Treat Me Well or Will Resist: Uptake of Mobile Genetic Elements Determine the Resistome of Corynebacterium striatum
Abstract
1. Introduction
2. Genome Organization of C. striatum
3. C. striatum Resistome
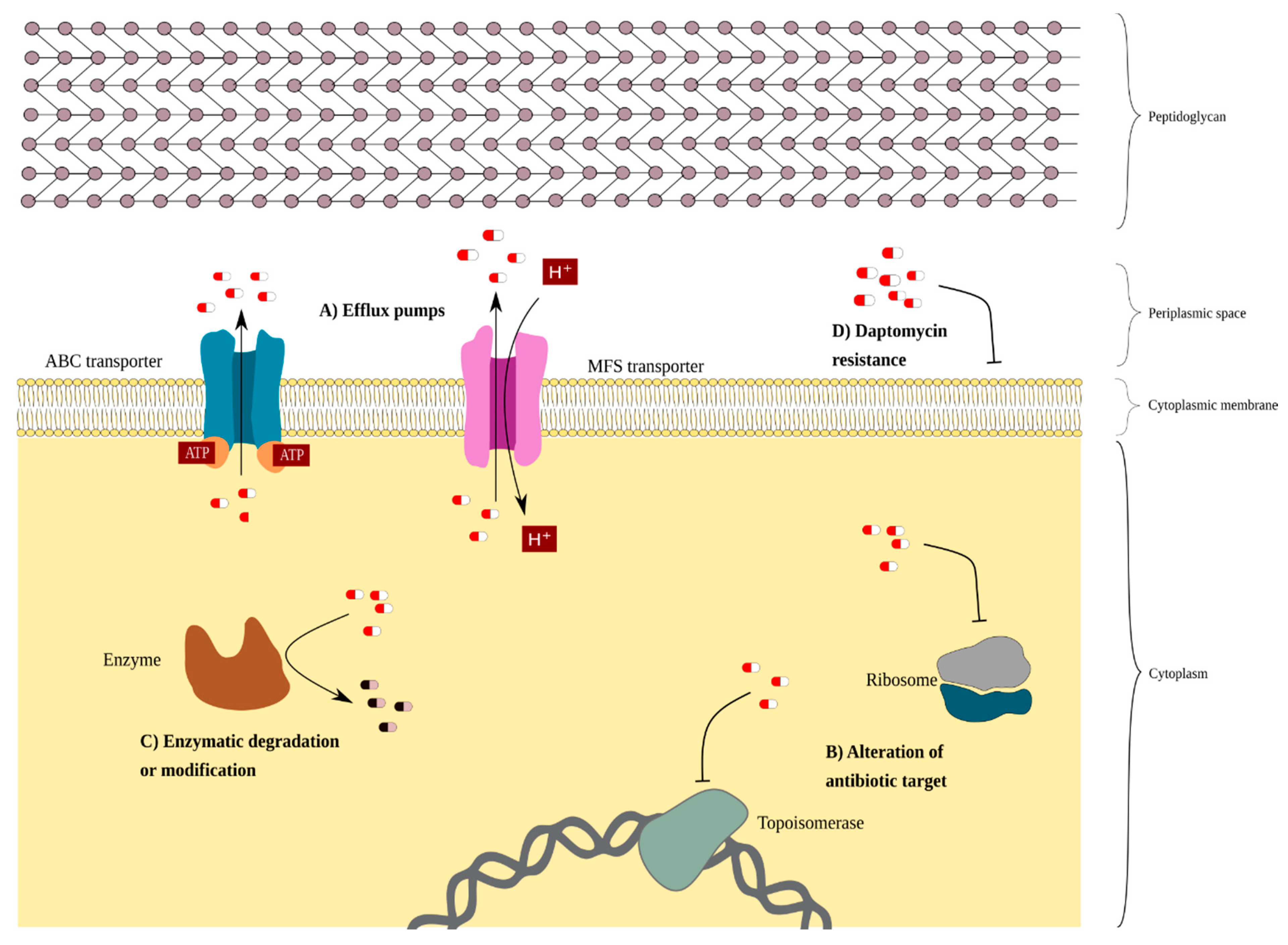
3.1. Intrinsic Resistance in C. striatum
3.2. Extrinsic Resistance Associated with Mobile Elements
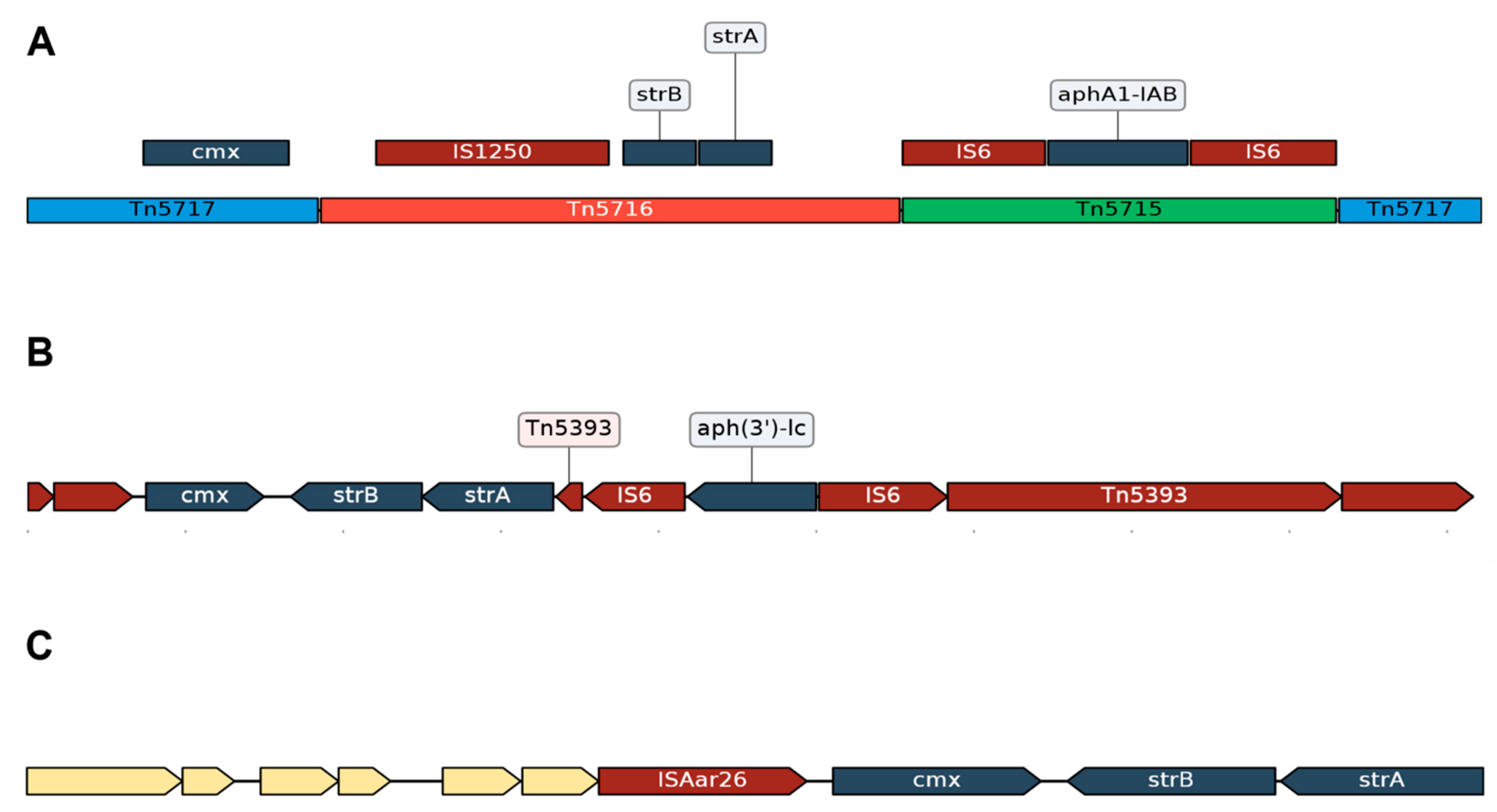
3.2.1. Resistance Due to Tn5432
3.2.2. Acquired Resistance Genes for Chloramphenicol (cmx) and Aminoglycosides (strA and strB) Are in the Same Genomic Context
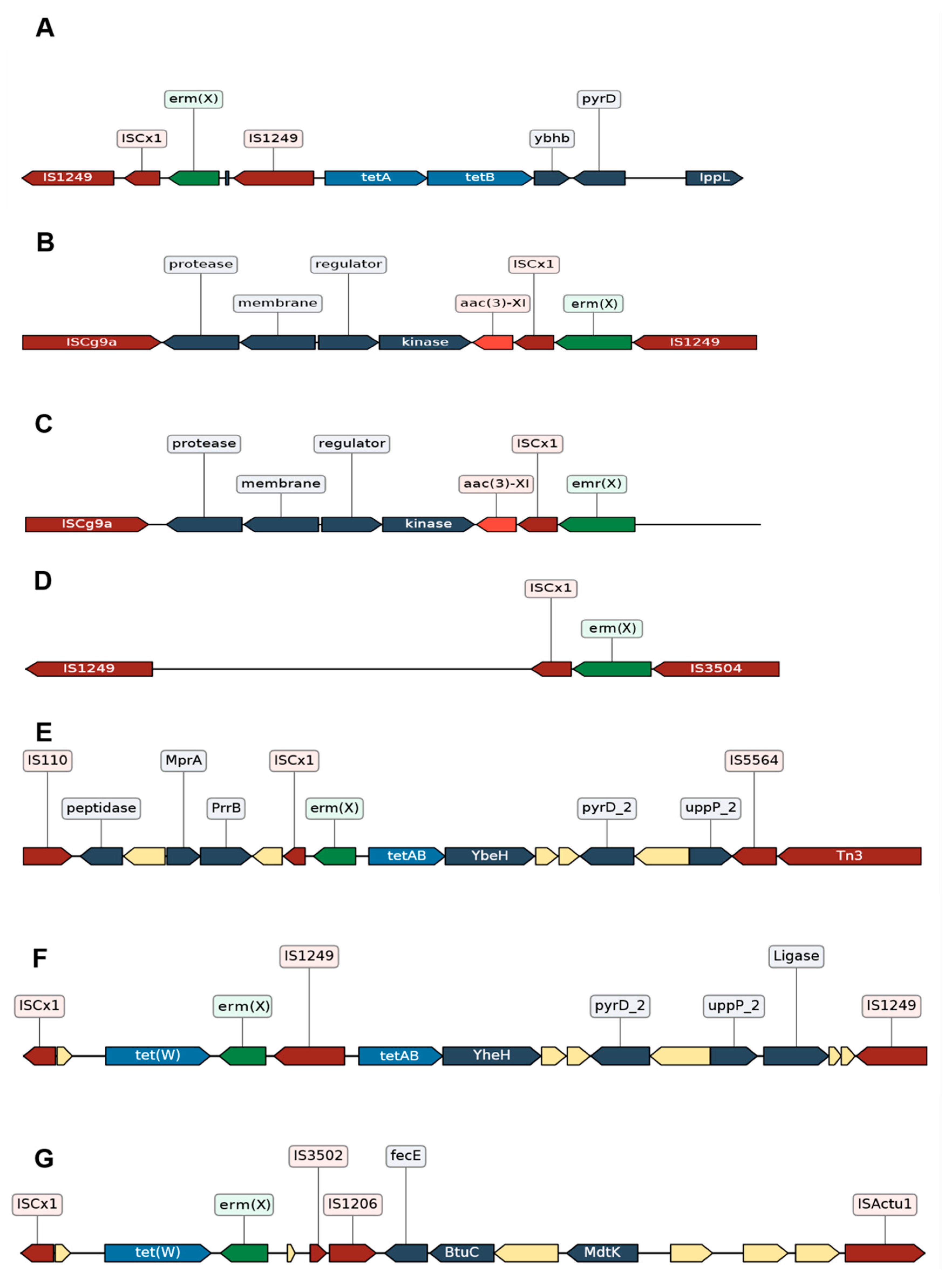
3.2.3. Different Genomic Contexts of the tet(W) Genes Suggests Possible Recombination Events
3.2.4. Impact of the Insertion Sequences in the Resistome of C. striatum
3.3. C. striatum Has Resistance Genes Probably Associated with Mobile Elements
4. Emergence of New C. striatum Clones
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baio, P.V.P.; Mota, H.F.; Freitas, A.D.; Gomes, D.L.R.; Ramos, J.N.; Sant’Anna, L.O.; Souza, M.C.; Camello, T.C.F.; Junior, R.H.; Vieira, V.V.; et al. Clonal multidrug-resistant Corynebacterium striatum within a nosocomial environment, Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz 2013, 108, 23–29. [Google Scholar] [CrossRef]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Zasada, A.A.; Mosiej, E. Contemporary microbiology and identification of Corynebacteria spp. causing infections in human. Lett. Appl. Microbiol. 2018, 66, 472–483. [Google Scholar] [CrossRef]
- Camello, T.C.F.; Mattos-Guaraldi, A.L.; Formiga, L.C.D.; Marques, E.A. Nondiphtherial Corynebacterium species isolated from clinical specimens of patients in a university hospital, Rio de Janeiro, Brazil. Braz. J. Microbiol. 2003, 34, 39–44. [Google Scholar] [CrossRef]
- Renom, F.; Garau, M.; Rubi, M.; Ramis, F.; Galmes, A.; Soriano, J.B. Nosocomial Outbreak of Corynebacterium striatum Infection in Patients with Chronic Obstructive Pulmonary Disease. J. Clin. Microbiol. 2007, 45, 2064–2067. [Google Scholar] [CrossRef]
- Schoen, C.; Unzicker, C.; Stuhler, G.; Elias, J.; Einsele, H.; Grigoleit, G.U.; Abele-Horn, M.; Mielke, S. Life-Threatening Infection Caused by Daptomycin-Resistant Corynebacterium jeikeium in a Neutropenic Patient. J. Clin. Microbiol. 2009, 47, 2328–2331. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, M.; Tsuboi, R.; Harada, K.; Cho, O.; Sugita, T. Skin microbiome of patients with interdigital tinea pedis: Corynebacterium striatum is more abundant in the patients (published online ahead of print, 2021 Apr 8). J. Dermatol. 2021. [Google Scholar] [CrossRef]
- Khan, D.; Shadi, M.; Mustafa, A.; Karam, B.; Munir, A.B.; Lafferty, J.; Glaser, A.; Mobarakai, N. A Wolf in Sheep’s clothing; Case reports and literature review of Corynebacterium striatum endocarditis. IDCases 2021, 24, e01070. [Google Scholar] [CrossRef]
- Shanbhag, S.S.; Shih, G.; Bispo, P.J.M.; Chodosh, J.; Jacobs, D.S.; Saeed, H.N. Diphtheroids as Corneal Pathogens in Chronic Ocular Surface Disease in Stevens-Johnson Syndrome/Toxic Epidermal Necrolysis. Cornea 2021, 40, 774–779. [Google Scholar] [CrossRef]
- Jagadeeshan, N.; Jayaprakash, S.; Ramegowda, R.T.; Manjunath, C.N.; Lavanya, V. An unusual case of Corynebacterium striatum endocarditis in a patient with congenital lymphedema and rheumatic heart disease. Indian Heart J. 2016, 68, S271–S273. [Google Scholar] [CrossRef] [PubMed][Green Version]
- de Carvalho, R.V.; Lima, F.F.D.S.; Santos, C.S.D.; de Souza, M.C.; da Silva, R.S.; de Mattos-Guaraldi, A.L. Central venous catheter-related infections caused by Corynebacterium amycolatum and other multiresistant non-diphtherial corynebacteria in paediatric oncology patients. Braz. J. Infect. Dis. 2018, 22, 347–351. [Google Scholar] [CrossRef]
- Ramos, J.N.; Souza, C.; Faria, Y.V.; da Silva, E.C.; Veras, J.F.C.; Baio, P.V.P.; Seabra, S.H.; de Oliveira Moreira, L.; Júnior, R.H.; Mattos-Guaraldi, A.L.; et al. Bloodstream and catheter-related infections due to different clones of multidrug-resistant and biofilm producer Corynebacterium striatum. BMC Infect. Dis. 2019, 19, 672. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Ohkusu, K.; Kawamura, Y.; Baba, S.; Ezaki, T.; Kimura, S. Emergence of multidrug-resistant Corynebacterium striatum as a nosocomial pathogen in long-term hospitalized patients with underlying diseases. Diagn. Microbiol. Infect. Dis. 2006, 54, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Severo, C.B.; Guazzelli, L.S.; Barra, M.B.; Hochhegger, B.; Severo, L.C. Nódulos pulmonares múltiplos causados por Corynebacterium striatum numa paciente imunocompetente. Rev. Inst. Med. Trop. Sao Paulo 2014, 56, 89–91. [Google Scholar] [CrossRef] [PubMed]
- Bowstead, T.T.; Santiago, S.M. Pleuropulmonary infection due to Corynebacterium striatum. Br. J. Dis. Chest 1980, 74, 198–200. [Google Scholar] [CrossRef]
- Nudel, K.; Zhao, X.; Basu, S.; Dong, X.; Hoffmann, M.; Feldgarden, M.; Allard, M.; Klompas, M.; Bry, L. Genomics of Corynebacterium striatum, an emerging multidrug-resistant pathogen of immunocompromised patients. Clin. Microbiol. Infect. 2018, 24, 1016.e7–1016.e13. [Google Scholar] [CrossRef]
- Asgin, N.; Otlu, B. Antimicrobial Resistance and Molecular Epidemiology of Corynebacterium striatum Isolated in a Tertiary Hospital in Turkey. Pathogens 2020, 9, 136. [Google Scholar] [CrossRef] [PubMed]
- Kalt, F.; Schulthess, B.; Sidler, F.; Herren, S.; Fucentese, S.F.; Zingg, P.O.; Berli, M.; Zinkernagel, A.S.; Zbinden, R.; Achermann, Y. Corynebacterium Species Rarely Cause Orthopedic Infections. J. Clin. Microbiol. 2018, 56, 1–8. [Google Scholar] [CrossRef]
- Mcmullen, A.R.; Anderson, N.; Wallace, M.A.; Shupe, A.; Burnham, C.D. When Good Bugs Go Bad: Epidemiology and Antimicrobial Resistance Profiles of Pathogen. Antimicrob. Agents Chemother. 2017, 61, e01111-17. [Google Scholar] [CrossRef] [PubMed]
- Dragomirescu, C.C.; Lixandru, B.E.; Coldea, I.L.; Corneli, O.N.; Pana, M.; Palade, A.M.; Cristea, V.C.; Suciu, I.; Suciu, G.; Manolescu, L.S.C.; et al. Antimicrobial susceptibility testing for Corynebacterium species isolated from clinical samples in Romania. Antibiotics 2020, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.P.; Ferguson, D.A.; Sarubbi, F.A. Corynebacterium striatum: An underappreciated community and nosocomial pathogen. J. Infect. 2005, 50, 338–343. [Google Scholar] [CrossRef]
- Campanile, F.; Carretto, E.; Barbarini, D.; Grigis, A.; Falcone, M.; Goglio, A.; Venditti, M.; Stefani, S. Clonal Multidrug-Resistant Corynebacterium striatum Strains, Italy. Emerg. Infect. Dis. 2009, 15, 75–78. [Google Scholar] [CrossRef]
- Martínez-Martínez, L.; Ortega, M.C.; Suárez, A.I. Comparison of E-test with broth microdilution and disk diffusion for susceptibility testing of coryneform bacteria. J. Clin. Microbiol. 1995, 33, 1318–1321. [Google Scholar] [CrossRef]
- Brandenburg, A.H.; van Belkum, A.; van Pelt, C.; Bruining, H.A.; Mouton, J.W.; Verbrugh, H.A. Patient-to-patient spread of a single strain of Corynebacterium striatum causing infections in a surgical intensive care unit. J. Clin. Microbiol. 1996, 34, 2089–2094. [Google Scholar] [CrossRef] [PubMed]
- Silva-Santana, G.; Silva, C.M.F.; Olivella, J.G.B.; Silva, I.F.; Fernandes, L.M.O.; Sued-Karam, B.R.; Santos, C.S.; Souza, C.; Mattos-Guaraldi, A.L. Worldwide survey of Corynebacterium striatum increasingly associated with human invasive infections, nosocomial outbreak, and antimicrobial multidrug-resistance, 1976–2020. Arch. Microbiol. 2021. [Google Scholar] [CrossRef]
- Wong, K.Y.; Chan, Y.C.; Wong, C.Y. Corynebacterium striatum as an emerging pathogen. J. Hosp. Infect. 2010, 76, 371–372. [Google Scholar] [CrossRef] [PubMed]
- de Souza, C.; Faria, Y.V.; Sant’Anna, L.D.O.; Viana, V.G.; Seabra, S.H.; de Souza, M.C.; Vieira, V.V.; Júnior, R.H.; Moreira, L.D.O.; de Mattos-Guaraldi, A.L. Biofilm production by multiresistant Corynebacterium striatum associated with nosocomial outbreak. Mem. Inst. Oswaldo Cruz 2015, 110, 242–248. [Google Scholar] [CrossRef] [PubMed]
- de Souza, C.; Mota, H.F.; Faria, Y.V.; Cabral, F.D.O.; de Oliveira, D.R.; Sant’Anna, L.D.O.; Nagao, P.E.; Santos, C.D.S.; Moreira, L.O.; Mattos-Guaraldi, A.L. Resistance to Antiseptics and Disinfectants of Planktonic and Biofilm-Associated Forms of Corynebacterium striatum. Microb. Drug Resist. 2020, 26, 1546–1558. [Google Scholar] [CrossRef]
- Leonard, R.B.; Nowowiejski, D.J.; Warren, J.J.; Finn, D.J.; Coyle, M.B. Molecular evidence of person-to-person transmission of a pigmented strain of Corynebacterium striatum in intensive care units. J. Clin. Microbiol. 1994, 32, 164–169. [Google Scholar] [CrossRef]
- Qin, L.; Sakai, Y.; Bao, R.; Xie, H.; Masunaga, K.; Miura, M.; Hashimoto, K.; Tanamachi, C.; Hu, B.; Watanabe, H. Characteristics of Multidrug-Resistant Corynebacterium spp. Isolated from Blood Cultures of Hospitalized Patients in Japan. Jpn. J. Infect. Dis. 2017, 70, 152–157. [Google Scholar] [CrossRef]
- Kang, S.J.; Choi, S.-M.; Choi, J.-A.; Choi, J.U.; Oh, T.-H.; Kim, S.E.; Kim, U.J.; Won, E.J.; Jang, H.-C.; Park, K.-H.; et al. Factors affecting the clinical relevance of Corynebacterium striatum isolated from blood cultures. PLoS ONE 2018, 13, e0199454. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, H.; Du, P.; Lan, R.; Chen, D.; Dong, A.; Lin, X.; Qiu, X.; Xu, S.; Ji, X.; et al. Genomic epidemiology of Corynebacterium striatum from three regions of China: An emerging national nosocomial epidemic. J. Hosp. Infect. 2020. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, H.; Chen, D.; Du, P.; Lan, R.; Qiu, X.; Hou, X.; Liu, Z.; Sun, L.; Xu, S.; et al. Whole-Genome Sequencing Reveals a Prolonged and Persistent Intrahospital Transmission of Corynebacterium striatum, an Emerging Multidrug-Resistant Pathogen. J. Clin. Microbiol. 2019, 57, e00683-19. [Google Scholar] [CrossRef]
- Ramos, J.N.; Rodrigues, I.D.S.; Baio, P.V.P.; Veras, J.F.C.; Ramos, R.T.J.; Pacheco, L.G.C.; Azevedo, V.A.; Júnior, R.H.; Marín, M.A.; de Mattos-Guaraldi, A.L.; et al. Genome sequence of a multidrug-resistant Corynebacterium striatum isolated from bloodstream infection from a nosocomial outbreak in Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz 2018, 113, e180051. [Google Scholar] [CrossRef]
- Tauch, A.; Krieft, S.; Kalinowski, J.; Pühler, A. The 51,409-bp R-plasmid pTP10 from the multiresistant clinical isolate Corynebacterium striatum M82B is composed of DNA segments initially identified in soil bacteria and in plant, animal, and human pathogens. Mol. Gen. Genet. 2000, 263, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Ding, T.; Li, M.; Wang, Y.; Zhang, X.; Ren, H.; Tong, Y. Complete genome analysis of a novel temperate bacteriophage induced from Corynebacterium striatum. Arch. Virol. 2019, 164, 2877–2880. [Google Scholar] [CrossRef]
- Peterson, E.; Kaur, P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Olender, A. Mechanisms of Antibiotic Resistance in Corynebacterium spp. Causing Infections in People. In Antibiotic Resistant Bacteria—A Continuous Challenge in the New Millennium; Pana, M., Ed.; InTech: Rijeka, Croatia, 2012; Volume 15, pp. 387–402. ISBN 978-953-51-0472-8. [Google Scholar]
- Goldner, N.K.; Bulow, C.; Cho, K.; Wallace, M.; Hsu, F.-F.; Patti, G.J.; Burnham, C.-A.; Schlesinger, P.; Dantas, G. Mechanism of High-Level Daptomycin Resistance in Corynebacterium striatum. mSphere 2018, 3, e00371-18. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.; Oliveira, L.C.; Aburjaile, F.; Benevides, L.; Tiwari, S.; Jamal, S.B.; Silva, A.; Figueiredo, H.C.P.; Ghosh, P.; Portela, R.W.; et al. Insight of Genus Corynebacterium: Ascertaining the Role of Pathogenic and Non-pathogenic Species. Front. Microbiol. 2017, 8, 1937. [Google Scholar] [CrossRef] [PubMed]
- Schröder, J.; Maus, I.; Meyer, K.; Wördemann, S.; Blom, J.; Jaenicke, S.; Schneider, J.; Trost, E.; Tauch, A. Complete genome sequence, lifestyle, and multi-drug resistance of the human pathogen Corynebacterium resistens DSM 45100 isolated from blood samples of a leukemia patient. BMC Genom. 2012, 13, 141. [Google Scholar] [CrossRef]
- Tauch, A.; Bischoff, N.; Brune, I.; Kalinowski, J. Insights into the genetic organization of the Corynebacterium diphtheriae erythromycin resistance plasmid pNG2 deduced from its complete nucleotide sequence. Plasmid 2003, 49, 63–74. [Google Scholar] [CrossRef]
- Redgrave, L.S.; Sutton, S.B.; Webber, M.A.; Piddock, L.J.V. Fluoroquinolone resistance: Mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014, 22, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.N.; Valadão, T.B.; Baio, P.V.P.; Mattos-Guaraldi, A.L.; Vieira, V.V. Novel mutations in the QRDR region gyrA gene in multidrug-resistance Corynebacterium spp. isolates from intravenous sites. Antonie van Leeuwenhoek 2020, 113, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Sierra, J.M.; Martinez-Martinez, L.; Vázquez, F.; Giralt, E.; Vila, J. Relationship between mutations in the gyrA gene and quinolone resistance in clinical isolates of Corynebacterium striatum and Corynebacterium amycolatum. Antimicrob. Agents Chemother. 2005, 49, 1714–1719. [Google Scholar] [CrossRef]
- Alibi, S.; Ferjani, A.; Boukadida, J.; Cano, M.E.; Fernández-Martínez, M.; Martínez-Martínez, L.; Navas, J. Occurrence of Corynebacterium striatum as an emerging antibiotic-resistant nosocomial pathogen in a Tunisian hospital. Sci. Rep. 2017, 7, 9704. [Google Scholar] [CrossRef]
- Huang, H.W. DAPTOMYCIN, its membrane-active mechanism vs. that of other antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183395. [Google Scholar] [CrossRef]
- Tran, T.T.; Jaijakul, S.; Lewis, C.T.; Diaz, L.; Panesso, D.; Kaplan, H.B.; Murray, B.E.; Wanger, A.; Arias, C.A. Native Valve Endocarditis Caused by Corynebacterium striatum with Heterogeneous High-Level Daptomycin Resistance: Collateral Damage from Daptomycin Therapy? Antimicrob. Agents Chemother. 2012, 56, 3461–3464. [Google Scholar] [CrossRef]
- TeKippe, E.M.; Thomas, B.S.; Ewald, G.A.; Lawrence, S.J.; Burnham, C.-A.D. Rapid emergence of daptomycin resistance in clinical isolates of Corynebacterium striatum… a cautionary tale. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 2199–2205. [Google Scholar] [CrossRef]
- Hagiya, H.; Kimura, K.; Okuno, H.; Hamaguchi, S.; Morii, D.; Yoshida, H.; Mitsui, T.; Nishi, I.; Tomono, K. Bacteremia due to high-level daptomycin-resistant Corynebacterium striatum: A case report with genetic investigation. J. Infect. Chemother. 2019, 25, 906–908. [Google Scholar] [CrossRef]
- Werth, B.J.; Hahn, W.O.; Butler-Wu, S.M.; Rakita, R.M. Emergence of High-Level Daptomycin Resistance in Corynebacterium striatum in Two Patients with Left Ventricular Assist Device Infections. Microb. Drug Resist. 2016, 22, 233–237. [Google Scholar] [CrossRef]
- Mitchell, K.F.; McElvania, E.; Wallace, M.A.; Droske, L.E.; Robertson, A.E.; Westblade, L.F.; Burnham, C.-A.D. Evaluating the Rapid Emergence of Daptomycin Resistance in Corynebacterium: A Multi-Center Study. J. Clin. Microbiol. 2021, 59, e02052-20. [Google Scholar] [CrossRef] [PubMed]
- Hines, K.M.; Waalkes, A.; Penewit, K.; Holmes, E.A.; Salipante, S.J.; Werth, B.J.; Xu, L. Characterization of the Mechanisms of Daptomycin Resistance among Gram-Positive Bacterial Pathogens by Multidimensional Lipidomics. mSphere 2017, 2, e00492-17. [Google Scholar] [CrossRef] [PubMed]
- Hachmann, A.-B.; Sevim, E.; Gaballa, A.; Popham, D.L.; Antelmann, H.; Helmann, J.D. Reduction in Membrane Phosphatidylglycerol Content Leads to Daptomycin Resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 2011, 55, 4326–4337. [Google Scholar] [CrossRef] [PubMed]
- Hennart, M.; Panunzi, L.G.; Rodrigues, C.; Gaday, Q.; Baines, S.L.; Barros-Pinkelnig, M.; Carmi-Leroy, A.; Dazas, M.; Wehenkel, A.M.; Didelot, X.; et al. Population genomics and antimicrobial resistance in Corynebacterium diphtheriae. Genome Med. 2020, 12, 107. [Google Scholar] [CrossRef] [PubMed]
- NeÅ¡vera, J.; Hochmannová, J.; Pátek, M. An integron of class 1 is present on the plasmid pCG4 from Gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol. Lett. 1998, 169, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Kono, M.; Sasatsu, M.; Aoki, T. R Plasmids in Corynebacterium xerosis Strains. Antimicrob. Agents Chemother. 1983, 23, 506–508. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Szemraj, M.; Kwaszewska, A.; Szewczyk, E.M. New Gene Responsible for Resistance of Clinical Corynebacteria to Macrolide, Lincosamide and Streptogramin B. Pol. J. Microbiol. 2018, 67, 237–240. [Google Scholar] [CrossRef]
- Szemraj, M.; Czekaj, T.; Kalisz, J.; Szewczyk, E.M. Differences in distribution of MLS antibiotics resistance genes in clinical isolates of staphylococci belonging to species: S. epidermidis, S. hominis, S. haemolyticus, S. simulans and S. warneri. BMC Microbiol. 2019, 19, 124. [Google Scholar] [CrossRef]
- Coyle, M.B.; Minshew, B.H.; Bland, J.A.; Hsu, P.C. Erythromycin and clindamycin resistance in Corynebacterium diphtheriae from skin lesions. Antimicrob. Agents Chemother. 1979, 16, 525–527. [Google Scholar] [CrossRef]
- Eady, E.A.; Coates, P.; Ross, J.I.; Ratyal, A.H.; Cove, J.H. Antibiotic resistance patterns of aerobic coryneforms and furazolidone-resistant Gram-positive cocci from the skin surface of the human axilla and fourth toe cleft. J. Antimicrob. Chemother. 2000, 46, 205–213. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rosato, A.E.; Lee, B.S.; Nash, K.A. Inducible Macrolide Resistance in Corynebacterium jeikeium. Antimicrob. Agents Chemother. 2001, 45, 1982–1989. [Google Scholar] [CrossRef] [PubMed]
- Guirao, G.Y.; Peris, B.M.; Martínez-Toldos, M.C.; González, T.R.; Guillén, P.L.V.; Hernández, M.S. Implicación de los genes ermX en la resistencia a los macrólidos y la telitromicina de Corynebacterium jeikeium y Corynebacterium amycolatum. Rev. Española Quimioter. 2005, 18, 236–242. [Google Scholar]
- Olender, A.; Niemcewicz, M. Macrolide, Lincosamide, and Streptogramin B–Constitutive-Type Resistance in Corynebacterium pseudodiphtheriticum Isolated from Upper Respiratory Tract Specimens. Microb. Drug Resist. 2010, 16, 119–122. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Zulkower, V.; Rosser, S. DNA features viewer, a sequence annotations formatting and plotting library for python. bioRxiv 2020. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Varani, A.; Ton-Hoang, B.; Chandler, M. Everyman’s Guide to Bacterial Insertion Sequences. In Mobile DNA III; ASM Press: Washington, DC, USA, 2015; pp. 555–590. [Google Scholar]
- Razavi, M.; Kristiansson, E.; Flach, C.-F.; Larsson, D.G.J. The Association between Insertion Sequences and Antibiotic Resistance Genes. mSphere 2020, 5, e00418-20. [Google Scholar] [CrossRef]
- He, S.; Hickman, A.B.; Varani, A.M.; Siguier, P.; Chandler, M.; Dekker, J.P.; Dyda, F. Insertion Sequence IS26 Reorganizes Plasmids in Clinically Isolated Multidrug-Resistant Bacteria by Replicative Transposition. MBio 2015, 6, e00762-15. [Google Scholar] [CrossRef]
- Harmer, C.J.; Hall, R.M. An analysis of the IS6/IS26 family of insertion sequences: Is it a single family? Microb. Genomics 2019, 5. [Google Scholar] [CrossRef]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef]
- De Boer, H.A.; Comstock, L.J.; Vasser, M. The tac promoter: A functional hybrid derived from the trp and lac promoters. Proc. Natl. Acad. Sci. USA 1983, 80, 21–25. [Google Scholar] [CrossRef]
- Ballouz, S.; Francis, A.R.; Lan, R.; Tanaka, M.M. Conditions for the Evolution of Gene Clusters in Bacterial Genomes. PLoS Comput. Biol. 2010, 6, e1000672. [Google Scholar] [CrossRef]
- Deng, Y.; Bao, X.; Ji, L.; Chen, L.; Liu, J.; Miao, J.; Chen, D.; Bian, H.; Li, Y.; Yu, G. Resistance integrons: Class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 45. [Google Scholar] [CrossRef] [PubMed]
- Rocha, D.J.P.; Azevedo, V.; Brenig, B.; Silva, A.; Blom, J.; Ramos, R.T.; Aguiar, E.R.G.; Chapartegui-González, I.; Fernández-Martínez, M.; Martínez-Martínez, L.; et al. Whole-genome sequencing reveals misidentification of a multidrug-resistant urine clinical isolate as Corynebacterium urealyticum. J. Glob. Antimicrob. Resist. 2020, 23, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Soares, M.; Pereira, C.; Leitao, N.; Henriques, I.; Correia, A. INTEGRALL: A database and search engine for integrons, integrases and gene cassettes. Bioinformatics 2009, 25, 1096–1098. [Google Scholar] [CrossRef]
- Barnass, S.; Holland, K.; Tabaqchali, S. Vancomycin-resistant Corynebacterium species causing prosthetic valve endocarditis successfully treated with imipenem and ciprofloxacin. J. Infect. 1991, 22, 161–169. [Google Scholar] [CrossRef]
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2019, 48, D561–D569. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020. [Google Scholar] [CrossRef]
- Navas, J.; Fernández-Martínez, M.; Salas, C.; Cano, M.E.; Martínez-Martínez, L. Susceptibility to Aminoglycosides and Distribution of aph and aac(3)-XI Genes among Corynebacterium striatum Clinical Isolates. PLoS ONE 2016, 11, e0167856. [Google Scholar] [CrossRef]
- Galimand, M.; Fishovitz, J.; Lambert, T.; Barbe, V.; Zajicek, J.; Mobashery, S.; Courvalin, P. AAC(3)-XI, a new aminoglycoside 3-N-acetyltransferase from Corynebacterium striatum. Antimicrob. Agents Chemother. 2015, 59, 5647–5653. [Google Scholar] [CrossRef] [PubMed]
- de Souza, C.; Simpson-Louredo, L.; Mota, H.F.; Faria, Y.V.; Cabral, F.D.O.; Colodette, S.D.S.; Canellas, M.E.F.C.; Cucinelli, A.D.E.S.; de Luna, M.D.G.; Santos, C.D.S.; et al. Virulence potential of Corynebacterium striatum towards Caenorhabditis elegans. Antonie Van Leeuwenhoek 2019, 112, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Alibi, S.; Ramos-Vivas, J.; Ben Selma, W.; Ben Mansour, H.; Boukadida, J.; Navas, J. Virulence of clinically relevant multi-drug resistant Corynebacterium striatum strains and their ability to adhere to human epithelial cells and inert surfaces. Microb. Pathog. 2021, 155, 104887. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Baruah, A.; Tomioka, M.; Iino, Y.; Kalita, M.C.; Khan, M. Caenorhabditis elegans: A model to understand host-microbe interactions. Cell. Mol. Life Sci. 2020, 77, 1229–1249. [Google Scholar] [CrossRef]
- Song, S.A.; Shin, J.H. Microbiological Characteristics of Corynebacterium striatum, an Emerging Pathogen. Hanyang Med. Rev. 2018, 38, 93. [Google Scholar] [CrossRef][Green Version]
- Shariff, M.; Beri, K. Corynebacterium striatum: An emerging respiratory pathogen. J. Infect. Dev. Ctries. 2018, 12, 581–586. [Google Scholar] [CrossRef]
- Neemuchwala, A.; Soares, D.; Ravirajan, V.; Marchand-Austin, A.; Kus, J.V.; Patel, S.N. In vitro antibiotic susceptibility pattern of non-diphtheriae Corynebacterium isolates in Ontario, Canada, from 2011 to 2016. Antimicrob. Agents Chemother. 2018, 62, e01776-17. [Google Scholar] [CrossRef]
- Abe, M.; Kimura, M.; Maruyama, H.; Watari, T.; Ogura, S.; Takagi, S.; Uchida, N.; Otsuka, Y.; Taniguchi, S.; Araoka, H. Clinical characteristics and drug susceptibility patterns of Corynebacterium species in bacteremic patients with hematological disorders. Eur. J. Clin. Microbiol. Infect. Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
| IS 1 | Group 1 | IS Family 1 | Tn 2 | Determinant Associated 3 | Origin 4 |
|---|---|---|---|---|---|
| IS6110 | IS51 | IS3 | tet(W), blaB | Mycobacterium tuberculosis | |
| IS3502 | tet(W), erm(X), mdtK | Corynebacterium jeikeium | |||
| IS3501 | erm(X), tetA, tetB, tet(W), yheH | Corynebacterium jeikeium | |||
| IS3504 | erm(X), aac(3)-XI, cmx | Corynebacterium diphtheriae | |||
| IS1600 | aac(6′)-la | Shigella sonnei | |||
| IS1206 | tet(W). erm(X), mdtK | Corynebacterium glutamicum | |||
| IS407 | IS407 | erm(X), tet(W) | Burkhoderia cepacia | ||
| ISAar26 | IS3 | sul1 ermE, cmx, strB, strA | Arthobacter arilaitensis | ||
| IS3503 | IS1249 | IS256 | tet(W), blab | Corynebacterium jeikeium | |
| ISActu1 | tet(W), erm(X), mdtK | Actinomyces turicensis | |||
| IS1249 | Tn5432 | erm(X), tetA, tetB, tet(W), yheH | Corynebacterium striatum 5 | ||
| ISCre1 | aac(6′)-la | Corynebacterium reistens | |||
| IS1250 | ND | Tn5716 | strB, strA | Corynebacterium striatum | |
| IS6100 | ND | IS6 | aac(6′)-la, aadA1 | Mycobacterium fortuitum | |
| IS26 | IS6 | Tn5715 | aphA1-IAB, aph(3′)-I, strA, strB | Proteus vulgaris | |
| IS1628 | ND | aac(6′)-la | Corynebacterium glutamicum | ||
| IS110 | ND | IS110 | erm(X) tetAB, yheH | Streptomyces coelicolor | |
| ISCg9a | ND | erm(X), aac(3)-XI | Corynebacterium glutamicum | ||
| IS5564 | ND | IS481 | Tn5564 | cmxA, cml, erm(X) tetA/B, yheH | Corynebacterium striatum |
| IS1513 | ND | IS30 | Tn5564 | cmx, cml | Corynebacterium striatum |
| Chemical Class | Drug 1 | Mechanism of Action | Genetic Factor | Associated Function | Mechanism of Resistance | References |
|---|---|---|---|---|---|---|
| Fluoroquinolones | Levofloxacin Ciprofloxacin Moxifloxacin | Inhibits DNA gyrase and DNA topoisomerase | gyrA | Negatively supercoils closed circular double-stranded DNA | Alteration of drug target due to mutation | [46] |
| Phenicols | Chloramphenicol | Inhibits protein elongation due to 23S ribosomal subunit binding | cmx | Encoding of a specific efflux protein of chloramphenicol | Transports chloramphenicol out of the membrane | [35] |
| Macrolides | Erythromycin Clindamycin | Inhibits protein synthesis due to 50S ribosomal subunit binding | erm(X) ermB | 23S ribosomal RNA methyltransferase | Alteration of drug target due to methylation | [35,47] |
| Tetracyclines | Tetracycline Doxycycline | Inhibits the initiation of translation by binding to 30S ribosomal subunit | tetA, tetB | Tetracycline efflux ABC transporter TetAB | Transports tetracyclines out of the membrane | [35] |
| tet(W) | Tetracycline resistance ribosomal protection protein | Binds to the ribosome and inhibits the binding of tetracyclines | [16] | |||
| Beta-Lactams | Penicillin Ampicillin Cefazolin Cefotiam Cefotaxime Meropenem Cefotaxime Imipenem Oxacillin Ceftriaxone | Inhibits cell wall biosynthesis | bla | Beta-lactamase class A (serine hydrolase) | Alteration of the drug due to enzymatic modification | [20,33,47] |
| tetA tetB | Tetracycline efflux ABC transporter TetAB | Transports beta-lactams out of the membrane | [35] | |||
| ampC | Beta-lactamase class C | Alteration of the drug due to enzymatic modification | [47] | |||
| Sulfonamides | Sulfamethoxazole -/trimethoprim | Acts by blocking the synthesis of folic acid and inhibits growth | sul1 | Dihydropteroate synthase | Antibiotic target replacement | [33] |
| Trimethoprim | ND | ND | ND | |||
| Aminoglycosides | Tobramycin Amikacin Streptomycin | Inhibits protein synthesis | aph(3′)-Ic aph(3”)-Ib (strA) aph(6)-Id (strB) | O-phosphotransferases | Catalyzes ATP-dependent phosphorylation of hydroxyl group | [47,82] |
| Gentamicin Kanamycin | aac(3)-XI | N-acetyltransferases | Catalyzes acetyl CoA-dependent adenylation of an amino group | [83] | ||
| Lipopeptides | Daptomycin | The aggregation of daptomycin alters the curvature of the membrane, which creates holes that leak ions | pgsA2 | Catalyzes the synthesis of phosphoglycerol (PG) | Inhibits membrane binding due to PG deficiency | [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leyton, B.; Ramos, J.N.; Baio, P.V.P.; Veras, J.F.C.; Souza, C.; Burkovski, A.; Mattos-Guaraldi, A.L.; Vieira, V.V.; Abanto Marin, M. Treat Me Well or Will Resist: Uptake of Mobile Genetic Elements Determine the Resistome of Corynebacterium striatum. Int. J. Mol. Sci. 2021, 22, 7499. https://doi.org/10.3390/ijms22147499
Leyton B, Ramos JN, Baio PVP, Veras JFC, Souza C, Burkovski A, Mattos-Guaraldi AL, Vieira VV, Abanto Marin M. Treat Me Well or Will Resist: Uptake of Mobile Genetic Elements Determine the Resistome of Corynebacterium striatum. International Journal of Molecular Sciences. 2021; 22(14):7499. https://doi.org/10.3390/ijms22147499
Chicago/Turabian StyleLeyton, Benjamin, Juliana Nunes Ramos, Paulo Victor Pereira Baio, João Flávio Carneiro Veras, Cassius Souza, Andreas Burkovski, Ana Luíza Mattos-Guaraldi, Verônica Viana Vieira, and Michel Abanto Marin. 2021. "Treat Me Well or Will Resist: Uptake of Mobile Genetic Elements Determine the Resistome of Corynebacterium striatum" International Journal of Molecular Sciences 22, no. 14: 7499. https://doi.org/10.3390/ijms22147499
APA StyleLeyton, B., Ramos, J. N., Baio, P. V. P., Veras, J. F. C., Souza, C., Burkovski, A., Mattos-Guaraldi, A. L., Vieira, V. V., & Abanto Marin, M. (2021). Treat Me Well or Will Resist: Uptake of Mobile Genetic Elements Determine the Resistome of Corynebacterium striatum. International Journal of Molecular Sciences, 22(14), 7499. https://doi.org/10.3390/ijms22147499






