Abstract
Studying the long-term impact of continuous-flow left ventricular assist device (CF-LVAD) offers an opportunity for a complex understanding of the pathophysiology of vascular changes in aortic tissue in response to a nonphysiological blood flow pattern. Our study aimed to analyze aortic mRNA/miRNA expression changes in response to long-term LVAD support. Paired aortic samples obtained at the time of LVAD implantation and at the time of heart transplantation were examined for mRNA/miRNA profiling. The number of differentially expressed genes (Pcorr < 0.05) shared between samples before and after LVAD support was 277. The whole miRNome profile revealed 69 differentially expressed miRNAs (Pcorr < 0.05). Gene ontology (GO) analysis identified that LVAD predominantly influenced genes involved in the extracellular matrix and collagen fibril organization. Integrated mRNA/miRNA analysis revealed that potential targets of miRNAs dysregulated in explanted samples are mainly involved in GO biological process terms related to dendritic spine organization, neuron projection organization, and cell junction assembly and organization. We found differentially expressed genes participating in vascular tissue engineering as a consequence of LVAD duration. Changes in aortic miRNA levels demonstrated an effect on molecular processes involved in angiogenesis.
1. Introduction
The use of continuous-flow left ventricular assist devices (CF-LVADs) in patients with end-stage heart failure has become a widely used and sustainable treatment strategy, both as a bridge to transplant (BTT) and as destination therapy (DT). CF-LVAD may induce pathological changes to the aortic wall and aortic valve [1]. Important histologic changes in the aortic wall, before and after CF-LVAD implantation, with degeneration of smooth muscle cells and elastic fibers, were previously reported [2]. CF-LVADs may contribute to the deterioration of aortic functional parameters (e.g., aortic stiffness) or structural changes (e.g., increase of wall thickness or collagen content) through adverse effects of the nonphysiological flow [3]. The nonpulsatile flow of CF-LVAD generates dynamic remodeling within the aorta. Remodeling within the aortic root and proximal ascending aorta may also contribute to the pathophysiology of aortic regurgitation with CF-LVAD. Studies have demonstrated that the proximal aorta dilates after chronic CF-LVAD support, and that an increasing aortic diameter was associated with the development of aortic regurgitation [4].
MiRNAs are endogenous small noncoding RNAs that regulate mRNA translation of target genes through the RNA interference pathway, strongly influencing a wide range of cellular processes and biological pathways [5]. As such, miRNAs are fine-tuners of gene expression patterns in response to pathophysiological stimuli. Most miRNAs are more ubiquitously expressed and are not cell-type specific. Thus, many miRNAs are expressed at relatively low levels under basal conditions, but during pathological stress, are strongly upregulated [6]. Changes in tissue miRNA expression levels due to blood flow can potentially affect networks of genes regulating endothelial and vascular smooth muscle cell (SMC) function, inflammation, and atherosclerosis [7].
Alteration in gene expression reflects changes in cellular function and behavior, in development, and disease states. The major cardiovascular diseases, including coronary artery disease, myocardial infarction, congestive heart failure and common congenital heart disease, are caused by multiple genetic and environmental factors, as well as the interactions between them. The underlying molecular pathogenic mechanisms for these disorders are still largely unknown, but gene expression may play a central role in the development and progression of cardiovascular disease [8]. Gene expression analysis can also contribute to understanding and discovering novel and sensitive biomarkers of cardiovascular disease. Over the past two decades, methods of measuring gene expression have improved dramatically with a plethora of hybridization arrays available, followed by RNA-Seq, the sequencing of short or long RNA reads using massively parallel sequencing technology [9].
Studying the long-term use of LVADs offers an opportunity for a complex understanding of the pathophysiology of vascular changes in patients with mechanical circulatory support, which produce nonphysiological blood flow patterns. The aim of the present study was therefore to detect accurate mRNA/miRNA associations in the aorta in response to long-term LVAD.
2. Results
The principal component analysis of expression profiles segregated samples before and after LVAD support (Supplementary Figure S1). The mRNAs profile (Figure 1A) demonstrated differentially expressed genes (DEGs). We identified a total of 277 DEGs (Pcorr < 0.05) after long-term LVAD support. 141 DEGs were upregulated in aortic tissue after LVAD support. Between twenty DEGs, we identified Collagen Type I Alpha 2 Chain (COL1A2); Chemokine-like receptor 1 (CMKLR1); S100 calcium-binding protein A4 (S100A4); Elastin (ELN); and Collagen Type III Alpha 1 Chain (COL3A1) to be upregulated (all Pcorr < 0.0001). Solute Carrier Family 12 Member 2 (SLC12A2); Pyruvate Dehydrogenase Kinase 4 (PDK4); Inhibitor of DNA Binding 1 (ID1); Arginine and Glutamate Rich 1 (ARGLU1) and noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) were downregulated (all Pcorr < 0.0001, Supplementary Figures S2 and S3). The Wald test performed on samples after support to assess the effect of HeartMate II (HM II), and HeartMate 3 (HM 3) devices did not show any significantly differentially expressed genes (Supplementary Data S1).
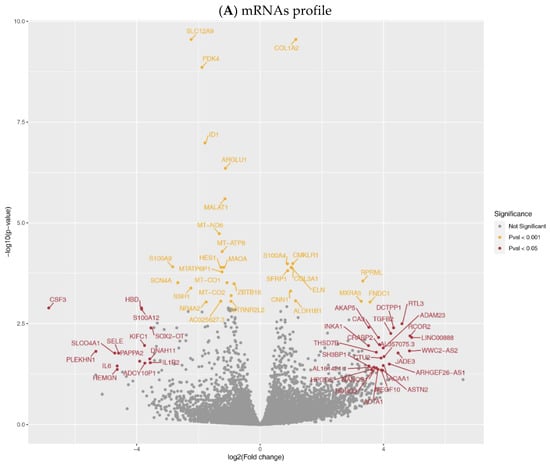
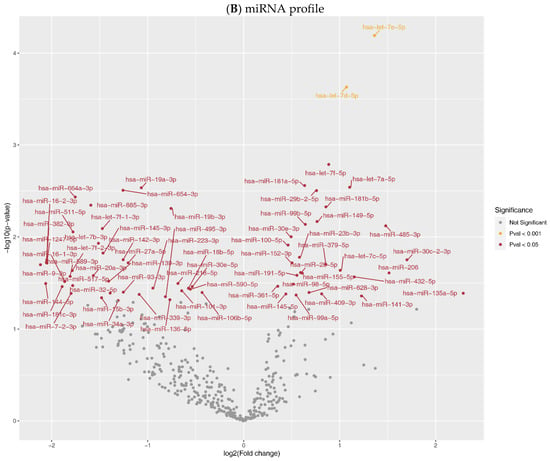
Figure 1.
Expression profiles in aortic tissue. Volcano plot of comparative mRNA resp. miRNAs expression profiles in samples before and after LVAD support; x-axis indicates difference in expression level on a log2 scale; y-axis represents corresponding P-values on a negative log scale. (A) mRNA profile: yellow points indicate mRNAs with Pcorr < 0.001; red points indicate mRNAs with Pcorr < 0.05, & Pcorr > 0.001, & |logfc| > 3.5.; (B) miRNA profile: yellow points indicate miRNAs with Pcorr < 0.001; red points indicate miRNAs with Pcorr < 0.05, & Pcorr > 0.001.
Gene ontology (GO) analysis identified 31 biological processes in GO terms enriched in explanted tissue (false discovery rate (FDR) q-value < 0.001), predominantly involved in extracellular matrix organization and collagen fibril organization. Moreover, 17 molecular functions of GO terms, and 10 cellular components of GO terms were also enriched (Supplementary Data S1).
The whole miRNome profile revealed 69 DE miRNAs (Pcorr<0.05, after Bonferroni correction (BFC), Figure 1B). A total of 30 miRNAs were upregulated (Supplementary Data S1), and the most increased expression was detected for let-7a/d/e/f; miR-181a/b; miR-29b; miR-149 and miR-99b (all Pcorr < 0.01). The opposite effect of LVAD was found on miR-19a/b; miR-654; miR-664a; miR-885; and -511 (all Pcorr < 0.01).
With the usage of miRWalk and miRDB, 170 DEGs were identified as potential targets of upregulated miRNAs and 159 DEGs as potential targets of downregulated miRNAs. From these, 138 DEGs were possible targets of both group miRNAs (Supplementary Data S1).
In order to discover the relationship between deregulated miRNAs and mRNAs, Pearson correlation between differentially expressed miRNAs and all potential target mRNAs was assessed (Figure 2). This integrated mRNA/miRNA analysis revealed that potential targets of miRNAs upregulated in explanted samples are mainly involved in GO biological process terms related to cell junction assembly and cell junction organization (FDR q-value < 0.05, Supplementary Data S1). Enriched GO terms in targets of downregulated miRNAs were related to dendritic spine organization and neuron projection organization (FDR q-value < 0.05, Supplementary Data S1). Moreover, an enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway related to endocytosis was found (FDR q-value < 0.05, Supplementary Data S1).
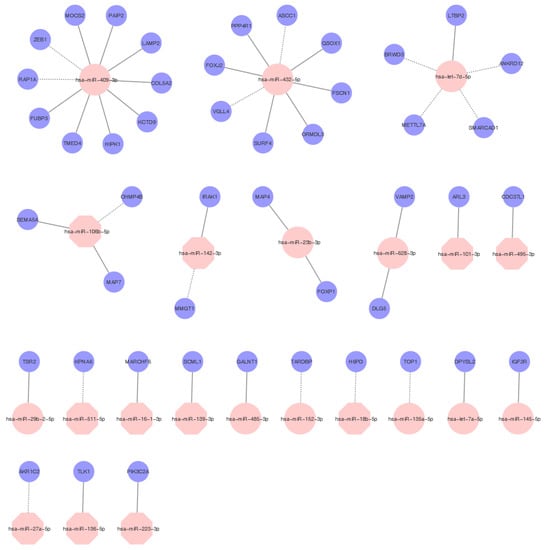
Figure 2.
MiRNAs regulatory network. Examples of flexible and selective regulatory networks between miRNAs and mRNAs in samples after LVAD support. Selected overexpressed miRNAs are highlighted in pink circles, and underexpressed miRNAs are highlighted in pink octagons. The targets are highlighted in blue circles. The full line indicates a positive correlation, and the discontinued line indicates a negative correlation with target mRNAs.
3. Discussion
Our multiple regression analysis is, to our best knowledge, the first study focused on changes in mRNA/miRNA expression in paired aortic samples collected before LVAD implantation and at the time of LVAD explantation, during heart transplantation (HTx).
We compared gene expression profiles using a robust and simple mRNA sequencing method. We found the significant DEGs were predominantly involved in extracellular matrix (ECM) and collagen fibril organization in aortic tissue after LVAD explantation. ECM is an active and dynamic structure with a fundamental role in regulating vascular function in normal and pathological conditions. Homeostasis of the vascular ECM may affect intrinsic properties of the arterial wall and arterial stiffness [10]. The ECM is a key component of the local cellular microenvironment. It comprises structural proteins (e.g., elastin and collagen), proteoglycans, and glycosaminoglycans. Among the different cell types, smooth muscle cells (SMCs) and fibroblasts are examples of cells that produce significant ECM. Interestingly, Coffey et al. [11], using analysis of the integrated miRNA/mRNA network, identified pathways predominantly involved in extracellular matrix function in patients affected by aortic stenosis.
The previously reported histologic analysis showed significant degenerative changes in the aortic wall, SMCs disorientation and depletion, elastic fiber fragmentation and depletion, medial fibrosis, and atherosclerosis changes in ascending aortic tissue at the follow-up than at device implantation [2]. Furthermore, there was evidence of structural remodeling within the aortas of CF-LVAD patients, including an increase in total wall thickness, an increase in collagen content, and reduced elastin content that may explain the increase in vessel stiffness [12]. We identified elevated expression of collagens COL1A1 and COL3A1. Collagen and elastin are the most abundant ECM proteins of the aortic wall, and they are responsible for characteristic mechanical properties—tensile strength and elasticity. Over-accumulated collagen in the aorta may lead to medial fibrosis, hypothetically resulting in decreased arterial distensibility [13]. Surprisingly, we also detected elevated expression of ELN, which is usually lowered in processes related to atherosclerosis, such as pathological flow. We may only hypothesize whether this phenomenon can be attributed to compensatory mechanisms reacting on the non-physiological flow pattern or should be a subject for further research, as the reason for this observation is unclear at this stage of research.
We found overexpressed S100A4, a member of the large family of S100 proteins, under LVAD support. S100A4 controls different cellular pathways, exerting numerous effects on processes that are cell- and tissue-type dependent. In activated fibroblasts, endothelial, dendritic, and mast cells, as well as in macrophages, monocytes, neutrophils, and T-lymphocytes, S100A4 has a significant role in stimulating invasion and migration, cytoskeletal dynamics and in promoting proinflammatory phenotypes [14]. S100A4 represents a well-known marker that characterizes a complex biological process where endothelial cells assume a mesenchymal phenotype, known as the “endothelial-to mesenchymal transition”, changing morphology and functions, acquiring accentuated motility and contractile properties, typical of fibrotic processes [15]. Furthermore, upregulated CMKLR1 is currently the only chemerin receptor. CMKLR1 receptor, and the proposed pro- and anti-inflammatory properties of chemerin, suggest a role of this adipokine in inflammatory states and possibly atherosclerosis. It was reported that foam cell CMKLR1 expression strongly and positively correlates with aortic atherosclerosis, but only marginally with coronary atherosclerosis [16]. Our findings of aortic increased expression levels of COL1A2; CMKLR1; S100A4; and COL3A1 may support previously mentioned morphological changes in the aorta under CF-LVAD.
Changes in aortic wall functional properties as the possible consequence of a pulsatility decrement caused by implantation of CF-LVAD were described [3]. Patel et al. reported that patients with CF-LVADs before heart transplant had an increase in proximal aortic stiffness compared with patients without an LVAD or with pulsatile flow LVADs before transplant [3,17]. DEGs involved in ECM organization could also suggest a potential link with the development of acquired aortic insufficiency (AI), a significant complication that develops following the implantation of CF-LVAD [18,19]. We detected downregulated MALAT1, a gene coding stiffness-sensitive long non-coding RNA. This non-coding RNA regulates stiffness-dependent VSMC proliferation and migration [20], which may influence aortic functional properties.
Despite the advantages and improving results of the CF-LVAD therapy, the loss of pulsatility may lead to different complications on the micro and macrovascular levels. Vascular changes may be linked with the occurrence of clinically adverse events related to CF-LVAD therapy, such as non-surgical bleeding, e.g., gastrointestinal bleeding related to arteriovenous malformations [21] or von Willebrand factor (vWF) deficiency [22], or other clinical complications such as cerebrovascular events [23,24], device thrombosis [25,26] or development of aortic insufficiency [27,28].
One of the pathogenetic mechanisms of cardiovascular complications with CF-LVADs may be endothelial dysfunction. Endothelial dysfunction is related to heart failure in general. After the implantation of the device, the endothelial dysfunction does not improve and may even deteriorate [29]. In our miRNAs profile, we identified deregulated multiple miRNAs involved in vascular remodeling (Table 1) which potentially involved endothelial dysfunction progression. Several miRNAs have been shown to control the varying mechanisms which govern SMC plasticity [30]. In response to LVAD, we found upregulated miRNAs that influence SMC dynamics and downregulated miRNAs known to stimulate apoptosis during atherosclerosis plaque development. Leeper et al. reported that chronic SMC apoptosis accelerates vascular disease progression, promotes calcification, and induces features of medial degeneration, like atrophy, elastin fragmentation, and enhanced glycosaminoglycan deposition, thus worsening endothelial dysfunction [30].

Table 1.
miRNAs participated in vascular remodeling in aortic tissue.
Several authors, including Morgan et al., observed worsened endothelial function in long-term CF-LVAD patients [31]. In our study, we observed dysregulation in miRNAs participating in the regulation of vascular development, growth, and differentiation [32], which may indicate the role of these miRNAs in the further development of endothelial dysfunction. Interestingly, we also identified upregulated miRNAs that regulate mRNAs encoded by genes in human endothelial cells related to vascular function and blood pressure regulation [33].
Correlation analysis revealed that miR-409-3p (upregulated in explanted tissue), one of the most potent fibrinogen downregulating miRs [45], potentially affects the expression of the highest number of genes (10 genes with a correlation coefficient higher than |7|). However, only two genes correlated negatively, suggesting they may be direct targets (ZEB1 and RAP1A). ZEB1 gene is associated with the regulation of vasculogenesis [46], whereas RAP1A promotes angiogenesis and dynamic regulation of endothelial barrier [47].
It should be mentioned that in our study, patients with two types of LVAD were studied—an axial-flow LVAD HeartMate II and the HeartMate 3, a centrifugal-flow pump with intrinsic artificial pulsatility. Nevertheless, this intrinsic pulsatility was originally designed to enhance pump washout and prevent blood stasis and thrombosis. Our previous work found that this pulsatility does not avert endothelial dysfunction [29,48]. Therefore, the HeartMate 3 should also be considered as CF-LVAD. We are aware that our population of patients is not exceedingly large; nevertheless, we did not observe any differences between the pump types.
The vasculature is one of the most dynamic tissues that encounter numerous mechanical cues derived from pulsatile blood flow, blood pressure, the activity of smooth muscle cells in the vessel wall, and the transmigration of immune cells [49]. Endothelial cell junction assembly and cell junction organization play pivotal roles in tissue integrity, barrier function, and cell–cell communication, respectively [50]. In this study, a multistep approach combining mRNA and miRNA expression profiles and bioinformatics analysis was adopted to identify the mRNA/miRNA regulatory network. Enriched GO terms in targets of upregulated miRNAs were related to cell junction assembly and cell junction organization.
4. Materials and Methods
4.1. Subjects
All examined individuals provided their informed consent, which the institution’s ethics committee approved together with the study protocol. The protocol of this study was conducted according to the principles of the Declaration of Helsinki [51].
A total of 16 patients (median age 57 years, range 18–65) who required mechanical circulatory support from HeartMate II (N = 4) and HeartMate 3 (N = 12) as a bridge to transplantation or destination therapy from July 2015 to March 2018 at the Institute for Clinical and Experimental Medicine in Prague, were enrolled in our study. The etiology of heart failure was predominantly nonischemic dilated cardiomyopathy (N = 13). The median LVAD support duration was 382 days (ranging from 162 to 887 days). The basic characteristics of the patients are summarized in Table 2.

Table 2.
Demographics of the patients selected for mRNA/miRNA analysis.
4.2. Sampling
Paired aortic tissue was obtained at the time of LVAD implantation and at the time of HTx from CF-LVAD patients. Approximately 30 mg of tissue was immediately after exclusion from aorta inserted into RNase/DNase-free tubes pre-filled with All Protect Tissue Reagent (Qiagen GmBH Strasse 1, Hilden, Germany). Samples were stored for 2–4 weeks at 4 °C and then at −80 °C before RNA extraction. The storage time of tissue ranged from 33 to 876 days.
4.3. mRNA and miRNA Analysis
Total RNA, including miRNA, was extracted from 10 mg of aortic tissue according to protocol using the miRCURYTM RNA isolation kit for tissue (Qiagen GmBH Strasse 1, Hilden, Germany). RNA quality and quantity were assessed using a Fragment Analyzer system (Agilent technologies, 301 Stevens Creek Blvd., Santa Clara, CA, USA).
Gene expression was measured in paired samples from 10 patients. QuantSeq 3′ mRNA sequencing for RNA quantification was performed using a high-throughput technique using 3′mRNA-Seq Library Prep Kit FWD and 3′mRNA-Seq Library Prep Kit REV (https://www.lexogen.com/quantseq-3mrna-sequencing/; accessed on 20 March 2020) at Lexogen (Campus Vienna Biocenter 5, 1030 Vienna, Austria). QuantSeq. 3′ mRNA library preparation predominantly produces fragments for sequencing close to the 3′ end of polyadenylated mRNA, generally from the last exon and the 3′ untranslated region (3 UTR) [52]. The total RNA input was 20 ng. There was no prior poly(A) enrichment or rRNA depletion. The QuantSeq Forward kit has an oligo (dT) primer containing the Illumina-specific Read 2 linker (P7), which is annealed to the 3′ end of the mRNA fragment to synthesize the first cDNA strand via reverse transcriptase. The second strand synthesis is commenced by random priming and DNA polymerase extension. The random primer contains the Illumina-specific Read 1 linker sequence (P5). Sequencing commences from the Read 1 sequencing primer and goes toward the poly(A) tail with only one fragment produced per transcript [9].
SYBR green-based real-time quantitative PCR (RT-PCR) for miRNA profiling (in a total of 16 patients) was performed using miRNome Panels (Qiagen GmBH Strasse 1, Hilden, Germany). Passive Reference Dye (ROXTM 30 nm) was included for all PCR reactions. Measurement was performed using the QuantStudio6 Flex instrument (ThermoFisher Scientific, 81 Wyman Street, Waltham, MA, USA). Inter-plate calibrators (IPC) for calibration between PCR plate runs, and spike-in controls to ensure the quality of RNA isolation, cDNA synthesis reaction, and PCR was included in each measurement [53].
4.4. Processing of mRNA Sequencing Data
QuantSeq 3′ mRNA sequencing produced 140 million reads with 35–76 bp length. Raw reads were trimmed of bases with Phred 33 quality lower than 30 using Cutadapt software v2.9 [54]. Reads mapping to rRNA and UniVec (common vector contaminations in RNA sequencing) databases were discarded. Mapping was performed using a bowtie aligner [55], with one mismatch allowed. The remaining reads were mapped with STAR aligner to the human genome (GRCh 38.95), with one mismatch allowed [56]. Only uniquely mapping reads were assigned to individual genes using featureCounts software [57].
4.5. DESeq2 Analysis of mRNA Expression
Genes with less than 10 counts per all samples were removed before further analysis. Raw counts were normalized using the median ratio method built in DESeq2 software [58]. To account for paired samples, parameters included in the DESeq2 model were patient IDs and conditions before (implant) and after (explant) LVAD. Differential expression between implant and explant samples was tested with one parameter Wald test built in DESeq2; p values were adjusted for multiple testing with Bonferroni correction (BFC). A p-value < 0.05 was considered statistically significant.
4.6. Gene Ontology Analysis
Gene ontology was performed using Gene Ontology enRIchment anaLysis and visuaLizAtion tool (Gorilla), a web-based tool [59]. All genes expressed in measured samples (threshold > 10 raw counts per all samples) were ranked using the following formula –log (p-value) * log2FC; the resulting list was used as input for calculating the p-value of the minimum hypergeometric score, as described in detail by [60]. KEGG pathway analysis was performed with clusterProfiler tool using 10,000 permutations and a gene set size between 3–800 genes. As input for KEGG pathway analysis, the same ranked list of genes was used for GO analysis [61,62].
4.7. miRNA Profile Analysis
Gene Expression software (GenEx SW, Multid Analysis AB, Göteborg, Sweden) was used for miRNA expression analysis. Ct values higher than 35 were replaced by 35. A total of 330 miRNAs with a call rate <40% (i.e., more than 60% data are invalid of that miRNA) were removed from further analysis. The missing data, exceedingly low miRNA levels, were replaced by deltaCt + 2 (representing at least 1/4 of the detectable miRNAs amount). Data were normalized with the mean expression of all miRNAs and converted to relative quantities and Log2. P values were corrected for multiple testing with BFC. A p-value < 0.05 was considered statistically significant.
4.8. Integrated mRNA/miRNA Analysis
Differentially expressed miRNAs were used to predict mRNA targets in miRWalk (http://mirwalk.umm.uni-heidelberg.de/search_mirnas/; accessed on 18 October 2020) and miRDB (http://www.mirdb.org/; accessed on 18 October 2020) databases. miRDB target prediction was restricted to gene targets with prediction scores less than 60, and miRNAs with more than 2000 genes in the genome were excluded. For prediction in the miRWalk database, no restrictions were used. All genes expressed in our samples, which appeared in the predicted targets in either of the databases, were used for correlation analysis. Pearson correlation was used to correlate log10 transformed normalized expression values of differentially expressed miRNAs with log10 transformed normalized expression values of expressed genes. Only pairs with a correlation better than −0.7/0.7 were considered for further GO analysis and KEGG pathway. GO analysis and KEGG pathway were performed using clusterProfiler with the same parameters described in Section 4.6.
5. Conclusions
The study provides additional insight into the pathophysiology of vascular changes observed in patients after LVAD implantation. Significant regulation of mRNAs involved in ECM and collagen fiber organization in response to the implantation of LVAD was observed, which may suggest infliction of ECM homeostasis resulting in changes of intrinsic properties of the vascular wall and arterial stiffness.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22147414/s1.
Author Contributions
P.I. and I.N. designed and directed the study, performed the clinical aspects of the project, contributed to the interpretation of the results, wrote and edited the article. D.D. designed and performed the experiments, analyzed the data, and wrote the article. Data curation was performed by Z.T. Data analysis conducted by S.B. Data review was performed by J.A.H. The original draft was written by D.D. and P.I. All authors reviewed the original draft and approved the final version of the manuscript.
Funding
This study is supported by the Ministry of Health of the Czech Republic under grant no. 16-27630A and by MH CZ-DRO (“Institute for Clinical and Experimental Medicine–IKEM, IN 00023001”). All rights reserved.
Institutional Review Board Statement
The study was approved on 10 June 2015 with approval number G-15-06-14 by the Human Ethics Review Board, Institute for Clinical and Experimental Medicine and Thomayer Hospital, Prague, Czech Republic, and was performed in accordance with the guidelines in the Declaration of Helsinki (2000) of the World Medical Association.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
P.I. is a consultant for Abbott and CARMAT S.A.; I.N. is a consultant, has received grant funds and is on advisory boards for Abbott, CARMAT S.A., Leviticus Cadio Ltd., and Eva Heart Inc.; P.I., I.N., and Z.T. have received institutional grant support from Abbott.
References
- Saito, T.; Wassilew, K.; Gorodetski, B.; Stein, J.; Falk, V.; Krabatsch, T.; Potapov, E. Aortic Valve Pathology in Patients Supported by Continuous-Flow Left Ventricular Assist Device. Circ. J. 2016, 80, 1371–1377. [Google Scholar] [CrossRef] [PubMed]
- Segura, A.M.; Gregoric, I.; Radovancevic, R.; Demirozu, Z.T.; Buja, L.M.; Frazier, O.H. Morphologic changes in the aortic wall media after support with a continuous-flow left ventricular assist device. J. Heart Lung Transplant. 2013, 32, 1096–1100. [Google Scholar] [CrossRef]
- Patel, A.C.; Dodson, R.B.; Cornwell, W.K., 3rd; Hunter, K.S.; Cleveland, J.C., Jr.; Brieke, A.; Lindenfeld, J.; Ambardekar, A.V. Dynamic Changes in Aortic Vascular Stiffness in Patients Bridged to Transplant With Continuous-Flow Left Ventricular Assist Devices. JACC Heart Fail. 2017, 5, 449–459. [Google Scholar] [CrossRef]
- Fine, N.M.; Park, S.J.; Stulak, J.M.; Topilsky, Y.; Daly, R.C.; Joyce, L.D.; Pereira, N.L.; Schirger, J.A.; Edwards, B.S.; Lin, G.; et al. Proximal thoracic aorta dimensions after continuous-flow left ventricular assist device implantation: Longitudinal changes and relation to aortic valve insufficiency. J. Heart Lung Transplant. 2016, 35, 423–432. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Thum, T. Facts and updates about cardiovascular non-coding RNAs in heart failure. ESC Heart Fail. 2015, 2, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Williams, D.; Sur, S.; Wang, J.Y.; Jo, H. Role of flow-sensitive microRNAs and long noncoding RNAs in vascular dysfunction and atherosclerosis. Vasc. Pharm. 2019, 114, 76–92. [Google Scholar] [CrossRef]
- Archacki, S.; Wang, Q. Expression profiling of cardiovascular disease. Hum. Genom. 2004, 1, 355–370. [Google Scholar] [CrossRef][Green Version]
- Corley, S.M.; Troy, N.M.; Bosco, A.; Wilkins, M.R. QuantSeq. 3′ Sequencing combined with Salmon provides a fast, reliable approach for high throughput RNA expression analysis. Sci. Rep. 2019, 9, 18895. [Google Scholar] [CrossRef] [PubMed]
- Junyan, X.; Guo-Ping, S. Vascular wall extracellular matrix proteins and vascular diseases. Mol. Basis Dis. 2014, 1842, 2106–2119. [Google Scholar] [CrossRef]
- Coffey, S.; Williams, M.J.; Phillips, L.V.; Galvin, I.F.; Bunton, R.W.; Jones, G.T. Integrated microRNA and messenger RNA analysis in aortic stenosis. Sci. Rep. 2016, 6, 36904. [Google Scholar] [CrossRef]
- Ambardekar, A.V.; Hunter, K.S.; Babu, A.N.; Tuder, R.M.; Dodson, R.B.; Lindenfeld, J. Changes in Aortic Wall Structure, Composition, and Stiffness With Continuous-Flow Left Ventricular Assist Devices: A Pilot Study. Circ. Heart Fail. 2015, 8, 944–952. [Google Scholar] [CrossRef]
- Wang, X.; LeMaire, S.A.; Chen, L.; Shen, Y.H.; Gan, Y.; Bartsch, H.; Carter, S.A.; Utama, B.; Ou, H.; Coselli, J.S.; et al. Increased collagen deposition and elevated expression of connective tissue growth factor in human thoracic aortic dissection. Circulation 2006, 114, I200–I205. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrosi, N.; Milani, M.; Apolloni, S. S100A4 in the Physiology and Pathology of the Central and Peripheral Nervous System. Cells 2021, 10, 798. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Liu, S.; Qin, Z. Extracellular S100A4 as a key player in fibrotic diseases. J. Cell. Mol. Med. 2020, 24, 5973–5983. [Google Scholar] [CrossRef]
- Kostopoulos, C.G.; Spiroglou, S.G.; Varakis, J.N.; Apostolakis, E.; Papadaki, H.H. Chemerin and CMKLR1 expression in human arteries and periadventitial fat: A possible role for local chemerin in atherosclerosis? BMC Cardiovasc. Disord. 2014, 14, 56. [Google Scholar] [CrossRef]
- Patel, C.B.; Cowger, J.A.; Zuckermann, A. A contemporary review of mechanical circulatory support. J. Heart Lung Transplant. 2014, 33, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Holtz, J.; Teuteberg, J. Management of aortic insufficiency in the continuous flow left ventricular assist device population. Curr. Heart Fail. Rep. 2014, 11, 103–110. [Google Scholar] [CrossRef]
- Imamura, T.; Kim, G.; Nitta, D.; Fujino, T.; Smith, B.; Kalantari, S.; Nguyen, A.; Narang, N.; Holzhauser, L.; Grinstein, J.; et al. Aortic Insufficiency and Hemocompatibility-related Adverse Events in Patients with Left Ventricular Assist Devices. J. Card. Fail. 2019, 25, 787–794. [Google Scholar] [CrossRef]
- Yu, C.K.; Xu, T.; Assoian, R.K.; Rader, D.J. Mining the Stiffness-Sensitive Transcriptome in Human Vascular Smooth Muscle Cells Identifies Long Noncoding RNA Stiffness Regulators. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Demirozu, Z.T.; Radovancevic, R.; Hochman, L.F.; Gregoric, I.D.; Letsou, G.V.; Kar, B.; Bogaev, R.C.; Frazier, O.H. Arteriovenous malformation and gastrointestinal bleeding in patients with the HeartMate II left ventricular assist device. J. Heart Lung Transplant. 2011, 30, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Uriel, N.; Pak, S.W.; Jorde, U.P.; Jude, B.; Susen, S.; Vincentelli, A.; Ennezat, P.V.; Cappleman, S.; Naka, Y.; Mancini, D. Acquired von Willebrand syndrome after continuous-flow mechanical device support contributes to a high prevalence of bleeding during long-term support and at the time of transplantation. J. Am. Coll. Cardiol. 2010, 56, 1207–1213. [Google Scholar] [CrossRef]
- Kato, T.S.; Schulze, P.C.; Yang, J.; Chan, E.; Shahzad, K.; Takayama, H.; Uriel, N.; Jorde, U.; Farr, M.; Naka, Y.; et al. Pre-operative and post-operative risk factors associated with neurologic complications in patients with advanced heart failure supported by a left ventricular assist device. J. Heart Lung Transplant. 2012, 31, 1–8. [Google Scholar] [CrossRef]
- Trachtenberg, B.H.; Cordero-Reyes, A.M.; Aldeiri, M.; Alvarez, P.; Bhimaraj, A.; Ashrith, G.; Elias, B.; Suarez, E.E.; Bruckner, B.; Loebe, M.; et al. Persistent blood stream infection in patients supported with a continuous-flow left ventricular assist device is associated with an increased risk of cerebrovascular accidents. J. Card. Fail. 2015, 21, 119–125. [Google Scholar] [CrossRef]
- Najjar, S.S.; Slaughter, M.S.; Pagani, F.D.; Starling, R.C.; McGee, E.C.; Eckman, P.; Tatooles, A.J.; Moazami, N.; Kormos, R.L.; Hathaway, D.R.; et al. HVAD Bridge to Transplant ADVANCE Trial Investigators. An analysis of pump thrombus events in patients in the HeartWare ADVANCE bridge to transplant and continued access protocol trial. J. Heart Lung Transplant. 2014, 33, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Starling, R.C.; Moazami, N.; Silvestry, S.C.; Ewald, G.; Rogers, J.G.; Milano, C.A.; Rame, J.E.; Acker, M.A.; Blackstone, E.H.; Ehrlinger, J.; et al. Unexpected abrupt increase in left ventricular assist device thrombosis. N. Engl. J. Med. 2014, 370, 33–40. [Google Scholar] [CrossRef]
- Cowger, J.; Pagani, F.D.; Haft, J.W.; Romano, M.A.; Aaronson, K.D.; Kolias, T.J. The development of aortic insufficiency in left ventricular assist device-supported patients. Circ. Heart Fail. 2010, 3, 668–674. [Google Scholar] [CrossRef]
- Pak, S.W.; Uriel, N.; Takayama, H.; Cappleman, S.; Song, R.; Colombo, P.C.; Charles, S.; Mancini, D.; Gillam, L.; Naka, Y.; et al. Prevalence of de novo aortic insufficiency during long-term support with left ventricular assist devices. J. Heart Lung Transplant. 2010, 29, 1172–1176. [Google Scholar] [CrossRef]
- Ivak, P.; Netuka, I.; Tucanova, Z.; Wohlfahrt, P.; Konarik, M.; Szarszoi, O.; Novakova, S.; Kubanek, M.; Lanska, V.; Pitha, J. The Effect of Artificial Pulsatility on the Peripheral Vasculature in Patients with a Continuous-Flow Ventricular Assist Device. Can. J. Cardiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Leeper, N.J.; Maegdefessel, L. Non-coding RNAs: Key regulators of smooth muscle cell fate in vascular disease. Cardiovasc. Res. 2018, 114, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Morgan, N.; Warner, P.; Kiernan, M.; Al-Quthami, A.; Rahban, Y.; Pham, D.T.; DeNofrio, D.; Karas, R.; Kuvin, J. Arterial Stiffness and Vascular Endothelial Function in Patients with Long-Term Continuous-Flow Left Ventricular Assist Devices. J. Card. Fail. 2013, 19, S18. [Google Scholar] [CrossRef]
- Fernández-Hernando, C.; Suárez, Y. MicroRNAs in endothelial cell homeostasis and vascular disease. Curr. Opin. Hematol. 2018, 25, 227–236. [Google Scholar] [CrossRef]
- Kriegel, A.J.; Baker, M.A.; Liu, Y.; Liu, P.; Cowley, A.W., Jr.; Liang, M. Endogenous microRNAs in human microvascular endothelial cells regulate mRNAs encoded by hypertension-related genes. Hypertension 2015, 66, 793–799. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Sit, A.; Feinberg, M.W. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc. Med. 2014, 24, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Ge, J.; Wang, Z.; Ren, J.; Wang, X.; Xiong, H.; Gao, J.; Zhang, Y.; Zhang, Q. Let-7e modulates the inflammatory response in vascular endothelial cells through ceRNA crosstalk. Sci. Rep. 2017, 7, 42498:1–42498:12. [Google Scholar] [CrossRef]
- Pin, A.L.; Houle, F.; Guillonneau, M.; Paquet, E.R.; Simard, M.J.; Huot, J. miR-20a represses endothelial cell migration by targeting MKK3 and inhibiting p38 MAP kinase activation in response to VEGF. Angiogenesis 2012, 15, 593–608. [Google Scholar] [CrossRef]
- Chamorro-Jorganes, A.; Araldi, E.; Rotllan, N.; Cirera-Salinas, D.; Suárez, Y. Autoregulation of glypican-1 by intronic microRNA-149 fine tunes the angiogenic response to FGF2 in human endothelial cells. J. Cell Sci. 2014, 127, 1169–1178. [Google Scholar] [CrossRef]
- Grundmann, S.; Hans, F.P.; Kinniry, S.; Heinke, J.; Helbing, T.; Bluhm, F.; Sluijter, J.P.; Hoefer, I.; Pasterkamp, G.; Bode, C.; et al. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation 2011, 123, 999–1009. [Google Scholar] [CrossRef]
- Kane, N.M.; Howard, L.; Descamps, B.; Meloni, M.; McClure, J.; Lu, R.; McCahill, A.; Breen, C.; Mackenzie, R.M.; Delles, C.; et al. Role of MicroRNAs 99b, 181a, and 181b in the Differentiation of Human Embryonic Stem Cells to Vascular Endothelial Cells. Stem Cells 2012, 30, 643–654. [Google Scholar] [CrossRef]
- Tang, X.; Yin, R.; Shi, H.; Wang, X.; Shen, D.; Wang, X.; Pan, C. LncRNA ZFAS1 confers inflammatory responses and reduces cholesterol efflux in atherosclerosis through regulating miR-654-3p-ADAM10/RAB22A axis. Int. J. Cardiol. 2020, 315, 72–80. [Google Scholar] [CrossRef]
- Chen, C.; Ponnusamy, M.; Liu, C.; Gao, J.; Wang, K.; Li, P. MicroRNA as a Therapeutic Target in Cardiac Remodeling. Biomed. Res. Int. 2017, 2017, 1278436. [Google Scholar] [CrossRef]
- Henn, D.; Abu-Halima, M.; Wermke, D.; Falkner, F.; Thomas, B.; Köpple, C.; Ludwig, N.; Schulte, M.; Brockmann, M.A.; Kim, Y.J. MicroRNA-regulated pathways of flow-stimulated angiogenesis and vascular remodeling in vivo. J. Transl. Med. 2019, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zeng, C.; Wei, M.; Hong, T. MicroRNA-664 regulates cell invasion and migration and epithelial-mesenchymal transition by targeting TGF-β signal in glioblastoma. Int. J. Clin. Exp. Pathol. 2016, 9, 12361–12370. [Google Scholar]
- Shi, M.J.; Xiao, H.M.; Xie, Y.B.; Jiang, J.M.; Zhao, P.T.; Cai, G.S.; Li, Y.X.; Li, S.; Zhang, C.Z.; Cao, M.L.; et al. Differences in MicroRNA Expression in Chronic Hepatitis B Patients with Early Liver Fibrosis Based on Traditional Chinese Medicine Syndromes. Evid. Based Complement. Alternat. Med. 2020, 2020, 5956940. [Google Scholar] [CrossRef] [PubMed]
- Fort, A.; Borel, C.; Migliavacca, E.; Antonarakis, S.E.; Fish, R.J.; Neerman-Arbez, M. Regulation of fibrinogen production by microRNAs. Blood 2010, 116, 2608–2615. [Google Scholar] [CrossRef]
- Jin, L.; Zhang, Y.; Liang, W.; Lu, X.; Piri, N.; Wang, W.; Kaplan, H.J.; Dean, D.C.; Zhang, L.; Liu, Y. Zeb1 promotes corneal neovascularization by regulation of vascular endothelial cell proliferation. Commun. Biol. 2020, 3, 349:1–349:10. [Google Scholar] [CrossRef]
- Singh, B.; Kosuru, R.; Lakshmikanthan, S.; Sorci-Thomas, M.G.; Zhang, D.X.; Sparapani, R.; Vasquez-Vivar, J.; Chrzanowska, M. Endothelial Rap1 (Ras-Association Proximate 1) Restricts Inflammatory Signaling to Protect From the Progression of Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 638–650. [Google Scholar] [CrossRef]
- Witman, M.A.; Garten, R.S.; Gifford, J.R.; Groot, H.J.; Trinity, J.D.; Stehlik, J.; Nativi, J.N.; Selzman, C.H.; Drakos, S.G.; Richardson, R.S. Further Peripheral Vascular Dysfunction in Heart Failure Patients With a Continuous-Flow Left Ventricular Assist Device: The Role of Pulsatility. JACC Heart Fail. 2015, 3, 703–711. [Google Scholar] [CrossRef]
- Dorland, Y.L.; Huveneers, S. Cell–cell junctional mechanotransduction in endothelial remodeling. Cell. Mol. Life Sci. 2017, 74, 279–292. [Google Scholar] [CrossRef]
- Wallez, Y.; Huber, P. Endothelial adherens and tight junctions in vascular homeostasis, inflammation and angiogenesis. Biochim. Biophys. Acta Biomembr. 2008, 1778, 794–809. [Google Scholar] [CrossRef]
- World Medical Association. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. JAMA 2013, 310, 2191–2194. [Google Scholar] [CrossRef]
- Moll, P.; Ante, M.; Seitz, A.; Reda, T. QuantSeq. 3′ mRNA sequencing for RNA quantification. Nat. Methods 2014, 11, 952. [Google Scholar] [CrossRef]
- Dlouha, D.; Blaha, M.; Blaha, V.; Fatorova, I.; Hubacek, J.A.; Stavek, P.; Lanska, V.; Parikova, A.; Pitha, J. Analysis of circulating miRNAs in patients with familial hypercholesterolaemia treated by LDL/Lp(a) apheresis. Atheroscler. Suppl. 2017, 30, 128–134. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef]
- Eden, E.; Lipson, D.; Yogev, S.; Yakhini, Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007, 3, e39. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.; Han, Y.; He, Q. Clusterprofiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).