [CrossRef]
- Khare, T.; Shriram, V.; Kumar, V. RNAi Technology: The Role in Development of Abiotic Stress-Tolerant Crops. In Biochemical, Physiological and Molecular Avenues for Combating Abiotic Stress in Plants; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar] [CrossRef]
- Li, J.L.; Chen, X.X.; Shi, C.C.; Liu, F.H.; Sun, J.; Ge, R.C. Effects of OsRPK1 gene overexpression and RNAi on the salt-tolerance at seedling stage in rice. Acta Agron. Sin. 2020, 46, 1217–1224. [Google Scholar] [CrossRef]
- Jagtap, U.B.; Gurav, R.G.; Vishwas, A.B. Role of RNA interference in plant improvement. Nat. Wiss. 2011, 98, 473–492. [Google Scholar] [CrossRef]
- Papadopoulou, N.; Devos, Y.; Álvarez-Alfageme, F.; Lanzoni, A.; Waigmann, E. Risk Assessment Considerations for Genetically Modified RNAi Plants: EFSA’s Activities and Perspective. Front. Plant Sci. 2020, 11, 445. [Google Scholar] [CrossRef] [Green Version]
- Dubelman, S.; Fischer, J.; Zapata, F.; Huizinga, K.; Jiang, C.; Uffman, J.; Levine, S.; Carson, D. Environmental Fate of Double-Stranded RNA in Agricultural Soils. PLoS ONE 2014, 9, e93155. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, E.H.; Hunter, C.P. Transport of dsRNA into cells by the transmembrane protein SID-1. Science 2003, 301, 1545–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winston, W.M.; Sutherlin, M.; Wright, A.J.; Feinberg, E.H.; Hunter, C.P. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA 2007, 104, 10565–10570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whangbo, J.S.; Hunter, C.P. Environmental RNA interference. Trends Genet. 2008, 24, 297–305. [Google Scholar] [CrossRef] [PubMed]
- McEwan, D.L.; Weisman, A.S.; Huntert, C.P. Uptake of extracellular double-Stranded RNA by SID-2. Mol. Cell. 2012, 47, 746–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolognesi, R.; Ramaseshadri, P.; Anderson, J.; Bachman, P.; Clinton, W.; Flannagan, R.; Ilagan, O.; Lawrence, C.; Levine, S.; Moar, W.; et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 2012, 7, e47534. [Google Scholar] [CrossRef]
- Ivashuta, S.I.; Zhang, Y.; Wiggins, B.E.; Ramaseshadri, P.; Segers, G.C.; Johnson, S.; Meyer, S.E.; Kerstetter, R.A.; McNulty, B.C.; Bolognesi, R.; et al. Environmental RNAi in herbivorous insects. RNA 2015, 21, 840–850. [Google Scholar] [CrossRef] [Green Version]
- Coleman, A.D.; Wouters, R.H.M.; Mugford, S.T.; Hogenhout, S.A. Persistence and transgenerational effect of plant-mediated RNAi in aphids. J. Exp. Bot. 2015, 66, 541–548. [Google Scholar] [CrossRef] [Green Version]
- Witwer, K.W.; Hirschi, K.D. Transfer and functional consequences of dietary microRNAs invertebrates: Concepts in search of corroboration. Bioessays 2014, 36, 394–406. [Google Scholar] [CrossRef]
- Vaucheret, H.; Chupeau, Y. Ingested plant miRNAs regulate gene expression in animals. Cell Res. 2012, 22, 3–5. [Google Scholar] [CrossRef] [Green Version]
- Biedenkopf, D.; Will, T.; Knauer, T.; Jelonek, L.; Furch, A.C.U.; Busche, T.; Koch, A. Systemic spreading of exogenous applied RNA biopesticides in the crop plant Hordeum vulgare. ExRNA 2020, 2, 12. [Google Scholar] [CrossRef]
- Šecic, E.; Kogel, K.-H. Requirements for fungal uptake of dsRNA and genesilencing in RNAi-based crop protection strategies. Curr. Opin. Biotechnol. 2021, 70, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.B.; Glick, E.; Paldi, N.; Bextine, B.R. Advances in RNA interference: dsRNA treatment in trees and grapevines for insect pest suppression. Southwest Entomol. 2012, 37, 85–87. [Google Scholar] [CrossRef]
- Yoo, B.C.; Kragler, F.; Varkonyi-Gasic, E.; Haywood, V.; Archer-Evans, S.; Lee, Y.M.; Lough, T.J.; Lucas, W.J. A systemic small RNA signaling system in plants. Plant Cell 2004, 16, 1979–2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, S.A.; Meyers, B.C. Small RNA-mediated epigenetic modifications in plants. Curr. Opin. Plant Biol. 2010, 14, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Henderson, I.R.; Lu, C.; Green, P.J.; Jacobsen, S.E. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA 2007, 104, 4536–4541. [Google Scholar] [CrossRef] [Green Version]
- Wierzbicki, A.T.; Haag, J.R.; Pikaard, C.S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 2008, 135, 635–648. [Google Scholar] [CrossRef] [Green Version]
- Vanderauwera, S.; De Block, M.; Van de Steene, N.; Van De Cotte, B.; Metzlaff, M.; Van Breusegem, F. Silencing of a poly (ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc. Natl. Acad. Sci. USA 2007, 104, 15150–15155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Younis, A.; Siddique, M.I.; Kim, C.; Lim, K. RNA Interference (RNAi) Induced Gene Silencing: A Promising Approach of Hi-Tech Plant Breeding. Int. J. Biol. Sci. 2014, 10, 1150–1158. [Google Scholar] [CrossRef]
- Wang, Y.; Beaith, M.; Chalifoux, M.; Ying, J.; Uchacz, T.; Sarvas, C.; Griffiths, R.; Kuzma, M.; Wan, J.; Huang, Y. Shoot-specific Down-regulation of Protein Farnesyltransferase (Alpha-subunit) for Yield Protection against Drought in Canola. Mol. Plant 2009, 2, 191–200. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Liu, H.; Yang, Y.; Zhen, P.; Liang, J. Down-Regulated Expression of RACK1 Gene by RNA Interference Enhances Drought Tolerance in Rice. Rice Sci. 2009, 16, 14–20. [Google Scholar] [CrossRef]
- Jian, X.; Zhang, L.; Li, G.; Zhang, L.; Wang, X.; Cao, X.; Fang, X.; Zha, F.C. Identification of novel stress-regulated microRNAs from Oryza sativa L. Genomics 2010, 95, 47–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Liu, Y.; Liu, Z.; Kong, D.; Duan, M.; Luo, L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 2010, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.T.; Liang, R.Q.; Ge, L.F.; Li, W.; Xiao, H.S.; Lin, H.X.; Ruan, K.C.; Jin, Y.X. Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 2007, 354, 585–590. [Google Scholar] [CrossRef]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef] [Green Version]
- Kantar, M.; Lucas, S.; Budak, H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 2010, 233, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Zhang, D.; Xiang, F.; Zhang, Z. Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. Int. J. Plant Sci. 2009, 170, 979–989. [Google Scholar] [CrossRef]
- Xia, K.; Wang, O.R.X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by gibberellin-regulated microRNA. Development. 2004, 131, 3357–3365. [Google Scholar] [CrossRef] [Green Version]
- Reyes, J.L.; Chua, N.H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007, 49, 592–606. [Google Scholar] [CrossRef]
- Xiong, H.; Yu, J.; Miao, J.; Li, J.; Zhang, H.; Wang, X.; Liu, P.; Zhao, Y.; Jiang, C.Z.Y.; Li, Y.; et al. Natural Variation in OsLG3 Increases Drought Tolerance in Rice by Inducing ROS Scavenging. Plant Physiol. 2018, 178, 451–467. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Liu, Z.; Liu, Y.; Kong, D.; Li, T.; Yu, S.; Mei, H.; Xu, X.; Liu, H.; Chen, L.; et al. A novel gene OsAHL1 improves both drought avoidance and drought tolerance in rice. Sci. Rep. 2016, 6, 30264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.S.; Yu, J.G.; Lee, G.H.; Park, Y.-D. Drought tolerance induction in transgenic tobacco through RNA interference of BrDST71, a drought-responsive gene from Chinese cabbage. Hortic. Environ. Biotechnol. 2018, 59, 749–757. [Google Scholar] [CrossRef]
- Hu, Y.; Wu, Q.; Peng, Z.; Sprague, S.A.; Wang, W.; Park, J.; Akhunov, E.; Jagadish, K.S.V.; Nakata, P.A.; Cheng, N.; et al. Silencing of OsGRXS17 in rice improves drought stress tolerance by modulating ROS accumulation and stomatal closure. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kantar, M.; Unver, T.; Budak, H. Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct. Integr. Genom. 2010, 10, 493–507. [Google Scholar] [CrossRef] [PubMed]
- Hamza, N.B.; Sharma, N.; Tripathi, A.; Sanan-Mishra, N. MicroRNA expression profiles in response to drought stress in Sorghum bicolor. Gene Expr. Patterns 2016, 20, 88–98. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Q.; Zhang, B. Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 2013, 530, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Jiang, G.; Ye, N.; Chu, Z.; Xu, X.; Zhang, J.; Zhu, G. A Key ABA Catabolic Gene, OsABA8ox3, Is Involved in Drought Stress Resistance in Rice. PLoS ONE 2015, 3, e0116646. [Google Scholar] [CrossRef]
- Zhang, F.; Luo, X.; Zhou, Y.; Xie, J. Genome-wide identification of conserved microRNA and their response to drought stress in Dongxiang wild rice (Oryza rufipogon Griff.). Biotechnol. Lett. 2016, 38, 711–721. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, J.; Wang, Z.; Wen, Y.; Wang, J.; He, W.; Liu, B.; Si, H.; Wang, D. Identification of novel and conserved microRNAs related to drought stress in potato by deep sequencing. PLoS ONE 2014, 9, e95489. [Google Scholar] [CrossRef]
- Giusti, L.; Mica, E.; Bertolini, E.; De Leonardis, A.M.; Faccioli, P.; Cattivelli, L.; Crosatti, C. Micro-RNAs differentially modulated in response to heat and drought stress in durum wheat cultivars with contrasting water use efficiency. Funct. Integr. Genom. 2017, 17, 293–309. [Google Scholar] [CrossRef]
- Ferdous, J.; Sanchez-Ferrero, J.C.; Langridge, P.; Milne, L.; Chowdhury, J.; Brien, C.; Tricker, P.J. Differential expression of microRNAs and potential targets under drought stress in barley. Plant Cell Environ. 2017, 40, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Li, S.P.; Dong, H.X.; Yang, G.; Wu, Y.; Su, S.Z.; Shan, X.H.; Liu, H.K.; Han, J.Y.; Liu, J.B.; Yuan, Y.P. Identification of microRNAs involved in chilling response of maize by high-throughput sequencing. Biol. Plant 2016, 60, 251–260. [Google Scholar] [CrossRef]
- Xin, M.; Wang, Y.; Yao, Y.; Xie, C.; Peng, H.; Ni, Z.; Sun, Q. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 2010, 10, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zou, Z.; Gong, P.; Zhang, J.; Ziaf, K.; Li, H.; Xiao, F.; Ye, Z. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol. Lett. 2011, 33, 403–409. [Google Scholar] [CrossRef]
- Anjali, N.; Nadiya, F.; Thomas, J.; Sabu, K.K. Discovery of MicroRNAs in cardamom (Elettaria cardamomum Maton) under drought stress. Dataset Pap. Sci. 2017, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Sunkar, R. MicroRNAs with macro-effects on plant stress responses. Semin. Cell Dev. Biol. 2010, 8, 805–811. [Google Scholar] [CrossRef]
- Tenllado, F.; Dıaz-Ruız, J.R. Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 2001, 75, 12288–12297. [Google Scholar] [CrossRef] [Green Version]
- Gan, D.; Zhang, J.; Jiang, H.; Jiang, T.; Zhu, S.; Cheng, B. Bacterially expressed dsRNA protects maize against SCMV infection. Plant Cell Rep. 2010, 29, 1261–1268. [Google Scholar] [CrossRef]
- Timmerman, G.M.; Frew, T.J.; Miller, A.L.; Weeden, N.F.; Jermyn, W.A. Effect of artificial dsRNA on infection of pea plants by pea seed-borne mosaic virus. Czech J. Genet. Plant Breeding. 2014, 50, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Lau, S.E.; Mazumdar, P.; Hee, T.W.; Song, A.L.A.; Othman, R.Y.; Harikrishna, J.A. Crude extracts of bacterially-expressed dsRNA protect orchid plants against Cymbidium mosaic virus during transplantation from in vitro culture. J. Hortic. Sci. Biotechnol. 2014, 89, 569–576. [Google Scholar] [CrossRef]
- Kaldis, A.; Berbati, M.; Melita, O.; Reppa, C.; Holeva, M.; Otten, P.; Voloudakis, A. Exogenously applied dsRNA molecules deriving from the Zucchini yellow mosaic virus (ZYMV) genome move systemically protect cucurbits against, Z.Y.M.V. Mol. Plant Pathol. 2018, 19, 883–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, W.; Tuo, D.; Yan, P.; Li, X.; Zhou, P. Detection of papaya leaf distortion mosaic virus by reverse-transcription loop-mediated isothermal amplification. J. Virol. Methods 2014, 195, 174–179. [Google Scholar] [CrossRef]
- Gong, L.; Chen, Y.; Hu, Z.; Hu, M. Testing Insecticidal Activity of Novel Chemically Synthesized siRNA against Plutella xylostella under Laboratory and Field Conditions. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Chen, Z.-Y.; Zhang, C.; Ganiger, M. Reduction of Phakopsora pachyrhizi infection on soybean through host- and spray-induced gene silencing. Mol. Plant Pathol. 2020, 21, 794–807. [Google Scholar] [CrossRef] [Green Version]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.-R.; Bäurle, I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell. 2014, 26, 1792–1807. [Google Scholar] [CrossRef] [Green Version]
- Pandey, R.; Joshi, G.; Bhardwaj, A.R.; Agarwal, M.; Katiyar-Agarwal, S. A Comprehensive Genome-Wide Study on Tissue Specific and Abiotic Stress-Specific miRNAs in Triticum aestivum. PLoS ONE 2014, 9, e95800. [Google Scholar] [CrossRef]
- Li, J.S.; Fu, F.L.; Ming, A.N.; Zhou, S.F.; She, Y.H.; Li, W.C. Differential Expression of MicroRNAs in Response to Drought Stress in Maize. J. Integr. Agric. 2013, 12, 1414–1422. [Google Scholar] [CrossRef]
- Ding, D.; Zhang, L.; Wang, H.; Liu, Z.; Zhang, Z.; Zheng, Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot. 2009, 103, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Barrera-Figueroa, B.; Gao, E.L.; Wu, Z.; Zhou, X.; Zhu, J.; Jin, H.; Liu, R.; Zhu, J. High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol. 2012, 12, 132. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Searle, I.R.; Watson-Haigh, N.S.; Baumann, U.; Mather, D.E.; Able, A.; Able, J.A. Genome-Wide Identification of MicroRNAs in Leaves and the Developing Head of Four Durum Genotypes during Water Deficit Stress. PLoS ONE 2015. [Google Scholar] [CrossRef]
- Li, J.; Wu, L.-Q.; Zheng, W.-Y.; Wang, R.-F.; Yang, L.-X. Genome-wide identification of micro RNAs responsive to high temperature in rice (Oryza sativa) by high-throughput deep sequencing. J. Agron. Crop. Sci. 2015, 201, 379–388. [Google Scholar] [CrossRef]
- Kansal, S.; Devi, R.M.; Balyan, S.C.; Arora, M.K.; Singh, A.K.; Mathur, S.; Raghuvanshi, S. Unique miRNome during anthesis in drought-tolerant indica rice var. Nagina 22. Planta 2015, 241, 1543–1559. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Elling, A.A.; Chen, B.; Deng, X. Differential expression of microRNAs in maize inbred hybrid lines during salt drought stress. Am. J. Plant Sci. 2010, 1, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.R.; Pathak, H.; Sharma, S.K.; Kala, Y.K.; Nirjal, M.K.; Singh, G.P.; Goswami, S.; Rai, R.D. Novel and conserved heat-responsive microRNAs in wheat (Triticum aestivum L.). Funct. Integr. Genom. 2015, 15, 323–348. [Google Scholar] [CrossRef]
- Eren, H.; Pekmezci, M.Y.; Okay, S.; Turktas, M.; Inal, B.; Ilhan, E.; Atak, M.; Erayman, M.; Unver, T. Hexaploid wheat (Triticum aestivum) root miRNome analysis in response to salt stress. Ann. Appl. Biol. 2015. [Google Scholar] [CrossRef]
- Gupta, O.P.; Meena, N.; Sharma, I.; Sharma, P. Differential regulation of microRNAs in response to osmotic, salt and cold stresses in wheat. Mol. Biol. Rep. 2014, 41, 4623–4629. [Google Scholar] [CrossRef]
- Lunardon, A.; Forestan, C.; Farinati, S.; Axtell, M.J.; Varotto, S. Author Notes. Genome-Wide Characterization of Maize Small RNA Loci and Their Regulation in the required to maintain repression6-1 (rmr6-1) Mutant and Long-Term Abiotic Stresses. Plant Physiol. 2016, 170, 1535–1548. [Google Scholar] [CrossRef] [Green Version]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and post-transcriptionally to promote drought resistance. Plant Cell. 2008, 20, 2238–2251. [Google Scholar] [CrossRef] [Green Version]
- Saurin, C.; Declerck, M.; Christ, A.; Blein, T.; Ma, L.; Lelandais-Brière, C.; Njo, M.F.; Beeckman, T.; Crespi, M.; Hartmann, C. A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 2014, 202, 1197–1211. [Google Scholar] [CrossRef] [PubMed]
- Luan, M.; Xu, M.; Lu, Y.; Zhang, L.; Fan, Y.; Wang, L. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene 2015, 555, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Shukla, L.I.; Chinnusamy, V.; Sunkar, R. The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim. Biophys. Acta 2008, 1779, 743–748. [Google Scholar] [CrossRef]
- Yang, C.; Li, D.; Mao, D.; Liu, X.; Ji, C.; Li, X.; Zhao, X.; Cheng, Z.; Chen, C.; Zhu, L. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 2013, 12, 2207–2218. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Xin, Z.; Wang, Z.; Yang, Q.; Guo, S.; Guo, X.; Cao, L.; Lin, T. Identification and comparative analysis of differentially expressed miRNAs in leaves of two wheat (Triticum aestivum L.) genotypes during dehydration stress. BMC Plant Biol. 2015, 15, 21. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Yang, T.; Yu, T.; Zhang, S.; Mao, X.; Zhao, J.; Wang, X.; Dong, J.; Liu, B. Integrating small RNA sequencing with QTL mapping for identification of miRNAs and their target genes associated with heat tolerance at the flowering stage in rice. Front. Plant Sci. 2017, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Wang, L.; Cui, L.; Feng, K.; Liu, F.; Du, X.; Tong, W.; Nie, X.; Ji, W.; Weining, S. Global identification of microRNAsand their targets in barley under salinity stress. PLoS ONE 2015, 10, e0137990. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Wang, H.; Hamera, S.; Chen, X.; Fang, R. MiR444a has multiple functions in therice nitrate-signaling pathway. Plant J. 2014, 78, 44–55. [Google Scholar] [CrossRef]
- Hackenber, M.; Gustafson, P.; Langridge, P.; Shi, B. Differential expression of microRNAs and other small RNAs in barley between water and drought conditions. Plant Biotechnol. J. 2014, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Akpinar, B.A.; Kantar, M.; Budak, H. Root precursors of microRNAs in wild emmer and modern wheats show major differences in response to drought stress. Funct. Integr. Genom. 2015, 5, 587–598. [Google Scholar] [CrossRef]
- Wang, B.; Fei Sun, Y.; Son, N.; Wei, J.; Wang, X.; Feng, H.; Yin, Z.; Kang, Z. MicroRNAs involving in cold, wounding and salt stresses in Triticum aestivum L. Plant Physiol. Biochem. 2014, 80, 90–96. [Google Scholar] [CrossRef]
- Joshi, R.; Singh, B.; Bohra, A.; Chinnusamy, V. Salt stress signalling pathways: Specificity and crosstalk. In Managing Salinity Tolerance in Plants: Molecular and Genomic Perspectives; Wani, S.H., Hossain, M.A., Eds.; CRC Press: Boca Raton, FL, USA, 2016; pp. 51–78. [Google Scholar]
- Kumar, V.; Khare, T. Differential growth and yield responses of salt-tolerant and susceptible rice cultivars to individual (Na+ and Cl_) and additive stress effects of NaCl. Acta Physiol. Plant 2016, 38, 170. [Google Scholar] [CrossRef]
- Wang, X.-J.; Gaaster, T.; Chua, N.-H. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005, 6, R30. [Google Scholar] [CrossRef] [Green Version]
- Marín-Sanz, M.; Giménez, M.J.; Barro, F.; Savin, R. Prolamin Content and Grain Weight in RNAi Silenced Wheat Lines under Different Conditions of Temperature and Nitrogen Availability. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Cao, X.; Wu, Z.; Jiang, F.; Zhou, R.; Yang, Z. Identification of chilling stress-responsive tomato microRNAs and their target genes by high-throughput sequencing and degradome analysis. BMC Genom. 2014, 15, 1130. [Google Scholar] [CrossRef] [Green Version]
- Hivrale, V.; Zheng, Y.; Puli, C.O.R.; Jagadeeswaran, G.; Gowdu, K.; Kakani, V.G.; Barakat, A.; Sunkar, R. Characterization of drought- and heat-responsive microRNAs in switchgrass. Plant Sci. 2016, 242, 214–223. [Google Scholar] [CrossRef]
- Mangrauthia, S.K.; Bhogireddy, S.; Agarwal, S.; Prasanth, V.V.; Voleti, S.R.; Neelamraju, S.; Subrahmanyam, D. Genome-wide changes in microRNA expression during short and prolongedheat stress and recovery in contrasting rice cultivars. J. Exp. Bot. 2017, 68, 2399–2412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Liu, N.; Mao, W.; Hu, Q.; Wang, G.; Gong, Y. Identification of chilling responsive micro RNAs and their targets in vegetable soybean (Glycine max L.). Sci. Rep. 2016, 6, 26619. [Google Scholar] [CrossRef] [Green Version]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Zlata Ogneva, V.; Dubrovina, A.S. Physiological Conditions and dsRNA Application Approaches for Exogenously induced RNA Interference in Arabidopsis thaliana. Plants 2021, 10, 264. [Google Scholar] [CrossRef]
- Das, P.R.; Sherif, S.M. Application of Exogenous dsRNAs-induced RNAi in Agriculture: Challenges and Triumphs. Front. Plant Sci. 2020, 11, 946. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Qin, H.; Zhou, J.; Quan, R.; Lu, X.; Huang, R.; Zhang, H. OsERF2 controls rice root growth and hormone responses through tuning expression of key genes involved in hormone signaling and sucrose metabolism. Plant Mol. Biol. 2016, 90, 293–302. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Li, M.; Gu, D.; Liu, X.; Zhang, J.; Wu, K.; Zhang, X.; da Silva, J.A.T.; Duan, J. Involvement of rice histone deacetylase HDA705 in seed germination and in response to ABA and abiotic stresses. Biochem. Biophys. Res. Commun. 2016, 470, 439–444. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdellatef, E.; Kamal, N.M.; Tsujimoto, H. Tuning Beforehand: A Foresight on RNA Interference (RNAi) and In Vitro-Derived dsRNAs to Enhance Crop Resilience to Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2021, 22, 7687. https://doi.org/10.3390/ijms22147687
Abdellatef E, Kamal NM, Tsujimoto H. Tuning Beforehand: A Foresight on RNA Interference (RNAi) and In Vitro-Derived dsRNAs to Enhance Crop Resilience to Biotic and Abiotic Stresses. International Journal of Molecular Sciences. 2021; 22(14):7687. https://doi.org/10.3390/ijms22147687
Chicago/Turabian StyleAbdellatef, Eltayb, Nasrein Mohamed Kamal, and Hisashi Tsujimoto. 2021. "Tuning Beforehand: A Foresight on RNA Interference (RNAi) and In Vitro-Derived dsRNAs to Enhance Crop Resilience to Biotic and Abiotic Stresses" International Journal of Molecular Sciences 22, no. 14: 7687. https://doi.org/10.3390/ijms22147687
APA StyleAbdellatef, E., Kamal, N. M., & Tsujimoto, H. (2021). Tuning Beforehand: A Foresight on RNA Interference (RNAi) and In Vitro-Derived dsRNAs to Enhance Crop Resilience to Biotic and Abiotic Stresses. International Journal of Molecular Sciences, 22(14), 7687. https://doi.org/10.3390/ijms22147687






