MicroRNAs in Epithelial–Mesenchymal Transition Process of Cancer: Potential Targets for Chemotherapy
Abstract
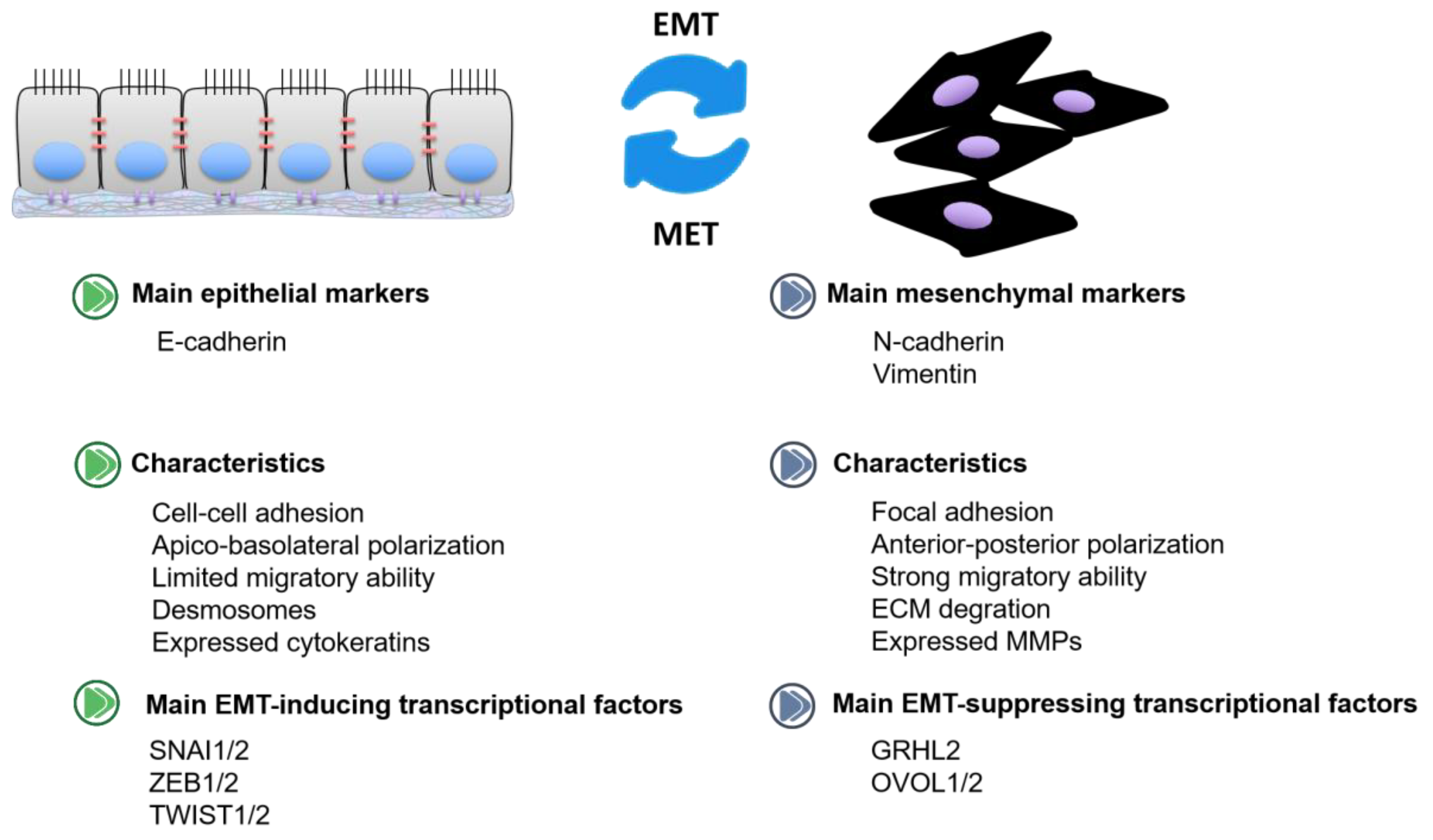
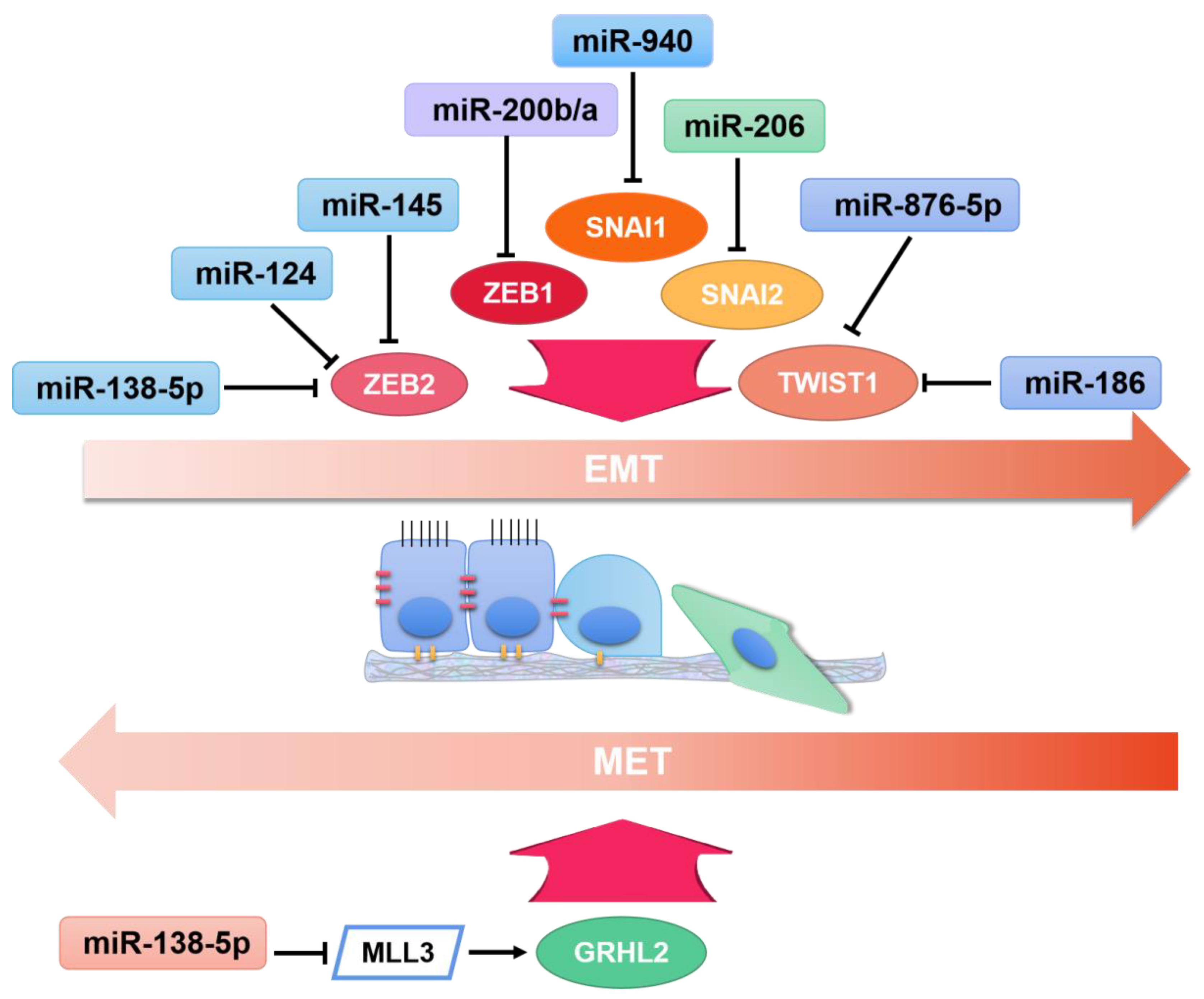
1. Introduction
2. OncomiRs in EMT
2.1. Breast Cancer
2.2. Colorectal Cancer
2.3. Cervical Cancer
2.4. Gastric Cancer
2.5. Nasopharyngeal Carcinoma
2.6. Lung Cancer
2.7. Osteosarcoma
2.8. Other Types of Cancer
3. TsmiRs in EMT
3.1. Breast Cancer
3.2. Colorectal Cancer
3.3. Gastric Cancer
3.4. Glioma
3.5. Lung Cancer
3.6. Pancreatic Cancer
3.7. Papillary Thyroid Cancer
3.8. Other Types of Cancer
4. Compounds Regulated miRNAs as Potential Therapeutics for Inhibiting EMT
4.1. Curcumin
4.2. Propofol
4.3. Shikonin
4.4. Other Drug Candidates
5. Conclusions and Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Demirkan, B. The Roles of Epithelial-to-Mesenchymal Transition (EMT) and Mesenchymal-to-Epithelial Transition (MET) in Breast Cancer Bone Metastasis: Potential Targets for Prevention and Treatment. J. Clin. Med. 2013, 2, 264–282. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Maitra, A. EMT: Matter of Life or Death? Cell 2016, 164, 840–842. [Google Scholar] [CrossRef]
- Biswas, K.H. Molecular Mobility-Mediated Regulation of E-Cadherin Adhesion. Trends Biochem. Sci. 2020, 45, 163–173. [Google Scholar] [CrossRef]
- Nakagawa, M.; Bando, Y.; Nagao, T.; Morimoto, M.; Takai, C.; Ohnishi, T.; Honda, J.; Moriya, T.; Izumi, K.; Takahashi, M.; et al. Expression of p53, Ki-67, E-cadherin, N-cadherin and TOP2A in triple-negative breast cancer. Anticancer Res. 2011, 31, 2389–2393. [Google Scholar]
- Wang, Y.; Shi, J.; Chai, K.; Ying, X.; Zhou, B.P. The Role of Snail in EMT and Tumorigenesis. Curr. Cancer Drug Targets 2013, 13, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.C.; Carneiro, F.; Hoefler, H.; Becker, K.F. Role of the epithelial-mesenchymal transition regulator Slug in primary human cancers. Front. Biosci. 2009, 14, 3041–3050. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.S.; Kang, H.E.; Kim, N.H.; Yook, J.I. Therapeutic implications of cancer epithelial-mesenchymal transition (EMT). Arch. Pharmacal Res. 2019, 42, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Yelle, N.; Venugopal, C.; Singh, S.K. EMT: Mechanisms and therapeutic implications. Pharmacol. Ther. 2018, 182, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulos-Soares, I.; Chartoumpekis, D.V.; Kyriazopoulou, V.; Zaravinos, A. EMT Factors and Metabolic Pathways in Cancer. Front. Oncol. 2020, 10, 499. [Google Scholar] [CrossRef]
- Chung, V.Y.; Tan, T.Z.; Ye, J.R.; Huang, R.L.; Lai, H.C.; Kappei, D.; Wollmann, H.; Guccione, E.; Huang, R.Y.J. The role of GRHL2 and epigenetic remodeling in epithelial-mesenchymal plasticity in ovarian cancer cells. Commun. Biol. 2019, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Mooney, S.M.; Talebian, V.; Jolly, M.K.; Jia, D.; Gromala, M.; Levine, H.; McConkey, B.J. The GRHL2/ZEB Feedback Loop-A Key Axis in the Regulation of EMT in Breast Cancer. J. Cell. Biochem. 2017, 118, 2559–2570. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, J. Ovol Proteins: Guardians against EMT during Epithelial Differentiation. Dev. Cell 2014, 29, 1–2. [Google Scholar] [CrossRef][Green Version]
- Khoshnaw, S.M.; Green, A.; Powe, D.G.; Ellis, I. MicroRNA involvement in the pathogenesis and management of breast cancer. J. Clin. Pathol. 2009, 62, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Grimaldi, A.M.; Salvatore, M.; Incoronato, M. miRNA-Based Therapeutics in Breast Cancer: A Systematic Review. Front. Oncol. 2021, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.; Tran, N. miRNA interplay: Mechanisms and consequences in cancer. Dis. Model. Mech. 2021, 14. [Google Scholar] [CrossRef]
- Berindan-Neagoe, I.; Calin, G.A. Molecular Pathways: MicroRNAs, Cancer Cells, and Microenvironment. Clin. Cancer Res. 2014, 20, 6247–6253. [Google Scholar] [CrossRef]
- D’Amato, N.C.; Howe, E.N.; Richer, J.K. MicroRNA regulation of epithelial plasticity in cancer. Cancer Lett. 2013, 341, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Li, Y.; Wang, J.; Wen, Z.; Lai, M.; Zhang, H. Molecular mechanisms of microRNAs in regulating epithelial–mesenchymal transitions in human cancers. Cancer Lett. 2016, 371, 301–313. [Google Scholar] [CrossRef]
- Peng, F.; Xie, X.; Peng, C. Chinese Herbal Medicine-Based Cancer Therapy: Novel Anticancer Agents Targeting MicroRNAs to Regulate Tumor Growth and Metastasis. Am. J. Chin. Med. 2019, 47, 1711–1735. [Google Scholar] [CrossRef]
- Ahmed, F.; Ijaz, B.; Ahmad, Z.; Farooq, N.; Sarwar, M.B.; Husnain, T. Modification of miRNA Expression through plant extracts and compounds against breast cancer: Mechanism and translational significance. Phytomedicine 2020, 68, 153168. [Google Scholar] [CrossRef] [PubMed]
- Sreekumar, R.; Sayan, B.S.; Mirnezami, A.H.; Sayan, A.E. MicroRNA Control of Invasion and Metastasis Pathways. Front. Genet. 2011, 2, 58. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.B.; Shi, W.W.; Fang, H.Y.; Zhang, X.H. miR-27a promotes human breast cancer cell migration by inducing EMT in a FBXW7-dependent manner. Mol. Med. Rep. 2018, 18, 5417–5426. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X.Y. miR-122-5p promotes aggression and epithelial-mesenchymal transition in triple-negative breast cancer by suppressing charged multivesicular body protein 3 through mitogen-activated protein kinase signaling. J. Cell. Physiol. 2020, 235, 2825–2835. [Google Scholar] [CrossRef]
- Liu, X.Y.; Li, Y.J.; Li, Z.; Hou, T. miR-155 promotes proliferation and epithelial-mesenchymal transition of MCF-7 cells. Exp. Ther. Med. 2021, 21, 7. [Google Scholar]
- Lei, B.; Wang, D.D.; Zhang, M.; Deng, Y.W.; Jiang, H.J.; Li, Y.W. miR-615-3p promotes the epithelial-mesenchymal transition and metastasis of breast cancer by targeting PICK1/TGFBRI axis. J. Exp. Clin. Cancer Res. 2020, 39, 14. [Google Scholar] [CrossRef]
- Hu, Y.; Su, Y.; He, Y.; Liu, W.; Xiao, B. Arginine methyltransferase PRMT3 promote tumorigenesis through regulating c-MYC stabilization in colorectal cancer. Gene 2021, 791, 145718. [Google Scholar] [CrossRef]
- Ma, Z.H.; Shi, P.D.; Wan, B.S. MiR-410-3p activates the NF-kappa B pathway by targeting ZCCHC10 to promote migration, invasion and EMT of colorectal cancer. Cytokine 2021, 140, 10. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.S.; Zhang, H.; Cui, M.M.; Chen, C.S.; Feng, Y. Hsa-miR-425-5p promotes tumor growth and metastasis by activating the CTNND1-mediated beta-catenin pathway and EMT in colorectal cancer. Cell Cycle 2020, 19, 1917–1927. [Google Scholar] [CrossRef]
- Wang, H.; Yan, B.; Zhang, P.; Liu, S.; Li, Q.; Yang, J.; Yang, F.; Chen, E. MiR-496 promotes migration and epithelial-mesenchymal transition by targeting RASSF6 in colorectal cancer. J. Cell. Physiol. 2020, 235, 1469–1479. [Google Scholar] [CrossRef]
- Zhu, J.; Han, S. miR-150-5p promotes the proliferation and epithelial-mesenchymal transition of cervical carcinoma cells via targeting SRCIN1. Pathol. Res. Pr. 2019, 215, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Yang, Y.; Huang, X. miR-199a-5p promotes proliferation and metastasis and epithelial-mesenchymal transition through targeting PIAS3 in cervical carcinoma. J. Cell. Biochem. 2019, 120, 13562–13572. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Jie, Z. MiR-21 Promotes the Invasion and Metastasis of Gastric Cancer Cells by Activating Epithelial-Mesenchymal Transition. Eur. Surg. Res. 2019, 60, 208–218. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, W.; Yue, P.; Li, X. M2 macrophage-derived extracellular vesicles promote gastric cancer progression via a microRNA-130b-3p/MLL3/GRHL2 signaling cascade. J. Exp. Clin. Cancer Res. 2020, 39, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Shimura, T.; Yin, C.; Okugawa, Y.; Kitajima, T.; Koike, Y.; Okita, Y.; Ohi, M.; Uchida, K.; Goel, A.; et al. Antitumor effects of Andrographis via ferroptosis-associated genes in gastric cancer. Oncol. Lett. 2021, 22, 1–8. [Google Scholar] [CrossRef]
- Wu, Z.-H.; Lin, C.; Liu, C.-C.; Jiang, W.-W.; Huang, M.-Z.; Liu, X.; Guo, W.-J. MiR-616-3p promotes angiogenesis and EMT in gastric cancer via the PTEN/AKT/mTOR pathway. Biochem. Biophys. Res. Commun. 2018, 501, 1068–1073. [Google Scholar] [CrossRef]
- Chen, H.; Luo, M.; Wang, X.; Liang, T.; Huang, C.; Huang, C.; Wei, L. Inhibition of PAD4 enhances radiosensitivity and inhibits aggressive phenotypes of nasopharyngeal carcinoma cells. Cell. Mol. Biol. Lett. 2021, 26, 1–12. [Google Scholar] [CrossRef]
- Cheng, Z.Q.; Qiang, H.L.; Wang, B.; Ding, Z.R.; Jiang, C.Y. mir-10b modulates the epithelial-mesenchymal transition, proliferation and migration of nasopharyngeal carcinoma cells. Acta Medica Mediterr. 2020, 36, 941–945. [Google Scholar]
- Zhang, P.; Lu, X.; Shi, Z.; Li, X.; Zhang, Y.; Zhao, S.; Liu, H. miR-205-5p regulates epithelial-mesenchymal transition by targeting PTEN via PI3K/AKT signaling pathway in cisplatin-resistant nasopharyngeal carcinoma cells. Gene 2019, 710, 103–113. [Google Scholar] [CrossRef]
- Dai, L.H.; Chen, F.H.; Zheng, Y.H.; Zhang, D.; Qian, B.; Ji, H.X.; Long, F.; Cretoiu, D. miR-21 regulates growth and EMT in lung cancer cells via PTEN/Akt/GSK3 beta signaling. Front. Biosci. 2019, 24, 1426–1439. [Google Scholar]
- Yang, F.M.; Shao, C.C.; Wei, K.; Jing, X.M.; Qin, Z.Q.; Shi, Y.N.; Shu, Y.Q.; Shen, H. miR-942 promotes tumor migration, invasion, and angiogenesis by regulating EMT via BARX2 in non-small-cell lung cancer. J. Cell. Physiol. 2019, 234, 23596–23607. [Google Scholar] [CrossRef]
- Marina, N.; Gebhardt, M.; Teot, L.; Gorlick, R. Biology and Therapeutic Advances for Pediatric Osteosarcoma. Oncologist 2004, 9, 422–441. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Xu, Y.; Sun, X.; Ma, Y.; Zhang, Y.; Wang, Y.; Guan, H.; Jia, Z.; Li, Y.; Wang, Y. miR-17-5p promotes proliferation and epithelial-mesenchymal transition in human osteosarcoma cells by targeting SRC kinase signaling inhibitor 1. J. Cell. Biochem. 2019, 120, 5495–5504. [Google Scholar] [CrossRef]
- Li, F.; Chen, Q.; Yang, Y.; Li, M.; Zhang, L.; Yan, Z.; Zhang, J.; Wang, K. ESR1 as a recurrence-related gene in intrahepatic cholangiocarcinoma: A weighted gene coexpression network analysis. Cancer Cell Int. 2021, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yang, J.R.; Wang, Y.G.; Tang, Z.Y.; Liu, S.L.; Tang, Y.T. MiR-19b-3p facilitates the proliferation and epithelial-mesenchymal transition, and inhibits the apoptosis of intrahepatic cholangiocarcinoma by suppressing coiled-coil domain containing 6. Arch. Biochem. Biophys. 2020, 686, 11. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Dai, Q.J.; Qi, H.B. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Discov. 2021, 7, 11. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Chen, Y.; Jia, J.; Zhu, X.; He, Y.; Wu, L.M. MiR-27a promotes EMT in ovarian cancer through active Wnt/beta-catenin signalling by targeting FOXO1. Cancer Biomark. 2019, 24, 31–42. [Google Scholar] [CrossRef]
- Coffey, K. Targeting the Hippo Pathway in Prostate Cancer: What’s New? Cancers 2021, 13, 611. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, J.D.; Qi, G.Y.; Wang, S.H.; Gao, J.J. miR-19a promotes the metastasis and EMT through CUL5 in prostate cancer cell line PC3. J. Buon 2020, 25, 2028–2035. [Google Scholar]
- Chou, S.-T.; Ho, B.-Y.; Tai, Y.-T.; Huang, C.-J.; Chao, W.-W. Bidirect effects from cisplatin combine with rosmarinic acid (RA) or hot water extracts of Glechoma hederacea (HWG) on renal cancer cells. Chin. Med. 2020, 15, 1–13. [Google Scholar] [CrossRef]
- Lei, Q.-Q.; Huang, Y.; Li, B.; Han, L.; Lv, C. MiR-155-5p promotes metastasis and epithelial–mesenchymal transition of renal cell carcinoma by targeting apoptosis-inducing factor. Int. J. Biol. Markers 2021, 36, 20–27. [Google Scholar] [CrossRef]
- Shen, J.L.; Lu, Y.Q.; Li, N.; Zhang, Y.; Hu, F.; Dai, H.; Cai, H.F.; Yan, J.Y. miR-34b Inhibits Breast Cancer Cell Epithelial-Mesenchymal Transition (EMT) and Invasion via Targeting Glioma-Associated Oncogene Protein 1 (Gli1). J. Biomater. Tissue Eng. 2020, 10, 1465–1470. [Google Scholar]
- Jia, H.; Sang, M.X.; Liu, F.; Ai, N.; Geng, C.Z. miR-124 regulates EMT based on ZEB2 target to inhibit invasion and metastasis in triple-negative breast cancer. Pathol. Res. Pract. 2019, 215, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Zhou, B.; Xiong, Y.; Cai, H. miR-135 regulated breast cancer proliferation and epithelial-mesenchymal transition acts by the Wnt/β-catenin signaling pathway. Int. J. Mol. Med. 2019, 43, 1623–1634. [Google Scholar] [CrossRef]
- Du, F.; Yu, L.; Wu, Y.; Wang, S.; Yao, J.; Zheng, X.; Xie, S.; Zhang, S.; Lu, X.; Liu, Y.; et al. miR-137 alleviates doxorubicin resistance in breast cancer through inhibition of epithelial-mesenchymal transition by targeting DUSP4. Cell Death Dis. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.J.; Zhang, Y.N.; Xue, P. miR-186 inhibits proliferation, migration, and epithelial-mesenchymal transition in breast cancer cells by targeting Twist1. J. Cell. Biochem. 2019, 120, 10001–10009. [Google Scholar] [CrossRef]
- Min, L.; Liu, C.; Kuang, J.; Wu, X.; Zhu, L. miR-214 inhibits epithelial–mesenchymal transition of breast cancer cells via downregulation of RNF8. Acta Biochim. Biophys. Sin. 2019, 51, 791–798. [Google Scholar] [CrossRef]
- Chi, Y.Y.; Wang, F.; Zhang, T.F.; Xu, H.; Zhang, Y.N.; Shan, Z.Z.; Wu, S.X.; Fan, Q.X.; Sun, Y. miR-516a-3p inhibits breast cancer cell growth and EMT by blocking the Pygo2/Wnt signalling pathway. J. Cell Mol. Med. 2019, 23, 6295–6307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Hu, Y.; Luo, N.; Li, X.; Chen, F.; Yuan, J.; Guo, L. miR-574-5p attenuates proliferation, migration and EMT in triple-negative breast cancer cells by targeting BCL11A and SOX2 to inhibit the SKIL/TAZ/CTGF axis. Int. J. Oncol. 2020, 56, 1240–1251. [Google Scholar] [CrossRef]
- Liu, G.; Wang, P.; Zhang, H. MiR-6838-5p suppresses cell metastasis and the EMT process in triple-negative breast cancer by targeting WNT3A to inhibit the Wnt pathway. J. Gene Med. 2019, 21, e3129. [Google Scholar] [CrossRef]
- Du, G.; Yu, X.; Chen, Y.; Cai, W. MiR-1-3p Suppresses Colorectal Cancer Cell Proliferation and Metastasis by Inhibiting YWHAZ-Mediated Epithelial–Mesenchymal Transition. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Wang, W.-X.; Yu, H.-L.; Liu, X. MiR-9-5p suppresses cell metastasis and epithelial-mesenchymal transition through targeting FOXP2 and predicts prognosis of colorectal carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6467–6477. [Google Scholar]
- Fan, Y.; Wang, K. miR-205 suppresses cell migration, invasion and EMT of colon cancer by targeting mouse double minute 4. Mol. Med. Rep. 2020, 22, 633–642. [Google Scholar] [CrossRef]
- Chen, Q.; Zhou, L.; Ye, X.; Tao, M.; Wu, J. miR-145-5p suppresses proliferation, metastasis and EMT of colorectal cancer by targeting CDCA3. Pathol. Res. Pr. 2020, 216, 152872. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Naghizadeh, S.; Gjerstorff, M.; Shanehbandi, D.; Shirjang, S.; Najafi, S.; Holmskov, U.; Khaze, V.; Duijf, P.H.; et al. miR-330 suppresses EMT and induces apoptosis by downregulating HMGA2 in human colorectal cancer. J. Cell. Physiol. 2020, 235, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.J.; Wang, D.X.; Qu, S.X.; Zhao, H.; Lin, Y. miR-370-3p Alleviates Ulcerative Colitis-Related Colorectal Cancer in Mice Through Inhibiting the Inflammatory Response and Epitheliale-Mesenchymal Transition. Drug Des. Dev. Ther. 2020, 14, 1127–1141. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Q.; Jiang, F.Q.; Ma, F.; Zhang, B. MiR-873-5p suppresses cell proliferation and epithelial-mesenchymal transition via directly targeting Jumonji domain-containing protein 8 through the NF-kappa B pathway in colorectal cancer. J. Cell Commun. Signal. 2019, 13, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Sun, G.; Zhang, D.; Li, Q.; Qian, H. MiR-3622a-3p acts as a tumor suppressor in colorectal cancer by reducing stemness features and EMT through targeting spalt-like transcription factor 4. Cell Death Dis. 2020, 11, 1–19. [Google Scholar] [CrossRef]
- Shang, J.-C.; Yu, G.-Z.; Ji, Z.-W.; Wang, X.-Q.; Xia, L. MiR-105 inhibits gastric cancer cells metastasis, epithelial-mesenchymal transition by targeting SOX9. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6160–6169. [Google Scholar]
- Wang, S.; Chen, Y.; Yu, X.; Lu, Y.; Wang, H.; Wu, F.; Teng, L. miR-129-5p attenuates cell proliferation and epithelial mesenchymal transition via HMGB1 in gastric cancer. Pathol. Res. Pract. 2019, 215, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Zhang, B.; Cui, J.Z.; Liang, Z.; Liu, K.X. miR-203 Inhibits the Invasion and EMT of Gastric Cancer Cells by Directly Targeting Annexin A4. Oncol. Res. 2019, 27, 789–799. [Google Scholar] [CrossRef]
- Xu, J.P.; You, Q.; Wei, Z.R.; Fu, H.P.; Zhang, Y.; Hu, Z.Q.; Cai, Q.P. miR-519 inhibits epithelial-mesenchymal transition and biologic behavior of gastric cancer cells by down-regulating FOXQ1. Int. J. Clin. Exp. Pathol. 2020, 13, 425–436. [Google Scholar] [PubMed]
- Li, D.; Tian, B.; Jin, X.S. miR-630 Inhibits Epithelial-to-Mesenchymal Transition (EMT) by Regulating the Wnt/beta-Catenin Pathway in Gastric Cancer Cells. Oncol. Res. 2019, 27, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.Z.; Zhang, C.D.; Zhang, C.; Pei, J.P.; Dai, D.Q. miR-665 Suppresses the Epithelial-Mesenchymal Transition and Progression of Gastric Cancer by Targeting CRIM1. Cancer Manag. Res. 2020, 12, 3489–3501. [Google Scholar] [CrossRef]
- Ostrom, Q.; Gittleman, H.; Liao, P.; Rouse, C.; Chen, Y.; Dowling, J.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007–2011. Neuro-Oncology 2014, 16, iv1–iv63. [Google Scholar] [CrossRef]
- Feng, S.Y.; Yao, J.; Zhang, Z.B.; Zhang, Y.Y.; Zhang, Z.Y.; Liu, J.L.; Tan, W.L.; Sun, C.H.; Chen, L.; Yu, X.G. miR-96 inhibits EMT by targeting AEG-1 in glioblastoma cancer cells. Mol. Med. Rep. 2018, 17, 2964–2972. [Google Scholar] [CrossRef]
- Dai, B.; Zhou, G.; Hu, Z.; Zhu, G.; Mao, B.; Su, H.; Jia, Q. MiR-205 suppresses epithelial–mesenchymal transition and inhibits tumor growth of human glioma through down-regulation of HOXD9. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Li, Z.; Qian, R.; Zhang, J.; Shi, X. MiR-218-5p targets LHFPL3 to regulate proliferation, migration, and epithelial–mesenchymal transitions of human glioma cells. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Huang, W.; Shi, Y.; Han, B.; Wang, Q.L.; Zhang, B.; Qi, C.J.; Liu, F. miR-802 inhibits the proliferation, invasion, and epithelial-mesenchymal transition of glioblastoma multiforme cells by directly targeting SIX4. Cell Biochem. Funct. 2020, 38, 66–76. [Google Scholar] [CrossRef]
- Ma, B.Y.; Xu, J.; Chen, G.; Wei, D.; Gu, P.Y.; Li, L.X.; Hu, W.X. miR-876-5p exerts tumor suppressor function by targeting TWIST1 and regulating the epithelial-mesenchymal transition in glioblastoma. Int. J. Clin. Exp. Med. 2020, 13, 1454–1463. [Google Scholar]
- Zhu, D.; Gu, L.; Li, Z.; Jin, W.; Lu, Q.; Ren, T. MiR-138-5p suppresses lung adenocarcinoma cell epithelial-mesenchymal transition, proliferation and metastasis by targeting ZEB2. Pathol. Res. Pr. 2019, 215, 861–872. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, J.; Wang, B.; Zheng, Y.; Wan, Y.; Wang, Y.; Zhou, L.; Liu, S.; Li, G.; Yan, Y. miR-145 modulates epithelial-mesenchymal transition and invasion by targeting ZEB2 in non–small cell lung cancer cell lines. J. Cell. Biochem. 2019, 120, 8409–8418. [Google Scholar] [CrossRef]
- Han, Q.; Cheng, P.; Yang, H.J.; Liang, H.P.; Lin, F.C. miR-146b Reverses epithelial-mesenchymal transition via targeting PTP1B in cisplatin-resistance human lung adenocarcinoma cells. J. Cell. Biochem. 2020, 121, 3901–3912. [Google Scholar] [CrossRef]
- Du, W.W.; Tang, H.C.; Lei, Z.; Zhu, J.J.; Zeng, Y.Y.; Liu, Z.Y.; Huang, J.A. miR-335-5p inhibits TGF-beta 1-induced epithelial-mesenchymal transition in non-small cell lung cancer via ROCK1. Respir. Res. 2019, 20, 11. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Gao, F.; Chu, H.; Lou, L.; Wang, H.; Chen, Y. miR-363-3p inhibits migration, invasion, and epithelial-mesenchymal transition by targeting NEDD9 and SOX4 in non-small-cell lung cancer. J. Cell. Physiol. 2020, 235, 1808–1820. [Google Scholar] [CrossRef]
- Liu, B.; Wang, Z.; Cheng, S.; Du, L.; Yin, Y.; Yang, Z.; Zhou, J. miR-379 inhibits cell proliferation and epithelial-mesenchymal transition by targeting CHUK through the NF-κB pathway in non-small cell lung cancer. Mol. Med. Rep. 2019, 20, 1418–1428. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Wu, Y.Y.; Yang, S.H.; Liu, X.C.; Lu, Y.; Liu, F.X.; Li, G.X.; Tian, G.R. miR-874 directly targets AQP3 to inhibit cell proliferation, mobility and EMT in non-small cell lung cancer. Thorac. Cancer 2020, 11, 1550–1558. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.Q.; Zhao, T.; Shen, M.J.; Zhang, F.Q.; Duan, S.Z.; Lei, Z.; Chen, Y.B. MiR-940 inhibits TGF-beta-induced epithelial-mesenchymal transition and cell invasion by targeting Snail in non-small cell lung cancer (vol 10, pg 2735, 2019). J. Cancer 2020, 11, 4897–4898. [Google Scholar] [CrossRef]
- An, N.; Zheng, B. MiR-203a-3p Inhibits Pancreatic Cancer Cell Proliferation, EMT, and Apoptosis by Regulating SLUG. Technol. Cancer Res. Treat. 2020, 19. [Google Scholar] [CrossRef]
- Sun, J.; Chen, L.; Dong, M. MiR-338-5p Inhibits EGF-Induced EMT in Pancreatic Cancer Cells by Targeting EGFR/ERK Signaling. Front. Oncol. 2021, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ji, W.; Li, T.; Liu, T.; Zhao, X. MiR-145 functions as a tumor suppressor in Papillary Thyroid Cancer by inhibiting RAB5C. Int. J. Med. Sci. 2020, 17, 1992–2001. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhao, Y. miR-451a inhibits cancer growth, epithelial-mesenchymal transition and induces apoptosis in papillary thyroid cancer by targeting PSMB8. J. Cell. Mol. Med. 2019, 23, 8067–8075. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.-M.; He, X.-Y.; Yang, Y.-L.; Jia, W.-J.; Yang, Z.-Q.; Yan, D.; Ma, J.-X. MiR-630 inhibits papillary thyroid carcinoma cell growth, metastasis, and epithelial-mesenchymal transition by suppressing JAK2/STAT3 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2453–2460. [Google Scholar] [PubMed]
- Gu, X.B.; Dong, M.L.; Liu, Z.Y.; Yang, J.; Shi, Y.G. MiR-499a-5p Inhibits Proliferation, Invasion, Migration, and Epithelial-Mesenchymal Transition, and Enhances Radiosensitivity of Cervical Cancer Cells via Targeting eIF4E. Oncotarget Ther. 2020, 13, 2913–2924. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.J.; Li, X.R.; Zuo, Y.Z.; Xu, Q.; Liu, L.; Wu, H.Y.; Chen, L.; Zhang, Y.; Liu, Y.; Li, Y.H. miR-373-3p inhibits epithelial-mesenchymal transition via regulation of TGF beta R2 in choriocarcinoma. J. Obstet. Gynaecol. Res. 2021, 47, 2417–2432. [Google Scholar] [CrossRef]
- Pan, H.; Hong, Y.; Yu, B.; Li, L.; Zhang, X. miR-4429 Inhibits Tumor Progression and Epithelial-Mesenchymal Transition Via Targeting CDK6 in Clear Cell Renal Cell Carcinoma. Cancer Biotherapy Radiopharm. 2019, 34, 334–341. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, Y.; Lu, Q.; Fei, X.; Lu, C.; Li, C.; Chen, H. MiR-34a-5p Inhibits Proliferation, Migration, Invasion and Epithelial-mesenchymal Transition in Esophageal Squamous Cell Carcinoma by Targeting LEF1 and Inactivation of the Hippo-YAP1/TAZ Signaling Pathway. J. Cancer 2020, 11, 3072–3081. [Google Scholar] [CrossRef]
- Wu, H.; Liu, J.; Zhang, Y.; Li, Q.; Wang, Q.; Gu, Z. miR-22 suppresses cell viability and EMT of ovarian cancer cells via NLRP3 and inhibits PI3K/AKT signaling pathway. Clin. Transl. Oncol. 2021, 23, 257–264. [Google Scholar] [CrossRef]
- Chung, V.Y.; Tan, T.Z.; Tan, M.; Wong, M.K.; Kuay, K.T.; Yang, Z.; Ye, J.R.; Muller, J.; Koh, C.M.; Guccione, E.; et al. GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci. Rep. 2016, 6, 15. [Google Scholar]
- Abdeyrim, A.; Cheng, X.; Lian, M.; Tan, Y. miR-490-5p regulates the proliferation, migration, invasion and epithelial-mesenchymal transition of pharyngolaryngeal cancer cells by targeting mitogen-activated protein kinase kinasekinase 9. Int. J. Mol. Med. 2019, 44, 240–252. [Google Scholar] [CrossRef]
- Ma, T.; Zhao, Z.G.; Wang, Z.M.; Wang, C.N.; Zhang, L.P. MiR-940 inhibits migration and invasion of tongue squamous cell carcinoma via regulating CXCR2/NF-kappa B system-mediated epithelial-mesenchymal transition. Naunyn-Schmiedebergs Arch. Pharmacol. 2019, 392, 1359–1369. [Google Scholar] [CrossRef]
- Peng, F.; Xiong, L.; Xie, X.; Tang, H.; Huang, R.; Peng, C. Isoliquiritigenin Derivative Regulates miR-374a/BAX Axis to Suppress Triple-Negative Breast Cancer Tumorigenesis and Development. Front. Pharmacol. 2020, 11, 378. [Google Scholar] [CrossRef]
- Coker-Gurkan, A.; Bulut, D.; Genc, R.; Arisan, E.D.; Obakan-Yerlikaya, P.; Palavan-Unsal, N. Curcumin prevented human autocrine growth hormone (GH) signaling mediated NF-κB activation and miR-183-96-182 cluster stimulated epithelial mesenchymal transition in T47D breast cancer cells. Mol. Biol. Rep. 2019, 46, 355–369. [Google Scholar] [CrossRef]
- Wang, H.; Cai, X.; Ma, L. Curcumin Modifies Epithelial–Mesenchymal Transition in Colorectal Cancer Through Regulation of miR-200c/EPM5. Cancer Manag. Res. 2020, 12, 9405–9415. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Zhang, C.; Xie, D.; Pei, M. Curcumin inhibits epithelial-mesenchymal transition in colorectal cancer cells by regulating miR-206/SNAI2 pathway. Trop. J. Pharm. Res. 2021, 18, 1405–1412. [Google Scholar] [CrossRef]
- Li, Y.; Zeng, Q.-G.; Qiu, J.-L.; Pang, T.; Wang, H.; Zhang, X.-X. Propofol suppresses migration, invasion, and epithelial-mesenchymal transition in papillary thyroid carcinoma cells by regulating miR-122 expression. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 5101–5110. [Google Scholar]
- Gao, M.; Guo, R.; Lu, X.H.; Xu, G.; Luo, S.X. Propofol suppresses hypoxia-induced esophageal cancer cell migration, invasion, andEMTthrough regulatinglncRNA TMPO-AS1/miR-498 axis. Thorac. Cancer 2020, 11, 2398–2405. [Google Scholar] [CrossRef] [PubMed]
- Bao, C.; Liu, T.; Qian, L.; Xiao, C.; Zhou, X.; Ai, H.; Wang, J.; Fan, W.; Pan, J. Shikonin inhibits migration and invasion of triple-negative breast cancer cells by suppressing epithelial-mesenchymal transition via miR-17-5p/PTEN/Akt pathway. J. Cancer 2021, 12, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Zeng, X.P. Shikonin suppresses progression and epithelial-mesenchymal transition in hepatocellular carcinoma (HCC) cells by modulating miR-106b/SMAD7/TGF-beta signaling pathway. Cell Biol. Int. 2020, 44, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhou, H.; Liu, J.; Xie, J.; Hu, R. Apigenin suppresses proliferation, invasion, and epithelial–mesenchymal transition of cervical carcinoma cells by regulation of miR -152/BRD4 axis. Kaohsiung J. Med. Sci. 2021, 37, 583–593. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Q.H.; Shang, J.J.; Lu, L.H.; Chen, G.Y. Crocin inhibits the migration, invasion, and epithelial-mesenchymal transition of gastric cancer cells via miR-320/KLF5/HIF-1 alpha signaling. J. Cell. Physiol. 2019, 234, 17876–17885. [Google Scholar] [CrossRef]
- Cheng, Z.; Xing, D. Ginsenoside Rg3 inhibits growth and epithelial-mesenchymal transition of human oral squamous carcinoma cells by down-regulating miR-221. Eur. J. Pharmacol. 2019, 853, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Tang, H.L.; Du, J.R.; Chen, J.P.; Peng, C. Isoliquiritigenin Suppresses EMT-Induced Metastasis in Triple-Negative Breast Cancer through miR-200c/C-JUN/beta-Catenin. Am. J. Chin. Med. 2021, 49, 505–523. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zhang, B.; Gu, G.; Yang, X.; Qian, Z. Metformin Increases the Chemosensitivity of Pancreatic Cancer Cells to Gemcitabine by Reversing EMT Through Regulation DNA Methylation of miR-663. OncoTargets Ther. 2020, 13, 10417–10429. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, K.; Xiao, J.; Song, Y.; Li, H. Neferine sensitized Taxol-resistant nasopharygeal carcinoma to Taxol by inhibiting EMT via downregulating miR-130b-5p. Biochem. Biophys. Res. Commun. 2020, 531, 573–580. [Google Scholar] [CrossRef]
- Zhou, Y.; Xue, R.; Wang, J.; Ren, H. Puerarin inhibits hepatocellular carcinoma invasion and metastasis through miR-21-mediated PTEN/AKT signaling to suppress the epithelial-mesenchymal transition. Braz. J. Med. Biol. Res. 2020, 53, e8882. [Google Scholar] [CrossRef]
- Wu, T.; Sun, L.; Wang, C.; Yu, P.; Cheng, L.; Chen, Y. Sevoflurane Suppresses the Migration, Invasion, and Epithelial-Mesenchymal Transition of Breast Cancer Cells Through the miR-139-5p/ARF6 Axis. J. Surg. Res. 2021, 258, 314–323. [Google Scholar] [CrossRef]
- Bakir, B.; Chiarella, A.M.; Pitarresi, J.R.; Rustgi, A.K. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol. 2020, 30, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Li, S.Y.; Zhu, S.L.; Yi, M.; Luo, S.X.; Wu, K.M. MiRNA-mediated EMT and CSCs in cancer chemoresistance. Exp. Hematol. Oncol. 2021, 10, 12. [Google Scholar] [CrossRef]
- De las Rivas, J.; Brozovic, A.; Izraely, S.; Casas-Pais, A.; Witz, I.P.; Figueroa, A. Cancer drug resistance induced by EMT: Novel therapeutic strategies. Arch. Toxicol. 2021, 95, 2279–2297. [Google Scholar] [CrossRef]
- Pan, G.; Liu, Y.; Shang, L.; Zhou, F.; Yang, S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun. 2021, 41, 199–217. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Wu, C.; Sun, W.; Liu, D.; Luo, M.; Su, B.; Zhang, L.; Mei, Q.; Hu, G. Snail-mediated cancer stem cell-like phenotype in human CNE2 nasopharyngeal carcinoma cell. Head Neck 2018, 40, 485–497. [Google Scholar] [CrossRef]
- Vesuna, F.; Lisok, A.; Kimble, B.; Raman, V. Twist Modulates Breast Cancer Stem Cells by Transcriptional Regulation of CD24 Expression. Neoplasia 2009, 11, 1318–1328. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Tillo, E.; Siles, L.; De Barrios, O.; Cuatrecasas, M.; Vaquero, E.C.; Castells, A.; Postigo, A. Expanding roles of ZEB factors in tumorigenesis and tumor progression. Am. J. Cancer Res. 2011, 1, 897–912. [Google Scholar] [PubMed]
- Roy, S.; Sunkara, R.R.; Parmar, M.Y.; Shaikh, S.; Waghmare, S.K. EMT imparts cancer stemness and plasticity: New perspectives and therapeutic potential. Front. Biosci. 2021, 26, 238–265. [Google Scholar] [CrossRef]
- Teeuwssen, M.; Fodde, R. Wnt Signaling in Ovarian Cancer Stemness, EMT, and Therapy Resistance. J. Clin. Med. 2019, 8, 1658. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, I.; Miele, L. Deadly crosstalk: Notch signaling at the intersection of EMT and cancer stem cells. Cancer Lett. 2013, 341, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Guen, V.J.; Chavarria, T.E.; Kröger, C.; Ye, X.; Weinberg, R.A.; Lees, J.A. EMT programs promote basal mammary stem cell and tumor-initiating cell stemness by inducing primary ciliogenesis and Hedgehog signaling. Proc. Natl. Acad. Sci. USA 2017, 114, E10532–E10539. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, F.; Wang, X.; Ouyang, G. The roles of microRNAs in the regulation of tumor metastasis. Cell Biosci. 2015, 5, 32. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Xiong, L.; Peng, C. (-)-Sativan Inhibits Tumor Development and Regulates miR-200c/PD-L1 in Triple Negative Breast Cancer Cells. Front. Pharmacol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Peng, F.; Peng, C.; Du, J.-R. Gut Microbiota in Tumor Microenvironment: A Critical Regulator in Cancer Initiation and Development as Potential Targets for Chinese Medicine. Am. J. Chin. Med. 2021, 49, 609–626. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, F.; Fan, H.; Li, S.; Peng, C.; Pan, X. MicroRNAs in Epithelial–Mesenchymal Transition Process of Cancer: Potential Targets for Chemotherapy. Int. J. Mol. Sci. 2021, 22, 7526. https://doi.org/10.3390/ijms22147526
Peng F, Fan H, Li S, Peng C, Pan X. MicroRNAs in Epithelial–Mesenchymal Transition Process of Cancer: Potential Targets for Chemotherapy. International Journal of Molecular Sciences. 2021; 22(14):7526. https://doi.org/10.3390/ijms22147526
Chicago/Turabian StylePeng, Fu, Huali Fan, Sui Li, Cheng Peng, and Xiaoqi Pan. 2021. "MicroRNAs in Epithelial–Mesenchymal Transition Process of Cancer: Potential Targets for Chemotherapy" International Journal of Molecular Sciences 22, no. 14: 7526. https://doi.org/10.3390/ijms22147526
APA StylePeng, F., Fan, H., Li, S., Peng, C., & Pan, X. (2021). MicroRNAs in Epithelial–Mesenchymal Transition Process of Cancer: Potential Targets for Chemotherapy. International Journal of Molecular Sciences, 22(14), 7526. https://doi.org/10.3390/ijms22147526





