The Antiviral Molecule 5-Pyridoxolactone Identified Post BmNPV Infection of the Silkworm, Bombyx mori
Abstract
:1. Introduction
2. Results
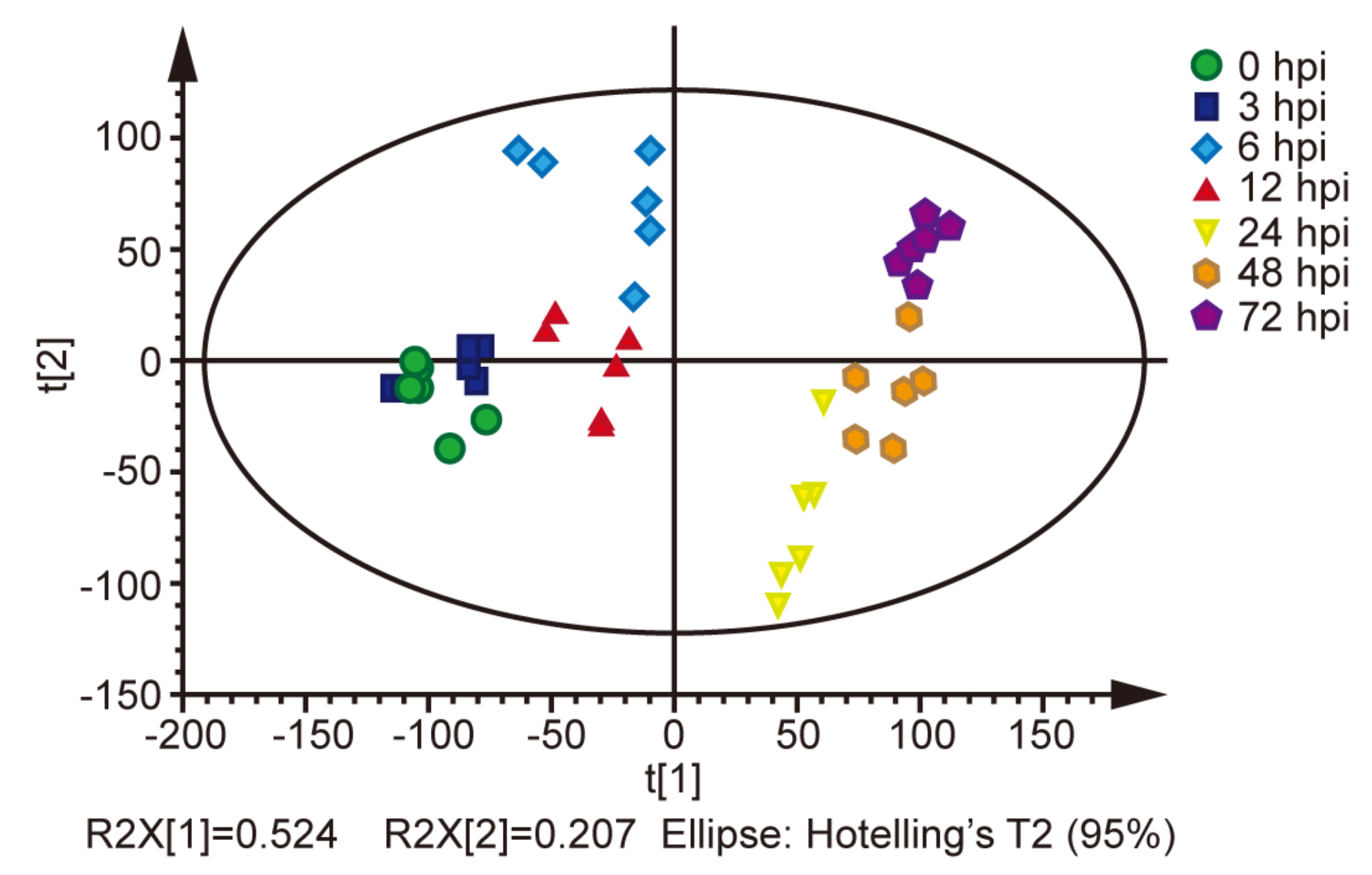
2.1. Metabolites in the BmE Cells Infected with BmNPV
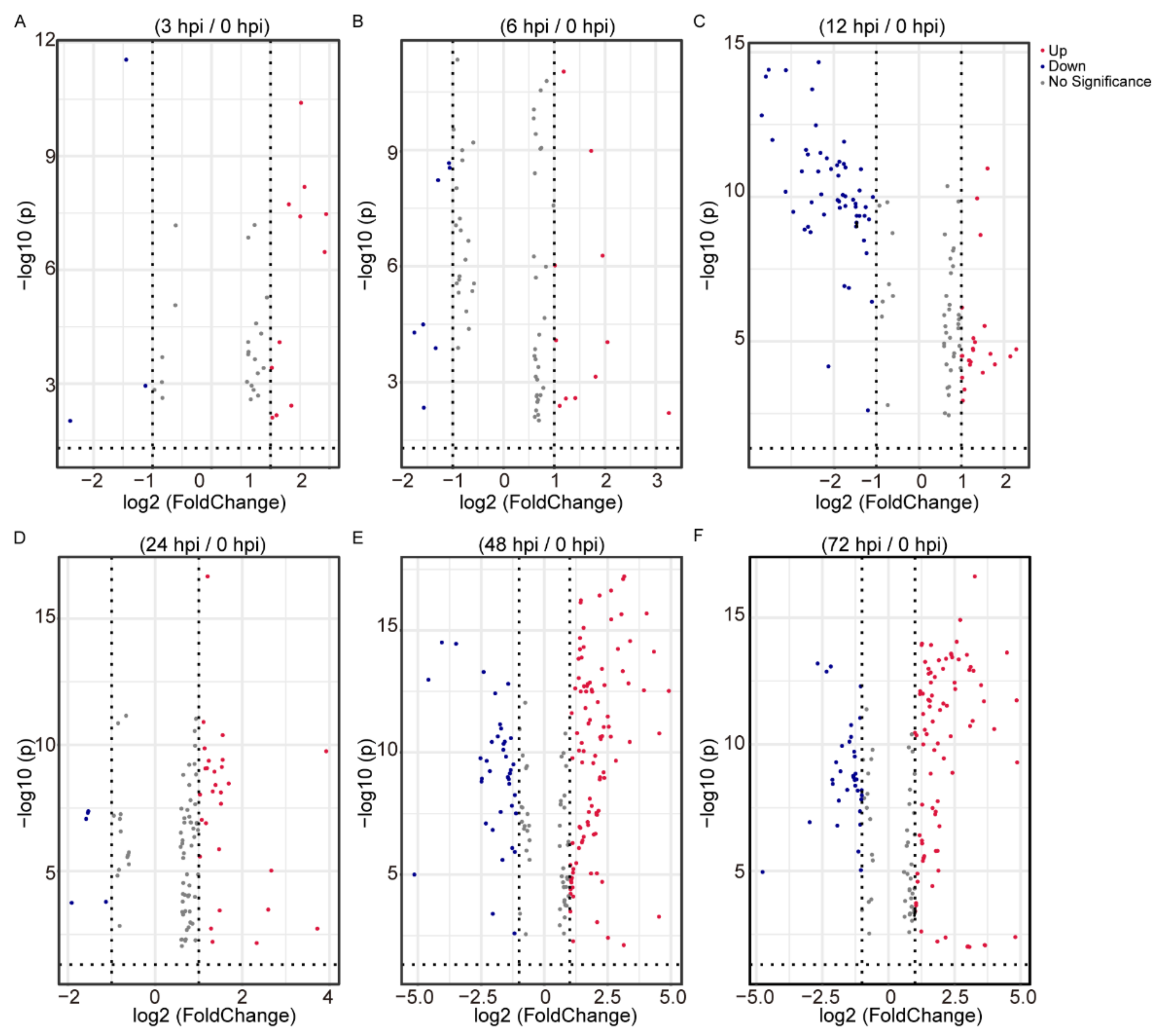
2.2. Differential Analysis of the Identified Metabolites Following BmNPV Stimulation
2.3. Pathway Analysis of the Identified Metabolites
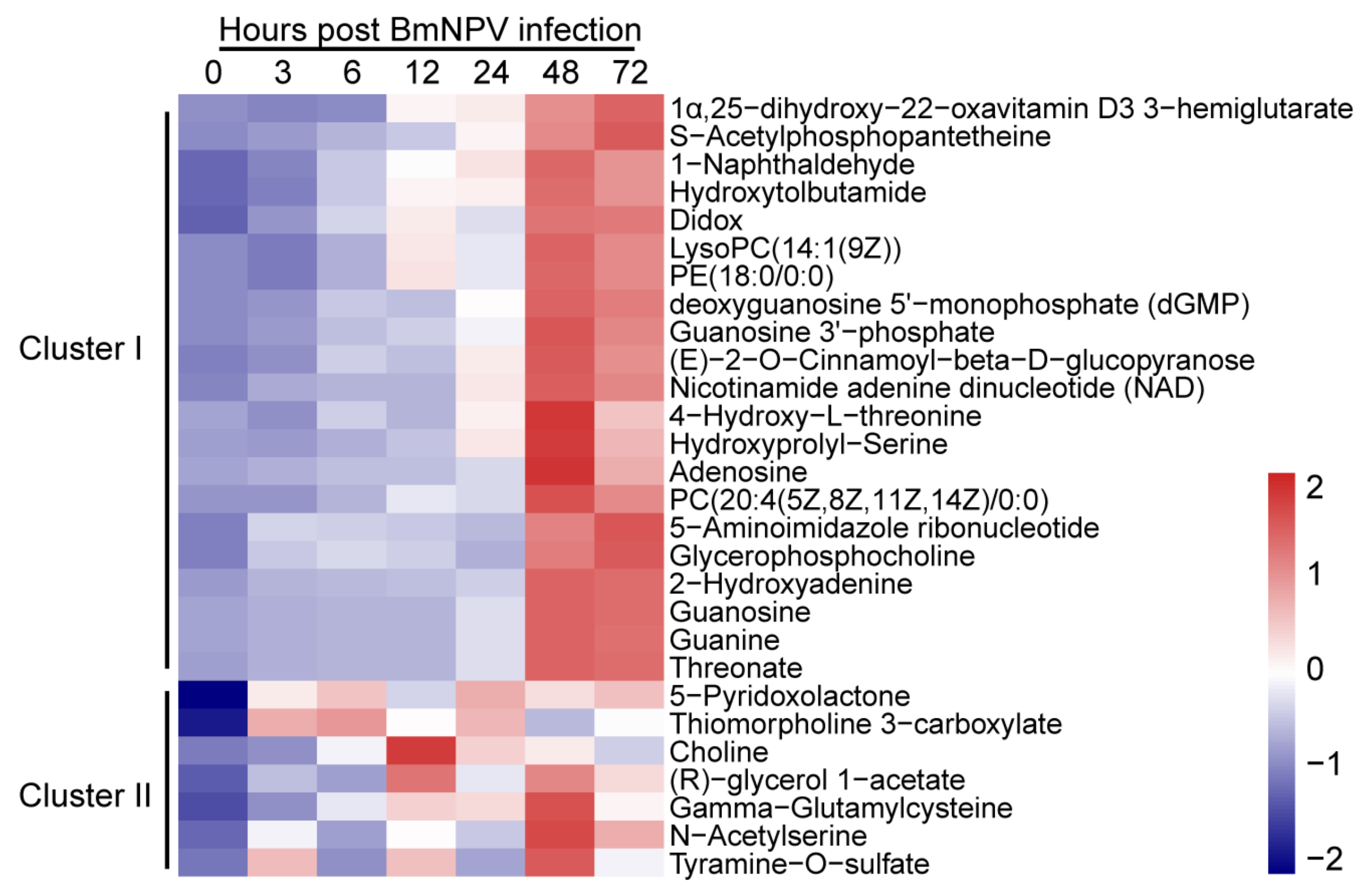
2.4. Patterns of Metabolites in BmE Cells Infected with BmNPV at Different Times
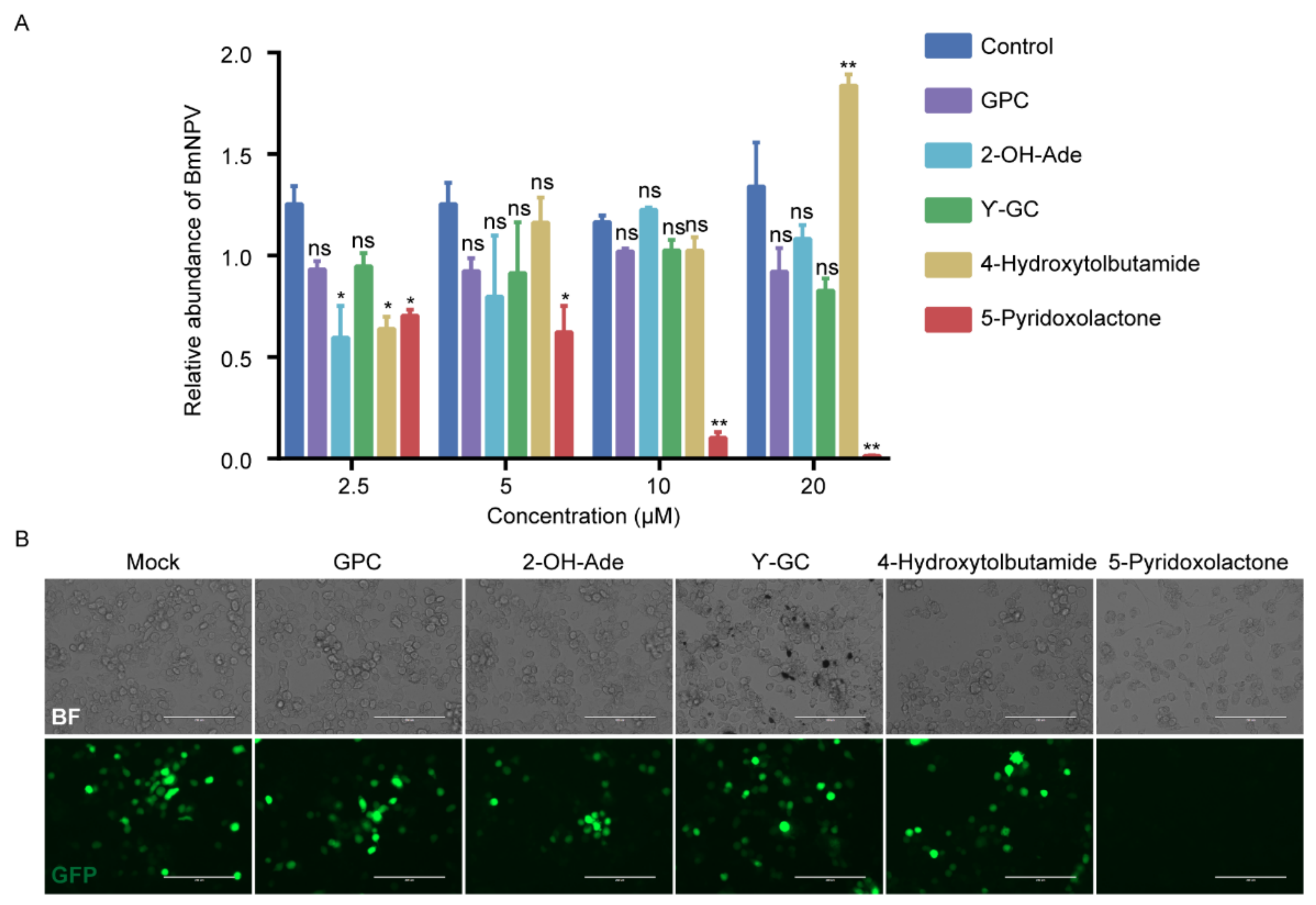
2.5. 5-Pyridoxolactone Is an Important Antiviral Molecule in Host
2.6. 5-Pyridoxolactone May Suppress the Invasion of BmNPV
3. Discussion
4. Materials and Methods
4.1. Cells and BmNPV
4.2. Sample Collection and Metabolite Extraction
4.3. LC-MS/MS Analysis
4.4. Data Preprocessing and Analysis
4.5. Pathway Analysis
4.6. Real-Time PCR
4.7. Measurement of Viral DNA Load
4.8. Cell Toxicity and Tests
4.9. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pujol-Lereis, L.M.; Rabossi, A.; Quesada-Allué, L.A. Lipid profiles as indicators of functional senescence in the medfly. Exp. Gerontol. 2012, 47, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, M.R.; Shimada, T.; Abe, H. The genetics and genomics of the silkworm. Bombyx mori. Annu. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [CrossRef]
- Nagaraju, J.; Goldsmith, M.R. Silkworm genomics—Progress and prospects. J. Curr. Sci. 2002, 83, 415–425. [Google Scholar]
- Li, M.; Shen, L.; Xu, A.; Miao, X.; Hou, C.; Sun, P.; Zhang, Y.; Huang, Y. Genetic diversity among silkworm (Bombyx mori L., Lep., Bombycidae) germplasms revealed by microsatellites. Genome 2005, 48, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Sagisaka, A.; Fujita, K.; Kaneko, Y.; Imanishi, S.; Yamakawa, M. Lipopolysaccharide elicits expression of immune-related genes in the silkworm, Bombyx mori. Insect Mol. Biol. 2009, 18, 71–75. [Google Scholar] [CrossRef]
- Jiang, L.; Xia, Q. The progress and future of enhancing antiviral capacity by transgenic technology in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2014, 48, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.Y.; Tang, X.D.; Lv, Z.Y.; Wang, X.Y.; Tian, C.H.; Xu, Y.P.; Zhang, C.X. Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics 2009, 94, 138–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.; Wang, R.; Yu, K.; Zhang, T.; Chen, X.; Liu, Y.; Shi, R.; Wang, X.; Xia, Q.; Ma, S. Genome-wide CRISPR screening reveals genes essential for cell viability and resistance to abiotic and biotic stresses in Bombyx mori. Genome Res. 2020, 30, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Goldsmith, M.R.; Xia, Q. Advances in the Arms Race Between Silkworm and Baculovirus. Front. Immunol. 2021, 12, 628151. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Li, B.; Song, L.; Hu, C.; Li, X.; Wang, D.; Xiong, Y.; Zhao, P.; He, H.; Xia, Q.; et al. Stimulator of interferon genes (STING) provides insect antiviral immunity by promoting Dredd caspase-mediated NF-κB activation. J. Biol. Chem. 2018, 293, 11878–11890. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Cheng, T.; Zhao, P.; Yang, Q.; Wang, G.; Jin, S.; Lin, P.; Xiao, Y.; Xia, Q. Resistance to BmNPV via overexpression of an exogenous gene controlled by an inducible promoter and enhancer in transgenic silkworm, Bombyx mori. PLoS ONE 2012, 7, e41838. [Google Scholar]
- Jiang, L.; Zhao, P.; Cheng, T.; Sun, Q.; Peng, Z.; Dang, Y.; Wu, X.; Wang, G.; Jin, S.; Lin, P.; et al. A transgenic animal with antiviral properties that might inhibit multiple stages of infection. Antivir. Res. 2013, 98, 171–173. [Google Scholar] [CrossRef]
- Lekha, G.; Gupta, T.; Awasthi, A.K.; Murthy, G.N.; Trivedy, K.; Ponnuvel, K.M. Genome wide microarray based expression profiles associated with BmNPV resistance and susceptibility in Indian silkworm races of Bombyx mori. Genomics 2015, 106, 393–403. [Google Scholar] [CrossRef]
- Sagisaka, A.; Fujita, K.; Nakamura, Y.; Ishibashi, J.; Noda, H.; Imanishi, S.; Mita, K.; Yamakawa, M.; Tanaka, H. Genome-wide analysis of host gene expression in the silkworm cells infected with Bombyx mori nucleopolyhedrovirus. Virus Res. 2010, 147, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhou, K.; Zhao, G.; Qian, H.; Xu, A. Transcriptome-wide analysis of the difference of alternative splicing in susceptible and resistant silkworm strains after BmNPV infection. Biotech 2019, 9, 152. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gao, L.; Shi, H.; Xia, H.; Gao, L.; Lian, C.; Chen, L.; Yao, Q.; Chen, K.; Liu, X. Microarray analysis of gene expression profile in resistant and susceptible Bombyx mori strains reveals resistance-related genes to nucleopolyhedrovirus. Genomics 2013, 101, 256–262. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.Y.; Wu, K.H.; Pang, H.L.; Xu, P.Z.; Li, M.W.; Zhang, G.Z. Study on the Role of Cytc in Response to BmNPV Infection in Silkworm, Bombyx mori (Lepidoptera). Int. J. Mol. Sci. 2019, 20, 4325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, L.; Shi, H.; Liu, X.; Zhang, C.; Yao, Q.; Wang, Y.; Chang, C.; Shi, J.; Cao, J.; Kong, J.; et al. Arginine kinase is highly expressed in a resistant strain of silkworm (Bombyx mori, Lepidoptera): Implication of its role in resistance to Bombyx mori nucleopolyhedrovirus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2011, 158, 230–234. [Google Scholar] [CrossRef]
- Qin, L.; Xia, H.; Shi, H.; Zhou, Y.; Chen, L.; Yao, Q.; Liu, X.; Feng, F.; Yuan, Y.; Chen, K. Comparative proteomic analysis reveals that caspase-1 and serine protease may be involved in silkworm resistance to Bombyx mori nuclear polyhedrosis virus. J. Proteom. 2012, 75, 3630–3638. [Google Scholar] [CrossRef]
- Nielsen, J. Systems Biology of Metabolism. Annu. Rev. Biochem. 2017, 86, 245–275. [Google Scholar] [CrossRef]
- Massie, J.P.; Reynolds, E.L.; Koestler, B.J.; Cong, J.P.; Agostoni, M.; Waters, C.M. Quantification of high-specificity cyclic diguanylate signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 12746–12751. [Google Scholar] [CrossRef] [Green Version]
- Sonkar, K.; Ayyappan, V.; Tressler, C.M.; Adelaja, O.; Cai, R.; Cheng, M.; Glunde, K. Focus on the glycerophosphocholine pathway in choline phospholipid metabolism of cancer. NMR Biomed. 2019, 32, e4112. [Google Scholar] [CrossRef]
- Barone, F.; McCulloch, S.D.; Macpherson, P.; Maga, G.; Yamada, M.; Nohmi, T.; Minoprio, A.; Mazzei, F.; Kunkel, T.A.; Karran, P.; et al. Replication of 2-hydroxyadenine-containing DNA and recognition by human MutSalpha. DNA Repair 2007, 6, 355–366. [Google Scholar] [CrossRef] [Green Version]
- Weimann, A.; McLeod, G.; Henriksen, T.; Cejvanovic, V.; Poulsen, H.E. Identification and quantification of isoguanosine in humans and mice. Scand. J. Clin. Lab. Investig. 2019, 79, 225–232. [Google Scholar] [CrossRef]
- Yang, Y.; Li, L.; Hang, Q.; Fang, Y.; Dong, X.; Cao, P.; Yin, Z.; Luo, L. γ-glutamylcysteine exhibits anti-inflammatory effects by increasing cellular glutathione level. Redox Biol. 2019, 20, 157–166. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Z.; Li, B.; Yao, H.; Zarka, M.; Welch, J.; Sachdev, P.; Bridge, W.; Braidy, N. Supplementation with γ-glutamylcysteine (γ-GC) lessens oxidative stress, brain inflammation and amyloid pathology and improves spatial memory in a murine model of AD. Neurochem. Int. 2021, 144, 104931. [Google Scholar] [CrossRef]
- Uehara, S.; Yoneda, N.; Higuchi, Y.; Yamazaki, H.; Suemizu, H. Methyl-hydroxylation and subsequent oxidation to produce carboxylic acid is the major metabolic pathway of tolbutamide in chimeric TK-NOG mice transplanted with human hepatocytes. Xenobiotica Fate Foreign Compd. Biol. Syst. 2021, 51, 582–589. [Google Scholar] [CrossRef]
- Jong, Y.J.; Snell, E.E. Enzymes of vitamin B6 degradation. Purification and properties of 4- and 5-pyridoxolactonases. J. Biol. Chem. 1986, 261, 15112–15114. [Google Scholar] [CrossRef]
- Gomi, S.; Majima, K.; Maeda, S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 1999, 80, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Huh, N.E.; Weaver, R.F. Categorizing some early and late transcripts directed by the Autographa californica nuclear polyhedrosis virus. J. Gen. Virol. 1990, 71, 2195–2200. [Google Scholar] [CrossRef]
- Wu, P.; Shang, Q.; Dweteh, O.A.; Huang, H.; Zhang, S.; Zhong, J.; Hou, Q.; Guo, X. Over expression of bmo-miR-2819 suppresses BmNPV replication by regulating the BmNPV ie-1 gene in Bombyx Mori. Mol. Immunol. 2019, 109, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Kamita, S.G.; Maeda, S. Inhibition of Bombyx mori nuclear polyhedrosis virus (NPV) replication by the putative DNA helicase gene of Autographa californica NPV. J. Virol. 1993, 67, 6239–6245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blissard, G.W.; Theilmann, D.A. Baculovirus Entry and Egress from Insect Cells. Annu. Rev. Virol. 2018, 5, 113–139. [Google Scholar] [CrossRef]
- Rahman, M.M.; Gopinathan, K.P. Characterization of the gene encoding the envelope fusion glycoprotein GP64 from Bombyx mori nucleopolyhedrovirus. Virus Res. 2003, 94, 45–57. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Zhao, S.; Wu, X. Identification of A functional region in Bombyx mori nucleopolyhedrovirus VP39 that is essential for nuclear actin polymerization. Virology 2020, 550, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Yu, H.Z.; Geng, L.; Xu, J.P.; Yu, D.; Zhang, S.Z.; Ma, Y.; Fei, D.Q. Comparative Transcriptome Analysis of Bombyx mori (Lepidoptera) Larval Midgut Response to BmNPV in Susceptible and Near-Isogenic Resistant Strains. PLoS ONE 2016, 11, e0155341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Wang, X.; Xu, J.; Ma, Y.; Zhang, S.; Yu, D.; Fei, D.; Muhammad, A. iTRAQ-based quantitative proteomics analysis of molecular mechanisms associated with Bombyx mori (Lepidoptera) larval midgut response to BmNPV in susceptible and near-isogenic strains. J. Proteom. 2017, 165, 35–50. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, D.; Zhao, Q.; Zhang, G.; Zhang, Y.; Qiu, Z.; Shen, D.; Lu, C. Label-free proteomic analysis of silkworm midgut infected by Bombyx mori nuclear polyhedrosis virus. J. Proteom. 2019, 200, 40–50. [Google Scholar] [CrossRef]
- Mao, F.; Lei, J.; Enoch, O.; Wei, M.; Zhao, C.; Quan, Y.; Yu, W. Quantitative proteomics of Bombyx mori after BmNPV challenge. J. Proteom. 2018, 181, 142–151. [Google Scholar] [CrossRef]
- Lü, P.; Xia, H.; Gao, L.; Pan, Y.; Wang, Y.; Cheng, X.; Lü, H.; Lin, F.; Chen, L.; Yao, Q.; et al. V-ATPase Is Involved in Silkworm Defense Response against Bombyx mori Nucleopolyhedrovirus. PLoS ONE 2013, 8, e64962. [Google Scholar] [CrossRef] [Green Version]
- Pan, M.H.; Xiao, S.Q.; Chen, M.; Hong, X.J.; Lu, C.J.V.C.; Animal, D.B. Establishment and characterization of two embryonic cell lines of Bombyx mori. Vitro Cell. Dev. Biol. Animal 2007, 43, 101–104. [Google Scholar] [CrossRef]
- Mon, H.; Kobayashi, I.; Ohkubo, S.; Tomita, S.; Lee, J.; Sezutsu, H.; Tamura, T.; Kusakabe, T. Effective RNA interference in cultured silkworm cells mediated by overexpression of Caenorhabditis elegans SID-1. RNA Biol. 2012, 9, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.; Cheng, T.; Guo, Y.; Lin, P.; Zhao, P.; Liu, C.; Kusakabe, T.; Xia, Q. Bombyx mori epidermal growth factor receptor is required for nucleopolyhedrovirus replication. Insect Mol. Biol. 2018, 27, 464–477. [Google Scholar] [CrossRef]
- Boccard, J.; Rutledge, D.N. A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multiblock Omics data fusion. Anal. Chim. Acta 2013, 769, 30–39. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, X.; Chen, Q.; Hou, Y.; Xia, Q.; Zhao, P. Metabolomics Analysis of the Larval Head of the Silkworm, Bombyx mori. Int. J. Mol. Sci. 2016, 17, 1460. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, Y.; Liu, Q.; Tan, X.; Xie, X.; Xia, Q.; Zhao, P. GC/MS-based metabolomics analysis reveals active fatty acids biosynthesis in the Filippi’s gland of the silkworm, Bombyx mori, during silk spinning. Insect Biochem. Mol. Biol. 2019, 105, 1–9. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, F.; Xu, S.; Wang, R.; Chen, W.; Hou, K.; Tian, C.; Wang, F.; Yu, L.; Lu, Z.; et al. Genetically engineered bi-functional silk material with improved cell proliferation and anti-inflammatory activity for medical application. Acta Biomater. 2019, 86, 148–157. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, X.; Zhang, Q.; Xu, W.; Wang, X.; Wang, F.; Zhao, P.; Xia, Q. The Antiviral Molecule 5-Pyridoxolactone Identified Post BmNPV Infection of the Silkworm, Bombyx mori. Int. J. Mol. Sci. 2021, 22, 7423. https://doi.org/10.3390/ijms22147423
Hua X, Zhang Q, Xu W, Wang X, Wang F, Zhao P, Xia Q. The Antiviral Molecule 5-Pyridoxolactone Identified Post BmNPV Infection of the Silkworm, Bombyx mori. International Journal of Molecular Sciences. 2021; 22(14):7423. https://doi.org/10.3390/ijms22147423
Chicago/Turabian StyleHua, Xiaoting, Quan Zhang, Wei Xu, Xiaogang Wang, Fei Wang, Ping Zhao, and Qingyou Xia. 2021. "The Antiviral Molecule 5-Pyridoxolactone Identified Post BmNPV Infection of the Silkworm, Bombyx mori" International Journal of Molecular Sciences 22, no. 14: 7423. https://doi.org/10.3390/ijms22147423
APA StyleHua, X., Zhang, Q., Xu, W., Wang, X., Wang, F., Zhao, P., & Xia, Q. (2021). The Antiviral Molecule 5-Pyridoxolactone Identified Post BmNPV Infection of the Silkworm, Bombyx mori. International Journal of Molecular Sciences, 22(14), 7423. https://doi.org/10.3390/ijms22147423





