Pectobacterium parmentieri SCC 3193 Mutants with Altered Synthesis of Cell Surface Polysaccharides Are Resistant to N4-Like Lytic Bacteriophage ϕA38 (vB_Ppp_A38) but Express Decreased Virulence in Potato (Solanum tuberosum L.) Plants
Abstract
1. Introduction
2. Results
2.1. Transposon Mutagenesis and Identification of Tn5 Disrupted Genes in ϕA38-Resistant P. parmentieri SCC 3193
2.2. Characterization of the Disrupted Genes, Transcriptional Organization, Biochemical Pathways and Cellular Enzymatic Networks Conferred by Tn5 Insertion
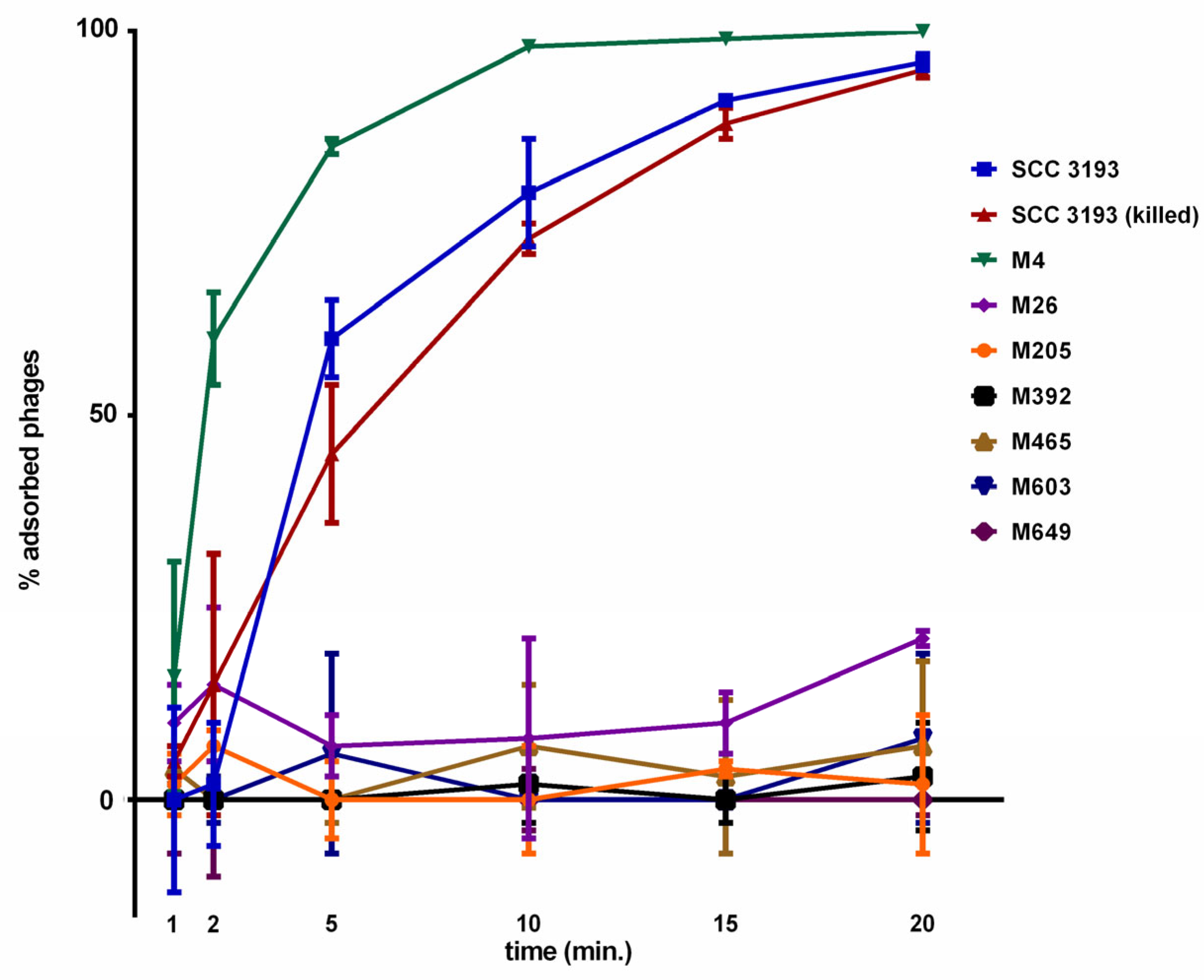
2.3. Adsorption of ϕA38 to Viable and Non-Viable Host Cells
2.4. Identification and Characterization of the Crude Lipopolysaccharide (LPS) Isolated from P. parmentieri Strain SCC 3193 Wild-Type and Tn5 Mutants
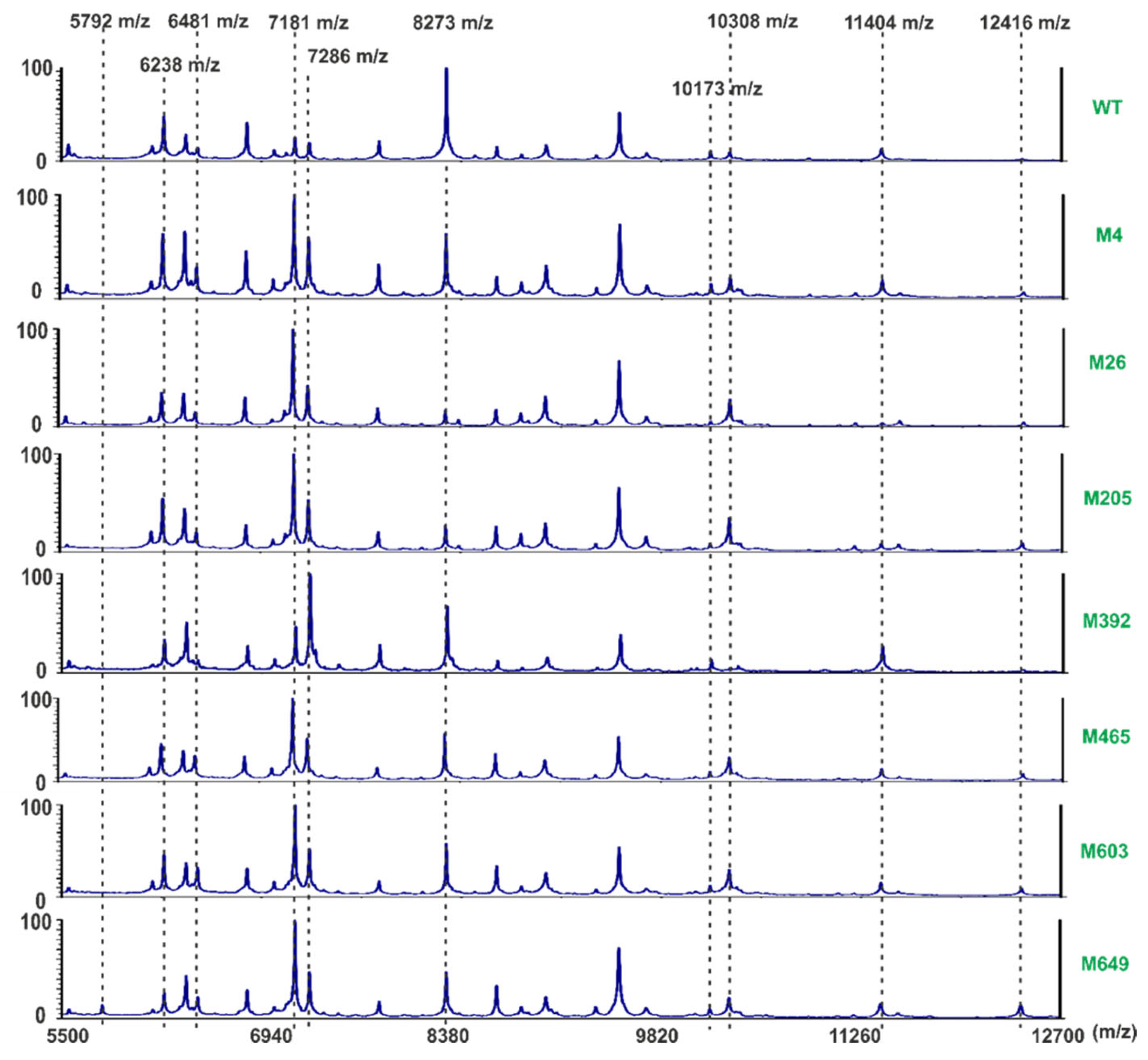
2.5. Mass Spectrometry Analysis of the P. parmentieri SCC 3193 and Tn5 Mutants’ Surface Proteins
2.6. Inactivation of ϕA38 by LPS
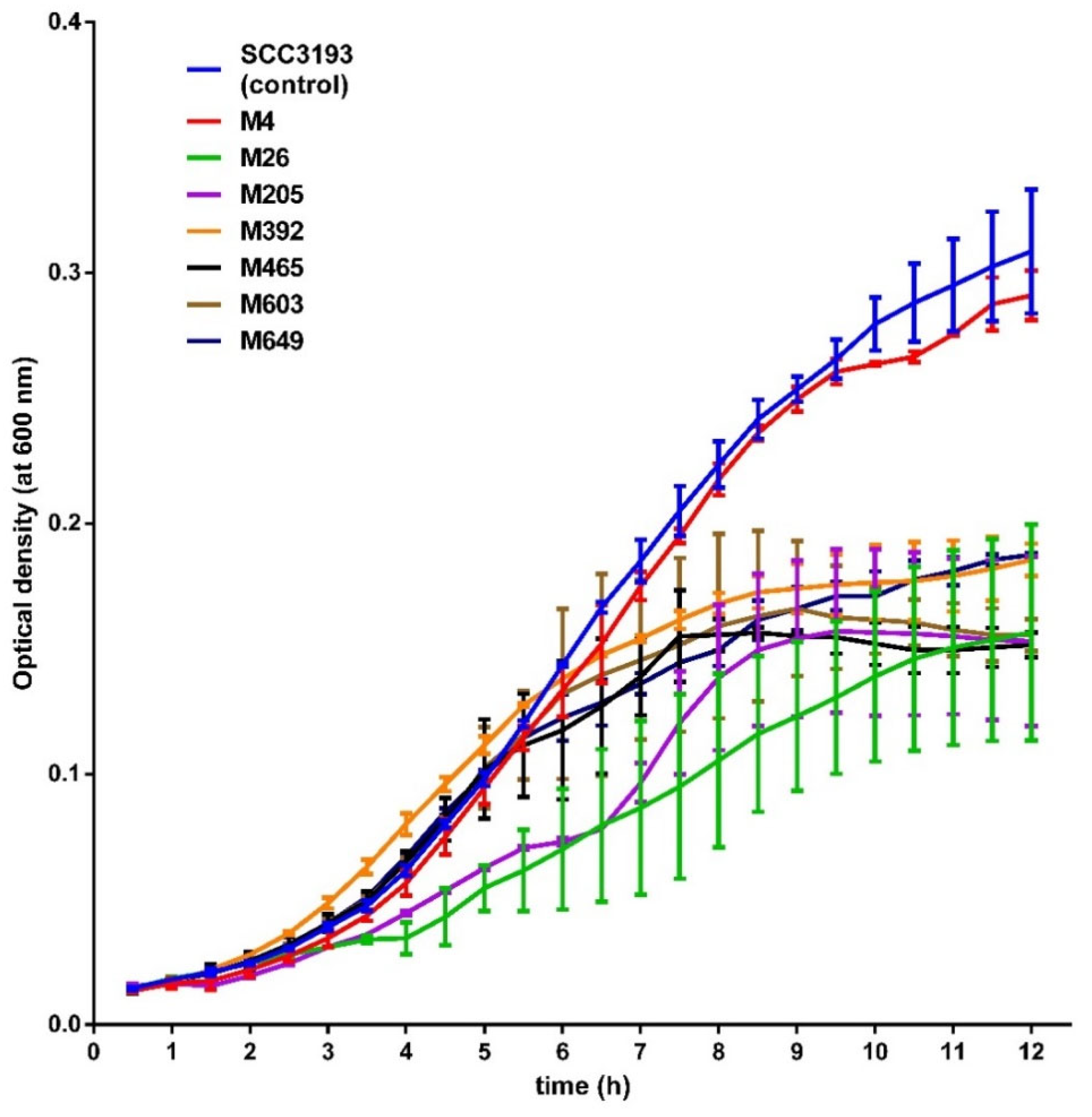
2.7. Phenotypes of P. parmentieri Phage-Resistant Mutants
2.8. Virulence of P. parmentieri Tn5 Mutants
3. Discussion
4. Materials and Methods
4.1. Bacteriophages, Bacterial Strains and Growth Media
4.2. Transposon Mutagenesis with Mini-Tn5
4.3. Verification of Tn5 Mutants by Plating on CVP Medium and Selection of ΦA38-Resistant P. parmentieri Mutants
4.4. Kinetics of ΦA38 Adsorption to Viable P. parmentieri SCC 3193 and P. parmentieri Tn5 Mutants
4.5. Kinetics of ΦA38 Adsorption to Chloramphenicol-Killed P. parmentieri SCC 3193
4.6. Identification of the Tn5 Insertion Sites in Phage-Resistant Mutants by Genome Sequencing
4.7. The Transcriptional Organization, Biochemical Pathways and Cellular Enzymatic Networks Affected by the Transposon Insertions into P. parmentieri Genomes
4.8. Extraction, Quantification, and Visualization of Lipopolysaccharide (LPS) from Wild-Type P. parmentieri Strain SCC 3193 and Selected Phage-Resistant P. parmentieri Tn5 Mutants
4.9. Inactivation of ΦA38 by Lipopolysaccharide (LPS) Isolated from Wild-Type P. parmentieri SCC 3193 Cells
4.10. Determination of the Generation Time of P. parmentieri Tn5 Mutants in Rich and Minimal Media
4.11. Determination of the Average Generation Time of P. parmentieri Tn5 Mutants at pH 5 and pH 10
4.12. Growth of Phage-Resistant Tn5 Mutants at Different Temperatures on Solid Media
4.13. Antibiotic Susceptibility of Tn5 Mutants
4.14. Phenotypic Characterization of P. parmentieri Tn5 Mutants Using BIOLOG Phenotypic Microarrays
4.15. Phenotypic Characterization of Tn5 Mutants Using Plate Assays
4.16. Assessment of Bacterial Cell and Colony Morphology of P. parmentieri Mutants
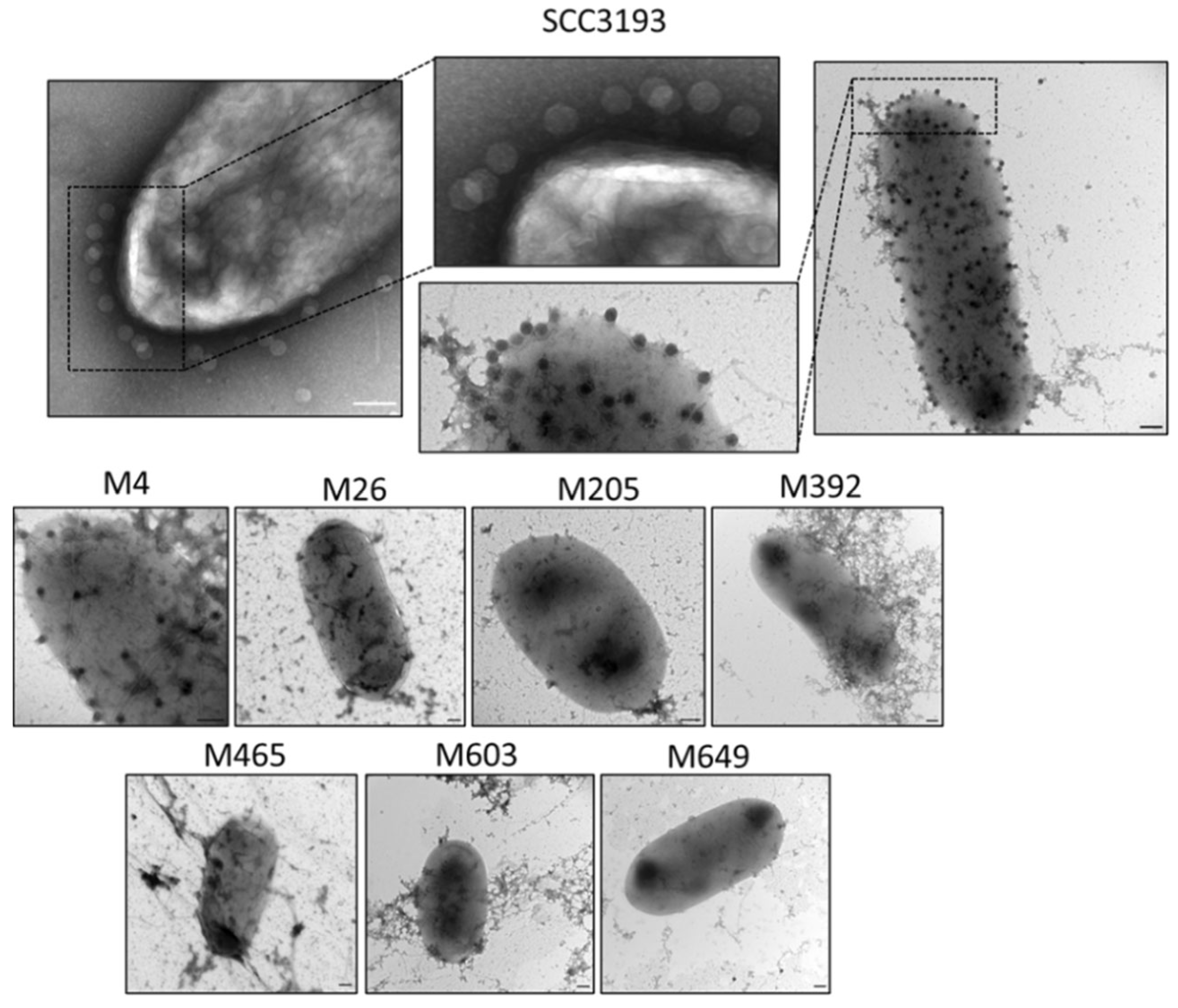
4.17. Interaction of ΦA38 with SCC 3193 and SCC 3193 Tn5 Mutants Assessed with Transmission Electron Microscopy (TEM)
4.18. Visualization of Capsule and Flagella of P. parmentieri Tn5 Mutants with Microscopic Techniques
4.19. Phenotypic Characterization of P. parmentieri Tn5 Mutants Using Whole-Cell MALDI-TOF MS Analyses
4.20. The Ability of P. parmentieri Tn5 Mutants to Macerate Potato Tuber Slices
4.21. Virulence of P. parmentieri Tn5 Mutants in Potato Plants Grown in a Growth Chamber
4.22. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perombelon, M.C.M. Potato diseases caused by soft rot Erwinias: An overview of pathogenesis. Plant Pathol. 2002, 51, 1–12. [Google Scholar]
- Charkowski, A.O. The changing face of bacterial soft-rot diseases. Annu. Rev. Phytopathol. 2018, 56, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Charkowski, A.O. The Soft Rot. Erwinia Plant Assoc. Bact. 2007, 423–505. [Google Scholar]
- Pérombelon, M.C.M. Potato blackleg: Epidemiology, host-pathogen interaction and control. Tijdschr. Over Plantenziekten 1992, 98, 135–146. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Pasanen, M.; Laurila, J.; Brader, G.; Palva, E.T.; Ahola, V.; van der Wolf, J.; Hannukkala, A.; Pirhonen, M. Characterisation of Pectobacterium wasabiae and Pectobacterium carotovorum subsp. carotovorum isolates from diseased potato plants in Finland. Ann. Appl. Biol. 2013, 163, 403–419. [Google Scholar] [CrossRef]
- Suárez, M.B.; Feria, F.J.; Martín-Robles, M.J.; del Rey, F.J.; Palomo, J.L. Pectobacterium parmentieri causing soft rot on potato tubers in Southern Europe. Plant Dis. 2017, 101, 1029. [Google Scholar] [CrossRef]
- Kastelein, P.; Förch, M.G.; Krijger, M.C.; van der Zouwen, P.S.; van den Berg, W.; van der Wolf, J.M. Systemic colonization of potato plants resulting from potato haulm inoculation with Dickeya solani or Pectobacterium parmentieri. Can. J. Plant Pathol. 2020, 43, 1–15. [Google Scholar] [CrossRef]
- Pitman, A.R.; Harrow, S.A.; Visnovsky, S.B. Genetic characterisation of Pectobacterium wasabiae causing soft rot disease of potato in New Zealand. Eur. J. Plant Pathol. 2010, 126, 423–435. [Google Scholar] [CrossRef]
- Khayi, S.; Cigna, J.; Chong, T.M.; Quetu-Laurent, A.; Chan, K.G.; Helias, V.; Faure, D. Transfer of the potato plant isolates of Pectobacterium wasabiae to Pectobacterium parmentieri sp. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5379–5383. [Google Scholar] [CrossRef]
- Van der Wolf, J.M.; de Haan, E.G.; Kastelein, P.; Krijger, M.; de Haas, B.H.; Velvis, H.; Mendes, O.; Kooman-Gersmann, M.; van der Zouwen, P.S. Virulence of Pectobacterium carotovorum subsp. brasiliense on potato compared with that of other Pectobacterium and Dickeya species under climatic conditions prevailing in the Netherlands. Plant Pathol. 2017, 66, 571–583. [Google Scholar] [CrossRef]
- Czajkowski, R.; Perombelon, M.C.M.; van Veen, J.A.; van der Wolf, J.M. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: A review. Plant Pathol. 2011, 60, 999–1013. [Google Scholar] [CrossRef]
- Van der Wolf, J.M.; de Boer, S.H. Bacterial pathogens of potato. In Potato Biology and Biotechnology, Advances and Perspectives; Elsevier: Oxford, UK, 2007; pp. 595–619. [Google Scholar]
- Jones, J.B.; Jackson, L.E.; Balogh, B.; Obradovic, A.; Iriarte, F.B.; Momol, M.T. Bacteriophages for plant disease control. Annu. Rev. Phytopathol. 2007, 45, 245–262. [Google Scholar] [CrossRef]
- Czajkowski, R.; Ozymko, Z.; Lojkowska, E. Isolation and characterization of novel soilborne lytic bacteriophages infecting Dickeya spp. biovar 3 (‘D. solani’). Plant Pathol. 2014, 63, 758–772. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Van Vaerenbergh, J.; Vandenheuvel, D.; Dunon, V.; Ceyssens, P.J.; De Proft, M.; Kropinski, A.M.; Noben, J.P.; Maes, M.; Lavigne, R. T4-related bacteriophage LIMEstone isolates for the control of soft rot on potato caused by ‘Dickeya solani’. PLoS ONE 2012, 7, e33227. [Google Scholar] [CrossRef]
- Czajkowski, R.; Ozymko, Z.; de Jager, V.; Siwinska, J.; Smolarska, A.; Ossowicki, A.; Narajczyk, M.; Lojkowska, E. Genomic, proteomic and morphological characterization of two novel broad host lytic bacteriophages phiPD10.3 and phiPD23.1 infecting pectinolytic Pectobacterium spp. and Dickeya spp. PLoS ONE 2015, 10, e0119812. [Google Scholar] [CrossRef]
- Kabanova, A.P.; Shneider, M.M.; Korzhenkov, A.A.; Bugaeva, E.N.; Miroshnikov, K.K.; Zdorovenko, E.L.; Kulikov, E.E.; Toschakov, S.V.; Ignatov, A.N.; Knirel, Y.A.; et al. Host specificity of the Dickeya bacteriophage PP35 Is directed by a tail spike interaction with bacterial O-antigen, enabling the infection of alternative non-pathogenic bacterial host. Front. Microbiol. 2018, 9, 3288. [Google Scholar] [CrossRef]
- Petrzik, K.; Vacek, J.; Brazdova, S.; Sevcik, R.; Koloniuk, I. Diversity of limestone bacteriophages infecting Dickeya solani isolated in the Czech Republic. Arch. Virol. 2021, 166, 1171–1175. [Google Scholar] [CrossRef]
- Djurhuus, A.M.; Carstens, A.B.; Neve, H.; Kot, W.; Hansen, L.H. Two new Dickeya dadantii phages with odd growth patterns expand the diversity of phages infecting Soft Rot Pectobacteriaceae. PHAGE 2020, 1, 251–259. [Google Scholar] [CrossRef]
- Czajkowski, R. Bacteriophages of Soft Rot Enterobacteriaceae—A minireview. FEMS Microbiol. Lett. 2016, 363, fnv230. [Google Scholar] [CrossRef]
- Jones, J.B.; Vallad, G.E.; Iriarte, F.B.; Obradović, A.; Wernsing, M.H.; Jackson, L.E.; Balogh, B.; Hong, J.C.; Momol, M.T. Considerations for using bacteriophages for plant disease control. Bacteriophage 2012, 2, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; Hendrix, H.; Lucid, A.; Neve, H.; Noben, J.-P.; Franz, C.; O’Mahony, J.; Lavigne, R.; Coffey, A. Novel N4-like bacteriophages of Pectobacterium atrosepticum. Pharmaceuticals 2018, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Smolarska, A.; Rabalski, L.; Narajczyk, M.; Czajkowski, R. Isolation and phenotypic and morphological characterization of the first Podoviridae lytic bacteriophages ϕA38 and ϕA41 infecting Pectobacterium parmentieri (former Pectobacterium wasabiae). Eur. J. Plant Pathol. 2017, 150, 413–425. [Google Scholar] [CrossRef]
- Czajkowski, R.; Rabalski, L.; Kosinski, M.; Smolarska, A. Complete genome sequence and phylogenomic analysis of the first N4-like lytic bacteriophage vB_Ppp_A38 (ϕA38) infecting Pectobacterium parmentieri. Eur. J. Plant Pathol. 2021. under review. [Google Scholar]
- Batinovic, S.; Wassef, F.; Knowler, S.A.; Rice, D.T.F.; Stanton, C.R.; Rose, J.; Tucci, J.; Nittami, T.; Vinh, A.; Drummond, G.R.; et al. Bacteriophages in natural and artificial environments. Pathogens 2019, 8, 100. [Google Scholar] [CrossRef]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Brockhurst, M.A.; Fenton, A.; Roulston, B.; Rainey, P.B. The impact of phages on interspecific competition in experimental populations of bacteria. BMC Ecol. 2006, 6, 19. [Google Scholar] [CrossRef]
- Sanchez, C. Bacterial evolution: Phage resistance comes at a cost. Nat. Rev. Microbiol. 2011, 9, 398–399. [Google Scholar] [CrossRef]
- Leon, M.; Bastias, R. Virulence reduction in bacteriophage resistant bacteria. Front. Microbiol. 2015, 6, 343. [Google Scholar] [CrossRef]
- Brussow, H.; Canchaya, C.; Hardt, W.D. Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004, 68, 560–602. [Google Scholar] [CrossRef]
- Leiman, P.G.; Kanamaru, S.; Mesyanzhinov, V.V.; Arisaka, F.; Rossmann, M.G. Structure and morphogenesis of bacteriophage T4. Cell. Mol. Life Sci. 2003, 60, 2356–2370. [Google Scholar] [CrossRef]
- Chaturongakul, S.; Ounjai, P. Phage-host interplay: Examples from tailed phages and Gram-negative bacterial pathogens. Front. Microbiol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Wiser, M.J.; Lenski, R.E. A comparison of methods to measure fitness in Escherichia coli. PLoS ONE 2015, 10, e0126210. [Google Scholar] [CrossRef]
- Mangalea, M.R.; Duerkop, B.A. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect. Immun. 2020, 88, e00926-19. [Google Scholar] [CrossRef]
- Koskinen, J.P.; Laine, P.; Niemi, O.; Nykyri, J.; Harjunpaa, H.; Auvinen, P.; Paulin, L.; Pirhonen, M.; Palva, T.; Holm, L. Genome sequence of Pectobacterium sp. strain SCC3193. J. Bacteriol. 2012, 194, 6004. [Google Scholar] [CrossRef]
- Kurowski, M.A.; Bujnicki, J.M. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003, 31, 3305–3307. [Google Scholar] [CrossRef]
- Chibani-Chennoufi, S.; Bruttin, A.; Dillmann, M.L.; Brussow, H. Phage-host interaction: An ecological perspective. J. Bacteriol. 2004, 186, 3677–3686. [Google Scholar] [CrossRef]
- Hargreaves, K.R.; Kropinski, A.M.; Clokie, M.R. Bacteriophage behavioral ecology: How phages alter their bacterial host’s habits. Bacteriophage 2014, 4, e29866. [Google Scholar] [CrossRef]
- Czajkowski, R. May the phage be with you? Prophage-like elements in the genomes of Soft Rot Pectobacteriaceae: Pectobacterium spp. and Dickeya spp. Front. Microbiol. 2019, 10, 138. [Google Scholar] [CrossRef]
- Kim, H.; Kim, M.; Bai, J.; Lim, J.A.; Heu, S.; Ryu, S. Colanic acid is a novel phage receptor of Pectobacterium carotovorum subsp. carotovorum phage POP72. Front. Microbiol. 2019, 10, 143. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef]
- Simpson, B.W.; Trent, M.S. Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 2019, 17, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Slauch, J.M.; Lee, A.A.; Mahan, M.J.; Mekalanos, J.J. Molecular characterization of the oafA locus responsible for acetylation of Salmonella typhimurium O-antigen: oafA is a member of a family of integral membrane trans-acylases. J. Bacteriol. 1996, 178, 5904–5909. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.; Herbert, S.; Jakob, A.; Vollmer, W.; Götz, F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 2005, 55, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.L.; Lin, Y.C.; Lin, R.H.; Wei, C.F.; Huang, Y.C.; Peng, H.L.; Huang, H.C. Effects of galU mutation on Pseudomonas syringae—plant interactions. Mol. Plant Microbe Interact. 2010, 23, 1184–1196. [Google Scholar] [CrossRef] [PubMed]
- Wandersman, C.; Letoffe, S. Involvement of lipopolysaccharide in the secretion of Escherichia coli alpha-haemolysin and Erwinia chrysanthemi proteases. Mol. Microbiol. 1993, 7, 141–150. [Google Scholar] [CrossRef]
- Schoonejans, E.; Expert, D.; Toussaint, A. Characterization and virulence properties of Erwinia chrysanthemi lipopolysaccharide-defective, phi EC2-resistant mutants. J. Bacteriol. 1987, 169, 4011–4017. [Google Scholar] [CrossRef]
- Guerry, P.; Ewing, C.P.; Schirm, M.; Lorenzo, M.; Kelly, J.; Pattarini, D.; Majam, G.; Thibault, P.; Logan, S. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 2006, 60, 299–311. [Google Scholar] [CrossRef]
- Beis, K.; Srikannathasan, V.; Liu, H.; Fullerton, S.W.; Bamford, V.A.; Sanders, D.A.; Whitfield, C.; McNeil, M.R.; Naismith, J.H. Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidised state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state. J. Mol. Biol. 2005, 348, 971–982. [Google Scholar] [CrossRef]
- Pandin, C.; Caroff, M.; Condemine, G. Antimicrobial peptide resistance genes in the plant pathogen Dickeya dadantii. Appl. Environ. Microbiol. 2016, 82, 6423–6430. [Google Scholar] [CrossRef]
- Costechareyre, D.; Chich, J.F.; Strub, J.M.; Rahbe, Y.; Condemine, G. Transcriptome of Dickeya dadantii infecting Acyrthosiphon pisum reveals a strong defense against antimicrobial peptides. PLoS ONE 2013, 8, e54118. [Google Scholar] [CrossRef]
- Noland, B.W.; Newman, J.M.; Hendle, J.; Badger, J.; Christopher, J.A.; Tresser, J.; Buchanan, M.D.; Wright, T.A.; Rutter, M.E.; Sanderson, W.E.; et al. Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide-modifying enzyme. Structure 2002, 10, 1569–1580. [Google Scholar] [CrossRef]
- Brown, K.L.; Hancock, R.E. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 2006, 18, 24–30. [Google Scholar] [CrossRef]
- Salah Ud-Din, A.I.M.; Roujeinikova, A. Flagellin glycosylation with pseudaminic acid in Campylobacter and Helicobacter: Prospects for development of novel therapeutics. Cell. Mol. Life Sci. 2018, 75, 1163–1178. [Google Scholar] [CrossRef]
- Merrick, M.; Filser, M.; Kennedy, C.; Dixon, R. Polarity of mutations induced by insertion of transposons Tn5, Tn7 and Tn10 into the nif gene cluster of Klebsiella pneumoniae. Mol. Gen. Genet. MGG 1978, 165, 103–111. [Google Scholar] [CrossRef]
- de Bruijn, F.J.; Lupski, J.R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids—A review. GENE 1984, 27, 131–149. [Google Scholar] [CrossRef]
- Chatterjee, A.K.; Thurn, K.K.; Feese, D.A. Tn5-induced mutations in the enterobacterial phytopathogen Erwinia chrysanthemi. Appl. Environ. Microbiol. 1983, 45, 644–650. [Google Scholar] [CrossRef]
- Osbourn, A.E.; Field, B. Operons. Cell. Mol. Life Sci. 2009, 66, 3755–3775. [Google Scholar] [CrossRef]
- Bohannan, B.J.M.; Lenski, R.E. Linking genetic change to community evolution: Insights from studies of bacteria and bacteriophage. Ecol. Lett. 2000, 3, 362–377. [Google Scholar] [CrossRef]
- Seed, K.D.; Faruque, S.M.; Mekalanos, J.J.; Calderwood, S.B.; Qadri, F.; Camilli, A. Phase variable O antigen biosynthetic genes control expression of the major protective antigen and bacteriophage receptor in Vibrio cholerae O1. PLoS Pathog. 2012, 8, e1002917. [Google Scholar] [CrossRef]
- Evans, T.J.; Ind, A.; Komitopoulou, E.; Salmond, G.P. Phage-selected lipopolysaccharide mutants of Pectobacterium atrosepticum exhibit different impacts on virulence. J. Appl. Microbiol. 2010, 109, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Lukianova, A.A.; Shneider, M.M.; Evseev, P.V.; Shpirt, A.M.; Bugaeva, E.N.; Kabanova, A.P.; Obraztsova, E.A.; Miroshnikov, K.K.; Senchenkova, S.N.; Shashkov, A.S.; et al. Morphologically different Pectobacterium brasiliense bacteriophages PP99 and PP101: Deacetylation of O-polysaccharide by the tail spike protein of phage PP99 accompanies the infection. Front. Microbiol. 2019, 10, 3147. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Turner, P.E. High-throughput discovery of phage receptors using transposon insertion sequencing of bacteria. Proc. Natl. Acad. Sci. USA 2020, 117, 18670–18679. [Google Scholar] [CrossRef]
- Hendrick, C.A.; Sequeira, L. Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl. Environ. Microbiol. 1984, 48, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.C.; McGhee, G.C.; Zhao, Y.; Sundin, G.W. Effect of a waaL mutation on lipopolysaccharide composition, oxidative stress survival, and virulence in Erwinia amylovora. FEMS Microbiol. Lett. 2009, 291, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Mannaa, M.; Kim, N.; Kim, J.; Choi, Y.; Kim, S.H.; Jung, B.; Lee, H.H.; Lee, J.; Seo, Y.S. Stress tolerance and virulence-related roles of lipopolysaccharide in Burkholderia glumae. Plant Pathol. J. 2019, 35, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Kutschera, A.; Ranf, S. The multifaceted functions of lipopolysaccharide in plant-bacteria interactions. Biochimie 2019, 159, 93–98. [Google Scholar] [CrossRef]
- Lisicka, W.; Fikowicz-Krosko, J.; Jafra, S.; Narajczyk, M.; Czaplewska, P.; Czajkowski, R. Oxygen availability influences expression of Dickeya solani genes associated with virulence in potato (Solanum tuberosum L.) and chicory (Cichorium intybus L.). Front. Plant Sci. 2018, 9, 374. [Google Scholar] [CrossRef]
- Czajkowski, R.; Kaczyńska, N.; Jafra, S.; Narajczyk, M.; Lojkowska, E. Temperature-responsive genetic loci in pectinolytic plant pathogenic Dickeya solani. Plant Pathol. 2017, 66, 584–594. [Google Scholar] [CrossRef]
- Xi, C.W.; Lambrecht, M.; Vanderleyden, J.; Michiels, J. Bi-functional gfp-and gusA-containing mini-Tn5 transposon derivatives for combined gene expression and bacterial localization studies. J. Microbiol. Methods 1999, 35, 85–92. [Google Scholar] [CrossRef]
- Pérez-Ortín, J.; LíDel Olmo, M.; Matallana, E.; Tordera, V. Making your own gene library. Biochem. Educ. 1997, 25, 237–242. [Google Scholar] [CrossRef]
- Czajkowski, R.; Marcisz, M.; Bartnik, P. Fast and reliable screening assay developed to preselect candidate Soft Rot Pectobacteriaceae Tn5 mutants showing resistance to bacteriophage infection. Eur. J. Plant Pathol. 2019, 155, 671–676. [Google Scholar] [CrossRef]
- Hélias, V.; Hamon, P.; Huchet, E.; Wolf, J.V.D.; Andrivon, D. Two new effective semiselective crystal violet pectate media for isolation of Pectobacterium and Dickeya. Plant Pathol. 2012, 61, 339–345. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, I.N. Bacteriophage adsorption rate and optimal lysis time. Genetics 2008, 180, 471–482. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Czajkowski, R.; Fikowicz-Krosko, J.; Maciag, T.; Rabalski, L.; Czaplewska, P.; Jafra, S.; Richert, M.; Krychowiak-Masnicka, M.; Hugouvieux-Cotte-Pattat, N. Genome-wide identification of Dickeya solani transcriptional units up-regulated in response to plant tissues from a crop-host Solanum tuberosum and a weed-host Solanum dulcamara. Front. Plant Sci. 2020, 11, 580330. [Google Scholar] [CrossRef]
- Altschul, S.F.; Koonin, E.V. Iterated profile searches with PSI-BLAST- a tool for discovery in protein databases. Trends Biochem. Sci. 1998, 23, 444–447. [Google Scholar] [CrossRef]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-mapper: A web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Letunic, I.; Yamada, T.; Kanehisa, M.; Bork, P. iPath: Interactive exploration of biochemical pathways and networks. Trends Biochem. Sci. 2008, 33, 101–103. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Bell, K.S.; Sebaihia, M.; Pritchard, L.; Holden, M.T.; Hyman, L.J.; Holeva, M.C.; Thomson, N.R.; Bentley, S.D.; Churcher, L.J.; Mungall, K.; et al. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA 2004, 101, 11105–11110. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Tsai, C.M.; Frasch, C.E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 1982, 119, 115–119. [Google Scholar] [CrossRef]
- Tomas, J.M.; Kay, W.W. Effect of bacteriophage P1 lysogeny on lipopolysaccharide composition and the lambda receptor of Escherichia coli. J. Bacteriol. 1984, 159, 1047–1052. [Google Scholar] [CrossRef]
- Roth, V. Doubling Time Computing. 2006. Available online: http://www.doubling-time.com/compute.php (accessed on 17 June 2021).
- Czajkowski, R.; de Boer, W.J.; van Veen, J.A.; van der Wolf, J.M. Characterization of bacterial isolates from rotting potato tuber tissue showing antagonism to Dickeya sp. biovar 3 in vitro and in planta. Plant Pathol. 2012, 61, 169–182. [Google Scholar] [CrossRef]
- Krzyzanowska, D.M.; Maciag, T.; Siwinska, J.; Krychowiak, M.; Jafra, S.; Czajkowski, R. Compatible mixture of bacterial antagonists developed to protect potato tubers from soft rot caused by Pectobacterium spp. and Dickeya spp. Plant Dis. 2019, 103, 1374–1382. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Shao, X.; Xie, Y.; Zhang, Y.; Deng, X. Biofilm formation assay in Pseudomonas syringae. Bio-Protocol 2019, 9, e3237. [Google Scholar] [CrossRef]
- Dickey, R.S. Erwinia chrysanthemi: A comparative study of phenotypic properties of strains from several hosts and other Erwinia species. Phytopathology 1979, 69, 324–329. [Google Scholar] [CrossRef]
- Perombelon, M.C.M.; van Der Wolf, J.M. Methods for the detection and quantification of Erwinia carotovora subsp. atroseptica (Pectobacterium carotovorum subsp. atrosepticum) on potatoes: A laboratory manual. Scott. Crop. Res. Inst. Annu. Rep. 2002, 10, 1–82. [Google Scholar]
- Ji, J.W.; Hugouvieux Cotte Pattat, N.; Robert Baudouy, J. Use of Mu-Lac insertions to study the secretion of pectate lyases by Erwinia chrysanthemi. J. Gen. Microbiol. 1987, 133, 793–802. [Google Scholar] [CrossRef][Green Version]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Maneval, W.E. Staining bacteria and yeasts with acid dyes. Stain. Technol. 2009, 16, 13–19. [Google Scholar] [CrossRef]
- Kodaka, H.; Armfield, A.Y.; Lombard, G.L.; Dowell, V.R., Jr. Practical procedure for demonstrating bacterial flagella. J. Clin. Microbiol. 1982, 16, 948–952. [Google Scholar] [CrossRef]
- Beveridge, T.J.; Lawrence, J.R.; Murray, R.G.E. Sampling and staining for light microscopy. In Methods for General and Molecular Microbiology; ASM Press: Washington, DC, USA, 2014; pp. 19–33. [Google Scholar]
- Czajkowski, R.; Smolarska, A.; Ozymko, Z. The viability of lytic bacteriophage phiD5 in potato-associated environments and its effect on Dickeya solani in potato (Solanum tuberosum L.) plants. PLoS ONE 2017, 12, e0183200. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Welch, B.L. The generalisation of student’s problems when several different population variances are involved. Biometrika 1947, 34, 28–35. [Google Scholar] [CrossRef]
- Box, G.E.P. Non-normality and tests on variances. Biometrika 1953, 40, 318–335. [Google Scholar] [CrossRef]
- Student. The probable error of a mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Czajkowski, R.; de Boer, W.J.; Velvis, H.; van der Wolf, J.M. Systemic colonization of potato plants by a soilborne, green fluorescent protein-tagged strain of Dickeya sp. biovar 3. Phytopathology 2010, 100, 134–142. [Google Scholar] [CrossRef]

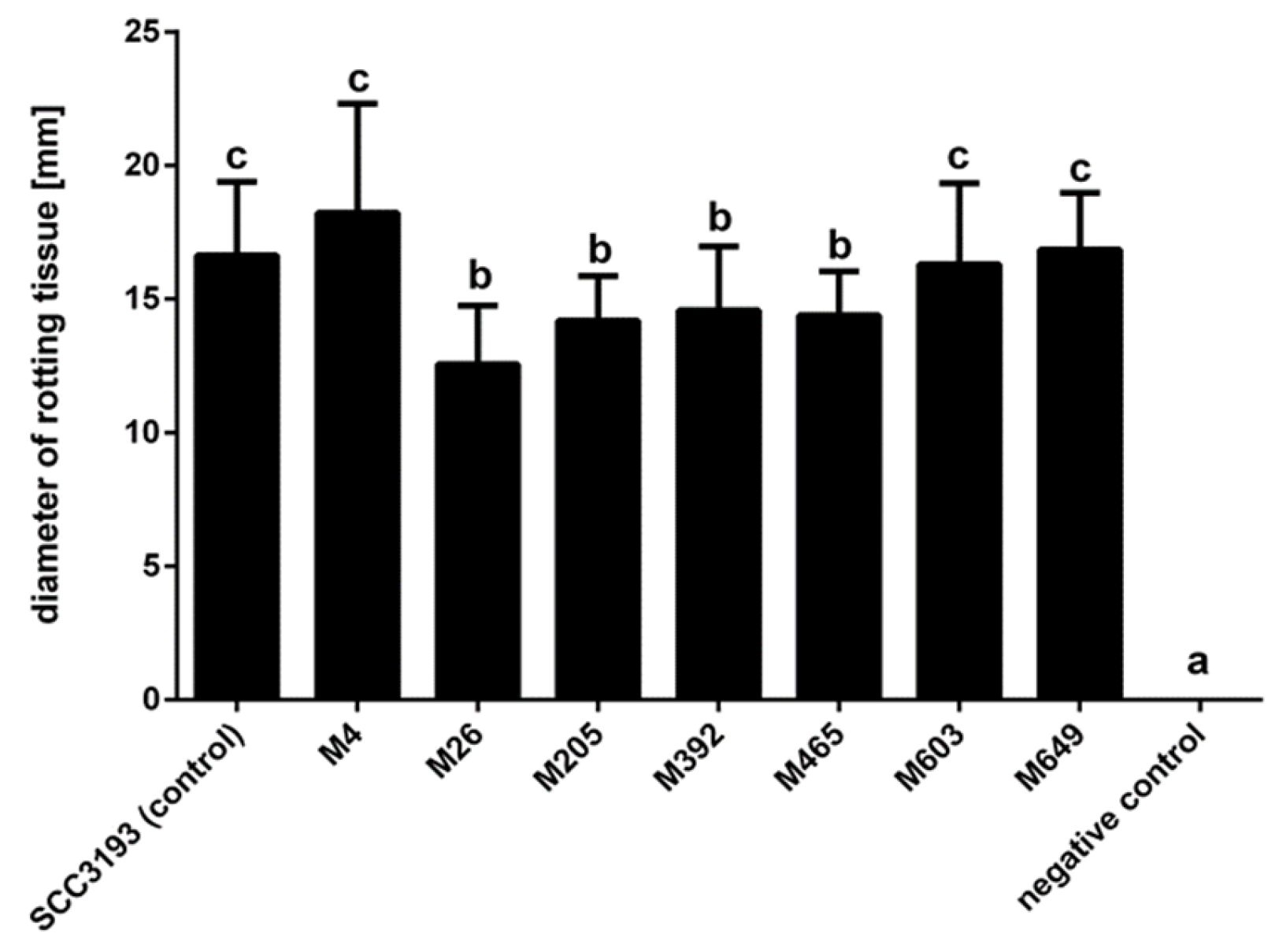
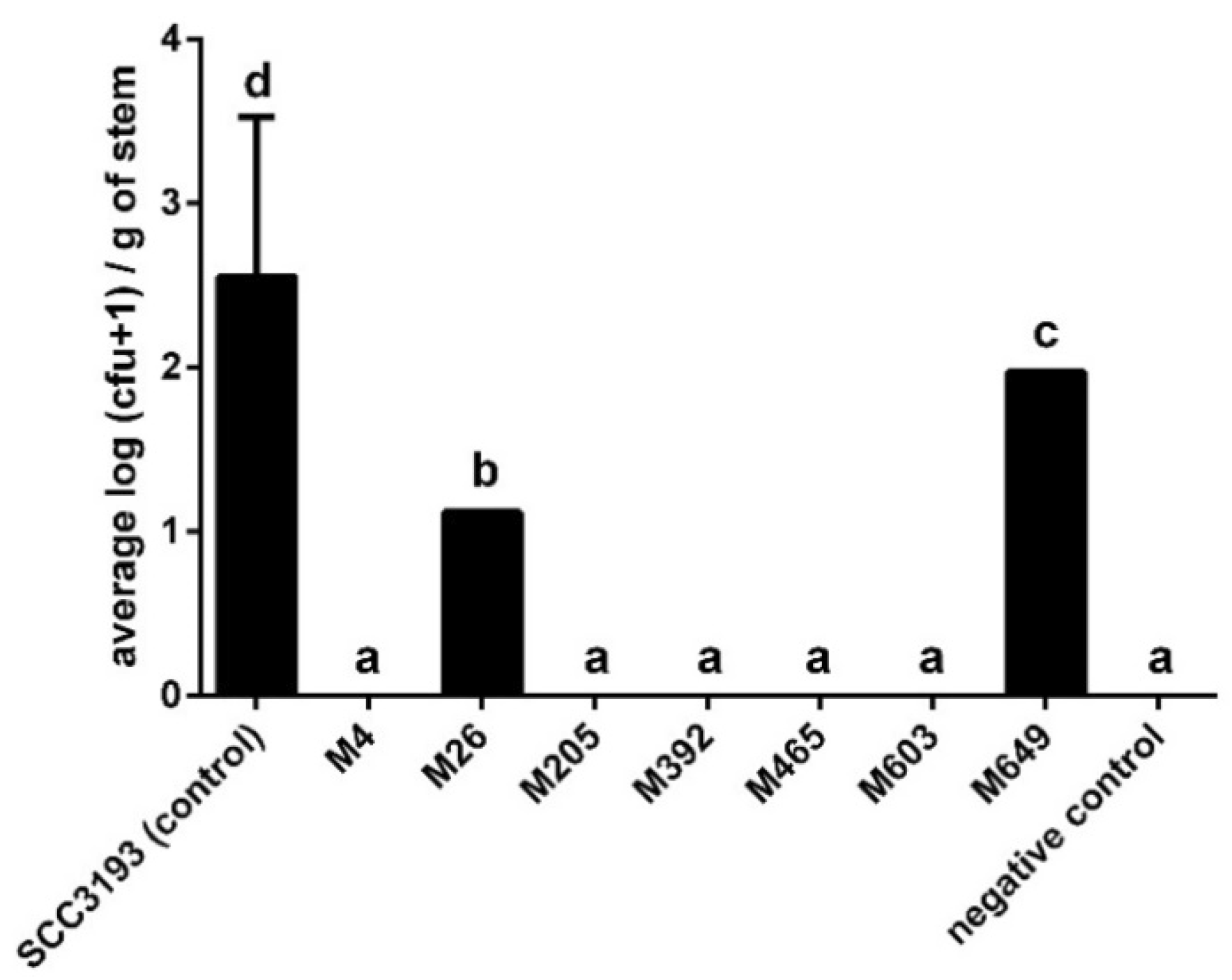
| No | Mutant a | Insertion Name, Tn5 Locus, Gene, CDC | Protein Name | Transcriptional Organization (Single Gene vs. Operon) b | Entry, KEGG Pathway, UniProt-Based Protein Function |
|---|---|---|---|---|---|
| 1 | M4 | p4, putative oafA, W5S_2104, AFI90193.1 | O-acetyl transferase (putative O-antigen LPS acetylase) | single gene | EC: entry not assigned, gene not included in the pathway |
| 2 | M26 | p26, galU, W5S_2225, AFI90313.1 | UTP-glucose-1-phosphate uridylyl transferase | single gene | EC: 2.7.7.9, O-antigen nucleotide sugar biosynthesis, amino sugar, and nucleotide sugar metabolism |
| 3 | M205 | p205, pseH, W5S_3016, AFI91099.1 | pseudaminic acid biosynthesis N-acetyl transferase | operon: contains 16 genes | EC: 23.1.202, O-antigen nucleotide sugar biosynthesis, amino sugar, and nucleotide sugar metabolism |
| 4 | M392 | p392, glf, W5S_3004, AFI91087.1 | UDP-galactopyranose mutase | operon: contains 2 genes | EC: 5.4.99.9, O-antigen nucleotide sugar biosynthesis, amino sugar and nucleotide sugar metabolism |
| 5 | M465 | p465, arnB, W5S_3019, AFI91102.1 | UDP-4-amino-4-deoxy-L-arabinose-oxoglutarate aminotransferase | operon: contains 8 genes | EC: 2.6.1.87, amino sugar and nucleotide sugar metabolism |
| 6 | M603 | p603, wbjB (synonyms: pseB, elgL), W5S_3021, AFI91104.1 | UDP-N-acetylglucosamine 4,6-dehydratase | operon: contains 16 genes | EC: 42.1.115, O-antigen nucleotide sugar biosynthesis |
| 7 | M649 | p649, W5S_3005, AFI91088.1 | hypothetical protein | operon: contains 2 genes | EC: 5.4.99.9, O-antigen nucleotide sugar biosynthesis, amino sugar and nucleotide sugar metabolism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartnik, P.; Jafra, S.; Narajczyk, M.; Czaplewska, P.; Czajkowski, R. Pectobacterium parmentieri SCC 3193 Mutants with Altered Synthesis of Cell Surface Polysaccharides Are Resistant to N4-Like Lytic Bacteriophage ϕA38 (vB_Ppp_A38) but Express Decreased Virulence in Potato (Solanum tuberosum L.) Plants. Int. J. Mol. Sci. 2021, 22, 7346. https://doi.org/10.3390/ijms22147346
Bartnik P, Jafra S, Narajczyk M, Czaplewska P, Czajkowski R. Pectobacterium parmentieri SCC 3193 Mutants with Altered Synthesis of Cell Surface Polysaccharides Are Resistant to N4-Like Lytic Bacteriophage ϕA38 (vB_Ppp_A38) but Express Decreased Virulence in Potato (Solanum tuberosum L.) Plants. International Journal of Molecular Sciences. 2021; 22(14):7346. https://doi.org/10.3390/ijms22147346
Chicago/Turabian StyleBartnik, Przemyslaw, Sylwia Jafra, Magdalena Narajczyk, Paulina Czaplewska, and Robert Czajkowski. 2021. "Pectobacterium parmentieri SCC 3193 Mutants with Altered Synthesis of Cell Surface Polysaccharides Are Resistant to N4-Like Lytic Bacteriophage ϕA38 (vB_Ppp_A38) but Express Decreased Virulence in Potato (Solanum tuberosum L.) Plants" International Journal of Molecular Sciences 22, no. 14: 7346. https://doi.org/10.3390/ijms22147346
APA StyleBartnik, P., Jafra, S., Narajczyk, M., Czaplewska, P., & Czajkowski, R. (2021). Pectobacterium parmentieri SCC 3193 Mutants with Altered Synthesis of Cell Surface Polysaccharides Are Resistant to N4-Like Lytic Bacteriophage ϕA38 (vB_Ppp_A38) but Express Decreased Virulence in Potato (Solanum tuberosum L.) Plants. International Journal of Molecular Sciences, 22(14), 7346. https://doi.org/10.3390/ijms22147346







