Study of the Expression and Function of Calcium-Sensing Receptor in Human Skeletal Muscle
Abstract
:1. Introduction
2. Results
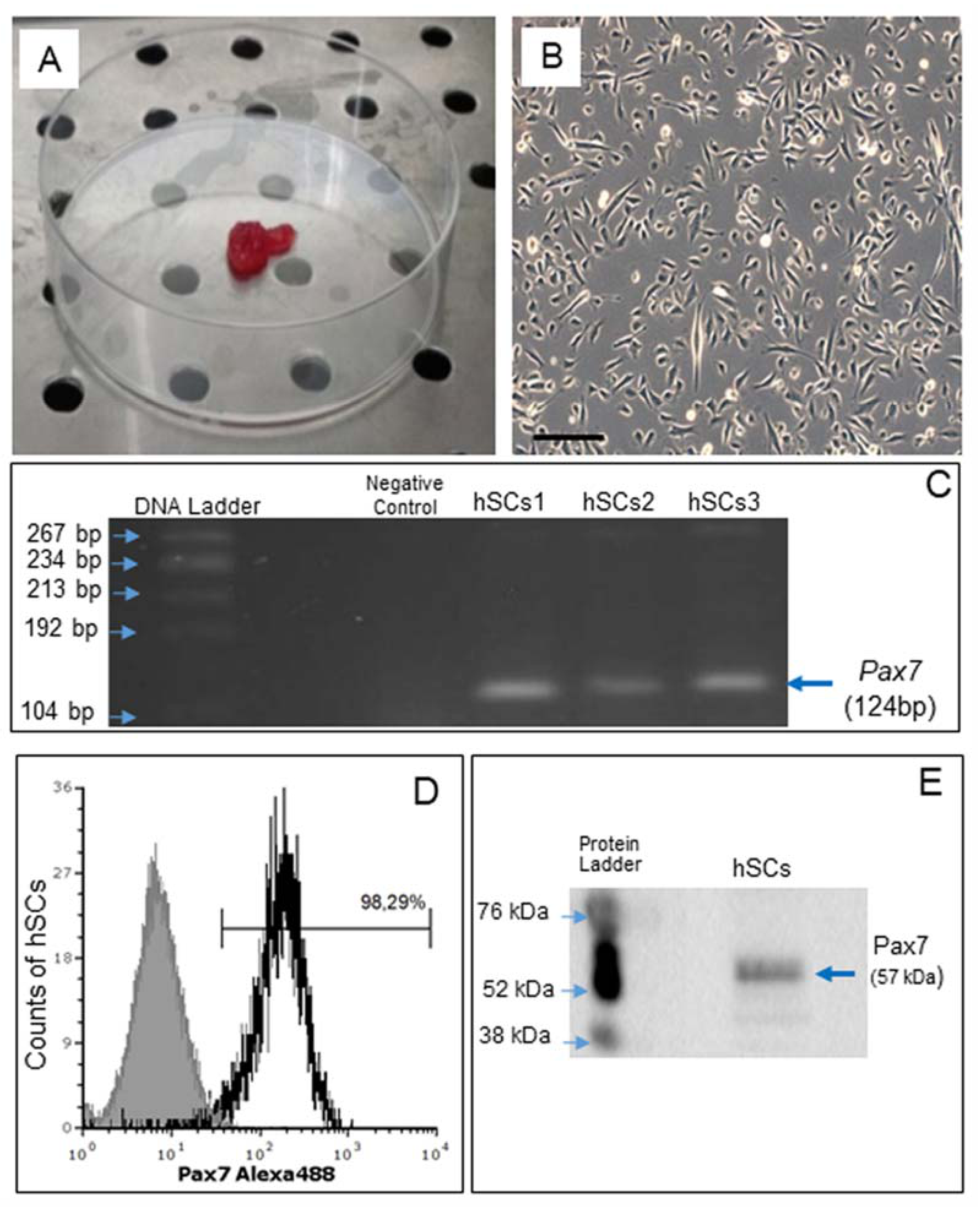
2.1. Isolation of Primary Culture of Human Satellite Cells (hSCs) and Characterization of Established hSC Lines
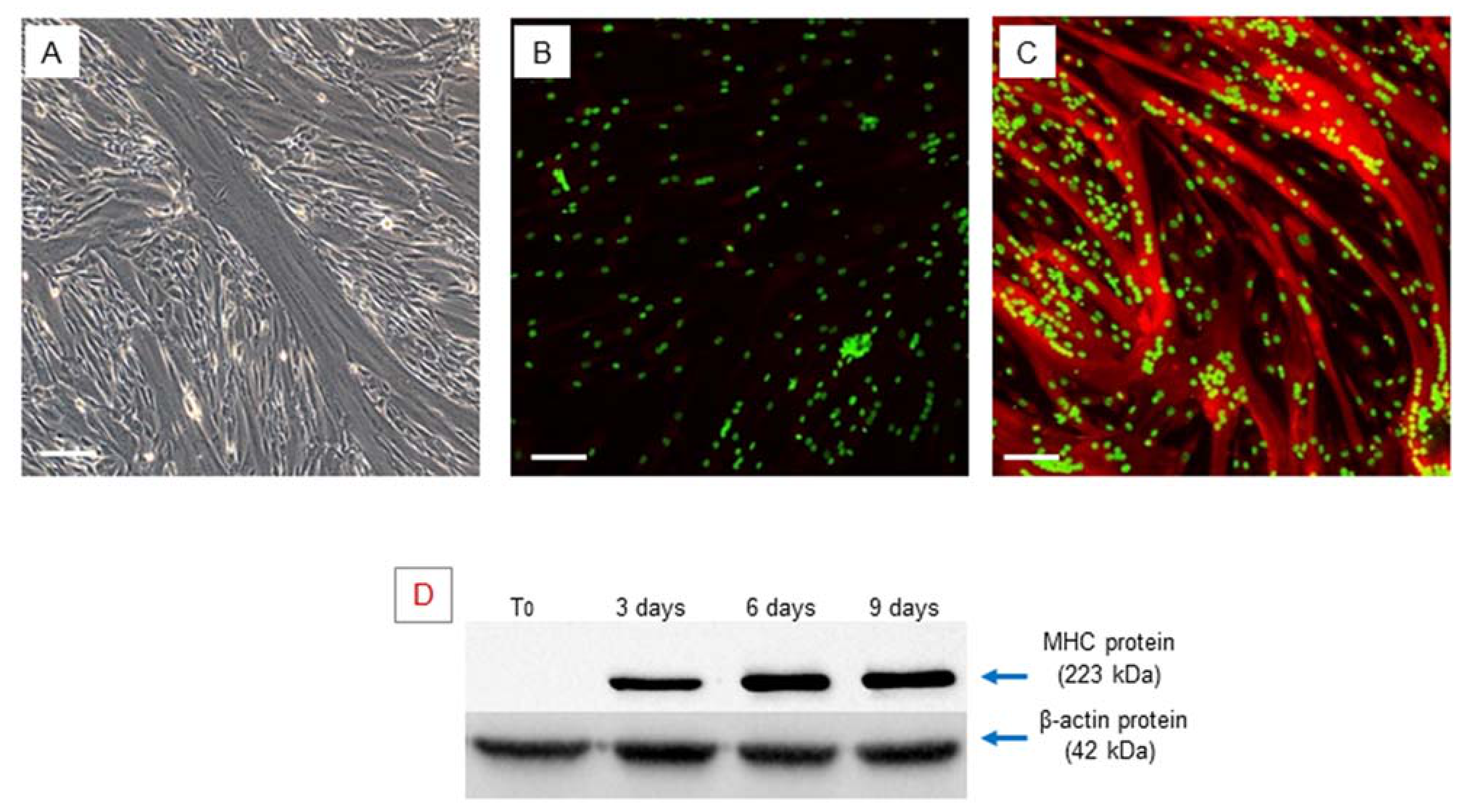
2.2. In Vitro Myogenic Differentiation of hSCs
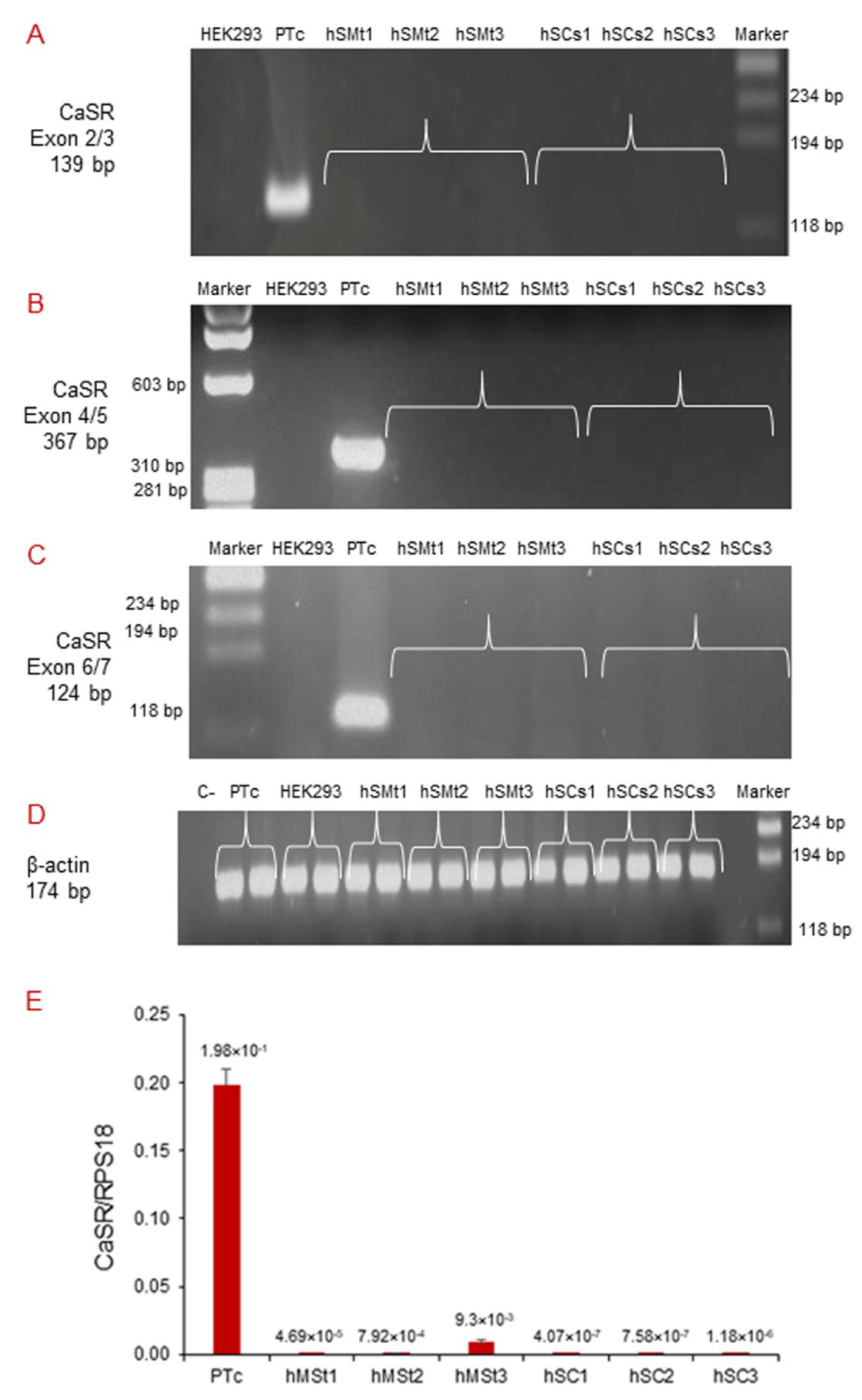
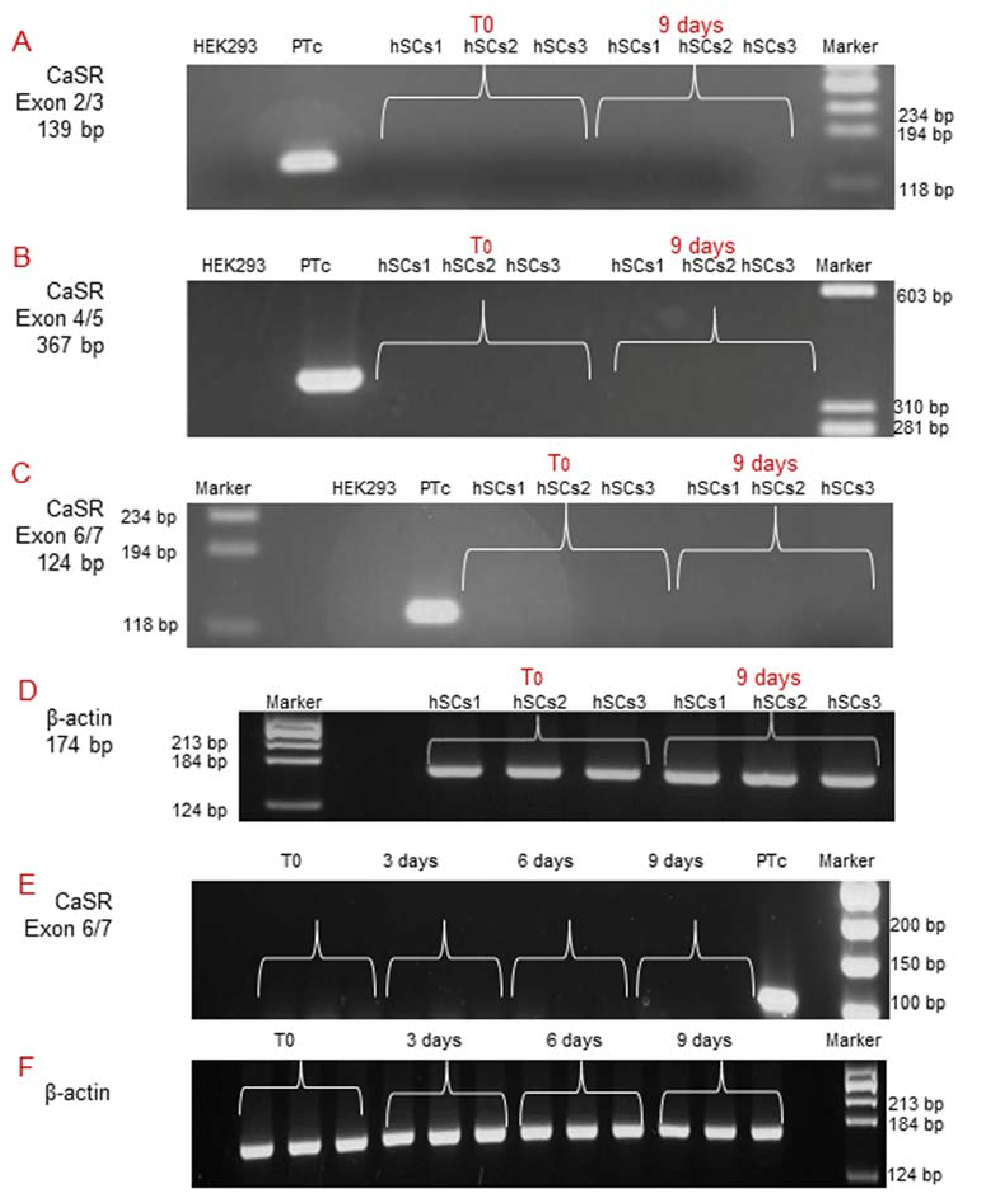
2.3. Analysis of CaSR Gene Expression in Human Skeletal Muscle Tissues (hSMts) and in the Derived Human Satellite Cell (hSC) Lines and during In Vitro Myogenesis
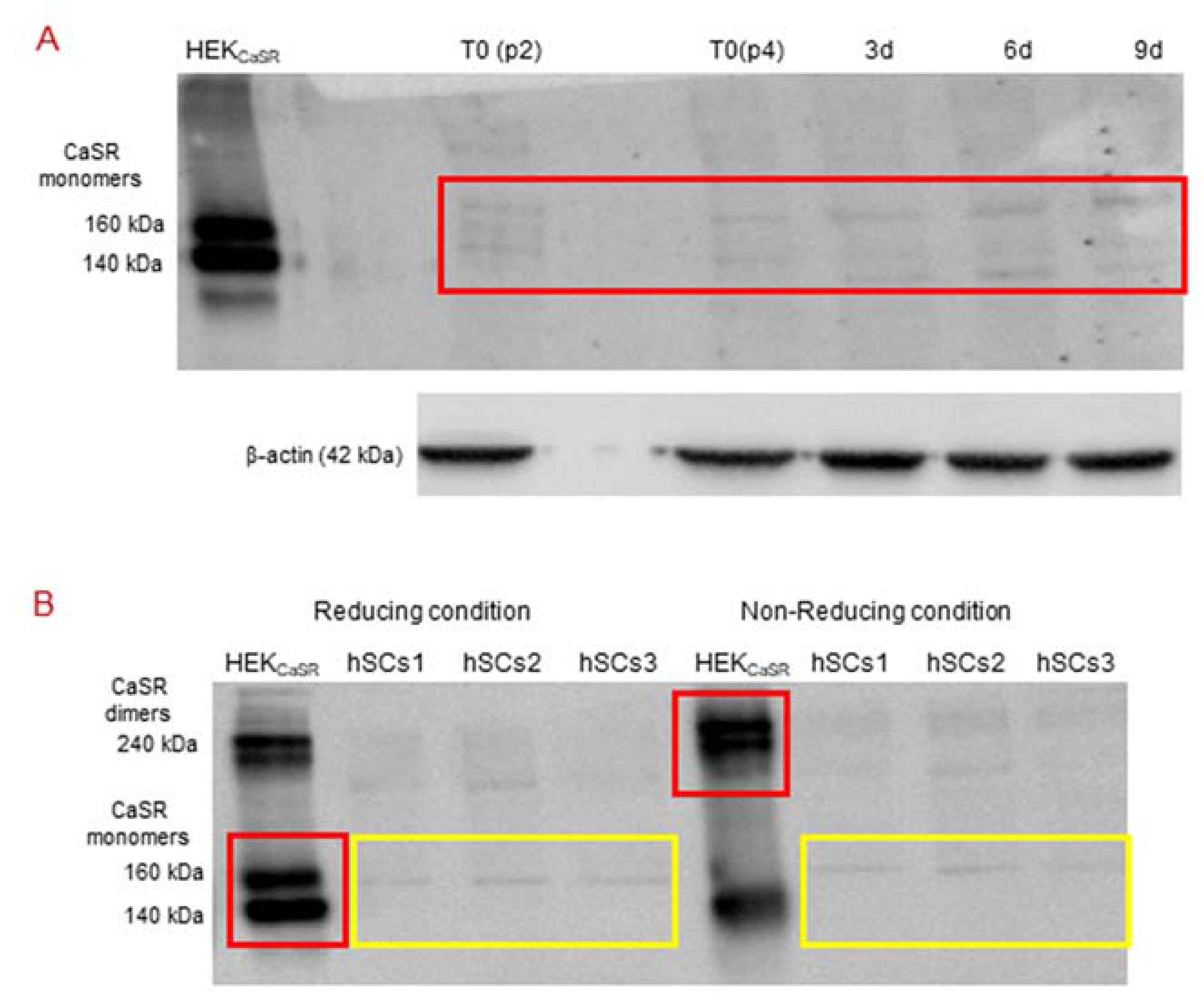
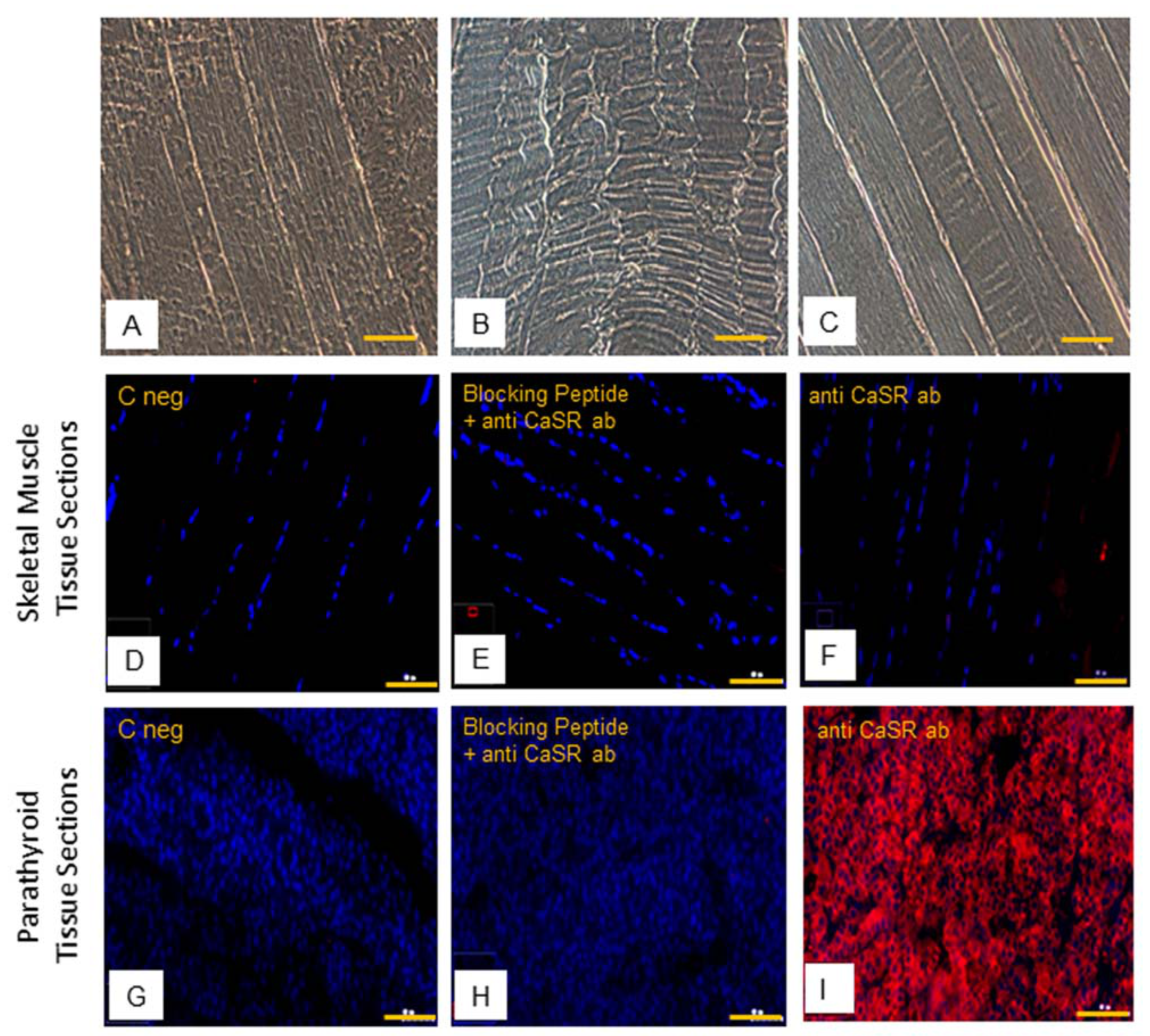
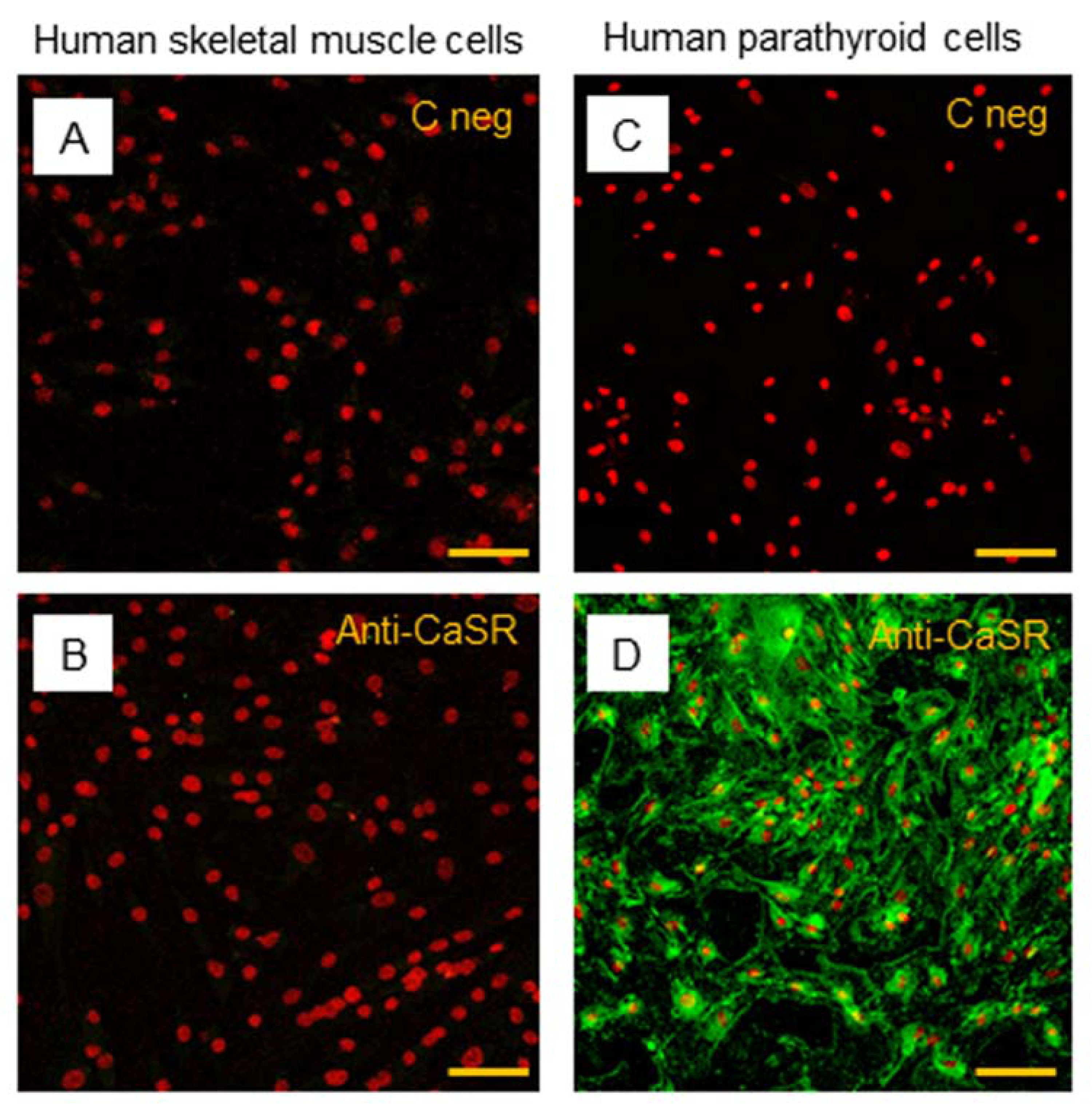
2.4. CaSR Protein Expression Analysis in Human Skeletal Muscle Tissues (hSMts), in hSCs, and during In Vitro Myogenesis
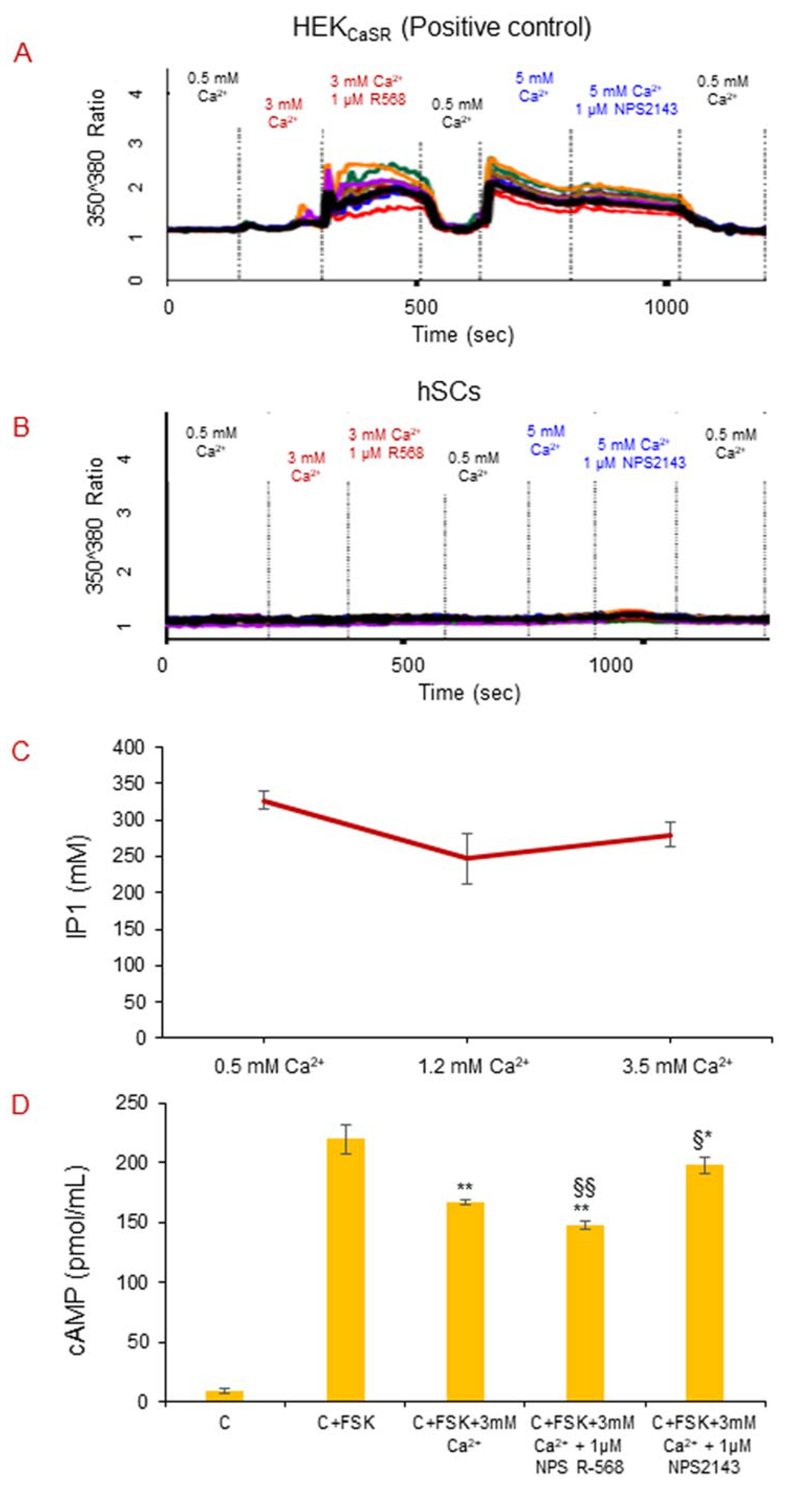
2.5. CaSR Secondary Messenger Analysis in hSC Lines
2.5.1. Intracellular Calcium Mobilization Imaging
2.5.2. CaSR Secondary Messenger Analysis in hSC Lines: Gαq/11 and Inositol Triphosphate (IP3)
2.5.3. CaSR Secondary Messenger Analysis in hSC Lines: Gαi/0 and Cyclic Adenosine Monophosphate (cAMP)
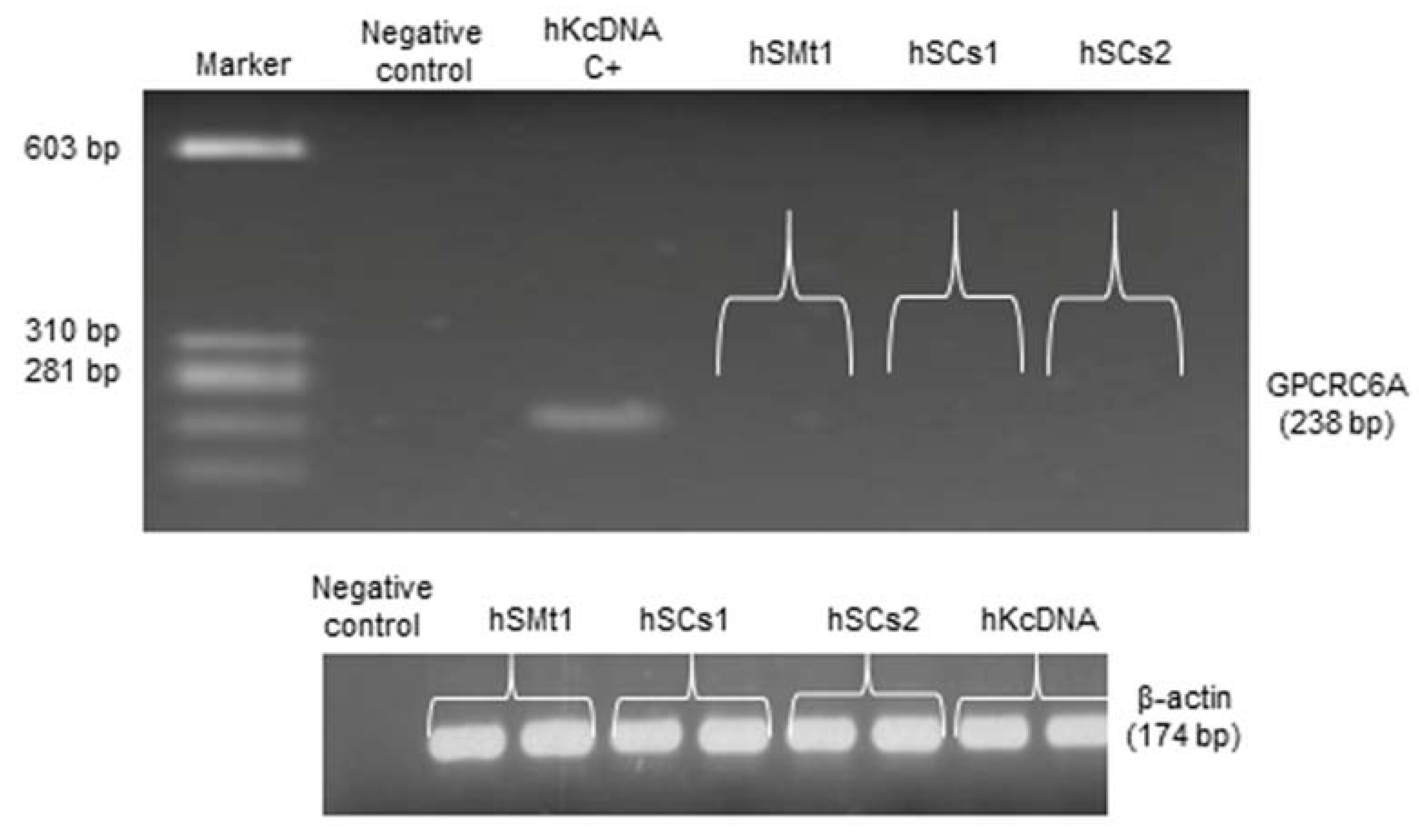
2.6. Analysis of a CaSR Homologous Protein in Human Skeletal Muscle: GPRC6A
3. Discussion
4. Materials and Methods
4.1. Primary Culture and Development of Lines of Human Satellite Cells (hSCs)
4.2. In Vitro Myogenic Differentiation
4.3. Flow Cytometry Analysis
4.4. Immunofluorescence Staining of Cells and Tissues
4.5. Gene Expression Analysis by Qualitative Reverse Transcriptase Polymerase Chain Reaction
4.6. Gene Expression Analysis by Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction
4.7. Protein Expression Analysis by Western Blot
4.8. Secondary Messenger Analysis
4.8.1. Inositol Monophosphate (IP1) ELISA Assay
4.8.2. Intracellular Calcium Mobilization Imaging
4.8.3. Cyclic Adenosine Monophosphate (cAMP) ELISA Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CaSR | Calcium-sensing receptor |
| PTH | Parathyroid hormone |
| SCs | Satellite cells |
| Pax7 | Paired box protein 7 |
| GPCRs | G protein-coupled receptors |
| MHC | Myosin heavy chain |
| MyoD1 | Myoblast determination protein 1 |
| Myf5 | Myogenic factor 5 |
| Myog | Myogenin |
| hSMT | Human skeletal muscle tissue |
| DTT | Dithiothreitol |
| IP3 | Inositol triphosphate |
| cAMP | Cyclic adenosine monophosphate |
| GPRC6A | G protein-coupled receptor class C group 6 member A |
| BSA | Bovine serum albumin |
| LSCM | Laser scanning confocal microscopy |
| FBS | Fetal bovine serum |
| FSK | Forskolin |
References
- Wang, Y.X.; Rudnicki, M.A. Satellite cells, the engines of muscle repair. Nat. Rev. Mol. Cell Biol. 2011, 13, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Frontera, W.R.; Ochala, J. Skeletal muscle: A brief review of structure and function. Calcif. Tissue Int. 2015, 96, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Marzetti, E.; Calvani, R.; Tosato, M.; Cesari, M.; Di Bari, M.; Cherubini, A.; Collamati, A.; D’Angelo, E.; Pahor, M.; Bernabei, R.; et al. Sarcopenia: An overview. Aging Clin. Exp. Res. 2017, 29, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.J.; Doherty, T.J. Sarcopenia: Prevalence, mechanisms, and functional consequences. Interdiscip. Top. Gerontol. 2010, 37, 94–114. [Google Scholar] [PubMed]
- Chang, N.C.; Chevalier, F.P.; Rudnicki, M.A. Satellite Cells in Muscular Dystrophy—Lost in Polarity. Trends Mol. Med. 2016, 22, 479–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumont, N.A.; Rudnicki, M.A. Targeting muscle stem cell intrinsic defects to treat Duchenne muscular dystrophy. NPJ Regen. Med. 2016, 1, 16006. [Google Scholar] [CrossRef] [Green Version]
- Morgan, J.; Partridge, T. Skeletal muscle in health and disease. Dis. Model. Mech. 2020, 13, dmm042192. [Google Scholar] [CrossRef] [Green Version]
- Alway, S.E.; Myers, M.J.; Mohamed, J.S. Regulation of satellite cell function in sarcopenia. Front. Aging Neurosci. 2014, 6, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.; Wang, H.; Hu, P. Stem cell activation in skeletal muscle regeneration. Cell. Mol. Life Sci. CMLS 2015, 72, 1663–1677. [Google Scholar] [CrossRef] [Green Version]
- Jaroch, K.; Jaroch, A.; Bojko, B. Cell cultures in drug discovery and development: The need of reliable in vitro-in vivo extrapolation for pharmacodynamics and pharmacokinetics assessment. J. Pharm. Biomed. Anal. 2018, 147, 297–312. [Google Scholar] [CrossRef]
- Gheller, B.J.; Blum, J.; Soueid-Baumgarten, S.; Bender, E.; Cosgrove, B.D.; Thalacker-Mercer, A. Isolation, Culture, Characterization, and Differentiation of Human Muscle Progenitor Cells from the Skeletal Muscle Biopsy Procedure. J. Vis. Exp. JoVE 2019, 150. [Google Scholar] [CrossRef]
- Riddle, E.S.; Bender, E.L.; Thalacker-Mercer, A.E. Expansion capacity of human muscle progenitor cells differs by age, sex, and metabolic fuel preference. Am. J. Physiol. Cell Physiol. 2018, 315, C643–C652. [Google Scholar] [CrossRef]
- Nehlin, J.O.; Just, M.; Rustan, A.C.; Gaster, M. Human myotubes from myoblast cultures undergoing senescence exhibit defects in glucose and lipid metabolism. Biogerontology 2011, 12, 349–365. [Google Scholar] [CrossRef]
- Romagnoli, C.; Zonefrati, R.; Sharma, P.; Innocenti, M.; Cianferotti, L.; Brandi, M.L. Characterization of Skeletal Muscle Endocrine Control in an In Vitro Model of Myogenesis. Calcif. Tissue Int. 2020, 107, 18–30. [Google Scholar] [CrossRef] [Green Version]
- Bareja, A.; Holt, J.A.; Luo, G.; Chang, C.; Lin, J.; Hinken, A.C.; Freudenberg, J.M.; Kraus, W.E.; Evans, W.J.; Billin, A.N. Human and mouse skeletal muscle stem cells: Convergent and divergent mechanisms of myogenesis. PLoS ONE 2014, 9, e90398. [Google Scholar] [CrossRef] [Green Version]
- Kallay, E. Editorial: Physiology and Pathophysiology of the Extracellular Calcium-Sensing Receptor. Front. Physiol. 2018, 9, 413. [Google Scholar] [CrossRef] [Green Version]
- Hannan, F.M.; Kallay, E.; Chang, W.; Brandi, M.L.; Thakker, R.V. The calcium-sensing receptor in physiology and in calcitropic and noncalcitropic diseases. Nat. Rev. Endocrinol. 2018, 15, 33–51. [Google Scholar] [CrossRef]
- Catterall, W.A. Voltage-gated calcium channels. Cold Spring Harb. Perspect. Biol. 2011, 3, a003947. [Google Scholar] [CrossRef]
- Tu, M.K.; Levin, J.B.; Hamilton, A.M.; Borodinsky, L.N. Calcium signaling in skeletal muscle development, maintenance and regeneration. Cell Calcium 2016, 59, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Avila, G. Disturbed Ca2+ Homeostasis in Muscle-Wasting Disorders. Adv. Exp. Med. Biol. 2018, 1088, 307–326. [Google Scholar]
- Cho, C.-H.; Woo, J.S.; Perez, C.F.; Lee, E.H. A focus on extracellular Ca2+ entry into skeletal muscle. Exp. Mol. Med. 2017, 49, e378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, S.C.; Thiem, U.; Roth, S.; Aggarwal, A.; Fetahu, I.S.; Tennakoon, S.; Gomes, A.R.; Brandi, M.L.; Bruggeman, F.; Mentaverri, R.; et al. Calcium sensing receptor signalling in physiology and cancer. Biochim. Biophys. Acta 2013, 1833, 1732–1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, E.F.; Shoback, D. Calcimimetic and calcilytic drugs for treating bone and mineral-related disorders. Best Pract. Res. Clin. Endocrinol. Metab. 2013, 27, 373–384. [Google Scholar] [CrossRef]
- Sun, J.; He, W.; Bai, S.; Peng, X.; Zhang, N.; Li, H.; Zhang, W.; Wang, L.; Shao, X.; He, Y.; et al. The expression of calcium-sensing receptor in mouse embryonic stem cells (mESCs) and its influence on differentiation of mESC into cardiomyocytes. Differ. Res. Biol. Divers. 2013, 85, 32–40. [Google Scholar] [CrossRef]
- Bikle, D.D.; Jiang, Y.; Nguyen, T.; Oda, Y.; Tu, C.-L. Disruption of Vitamin D and Calcium Signaling in Keratinocytes Predisposes to Skin Cancer. Front. Physiol. 2016, 7, 296. [Google Scholar] [CrossRef] [Green Version]
- Mateo-Lozano, S.; García, M.; Rodríguez-Hernández, C.J.; de Torres, C. Regulation of Differentiation by Calcium-Sensing Receptor in Normal and Tumoral Developing Nervous System. Front. Physiol. 2016, 7, 169. [Google Scholar] [CrossRef] [Green Version]
- Tharmalingam, S.; Hampson, D.R. The Calcium-Sensing Receptor and Integrins in Cellular Differentiation and Migration. Front. Physiol. 2016, 7, 190. [Google Scholar] [CrossRef] [Green Version]
- Roesler, A.M.; Wicher, S.A.; Ravix, J.; Britt, R.D.; Manlove, L.; Teske, J.J.; Cummings, K.; Thompson, M.A.; Farver, C.; MacFarlane, P.; et al. Calcium sensing receptor in developing human airway smooth muscle. J. Cell. Physiol. 2019, 234, 14187–14197. [Google Scholar] [CrossRef]
- Sambasivan, R.; Yao, R.; Kissenpfennig, A.; Van Wittenberghe, L.; Paldi, A.; Gayraud-Morel, B.; Guenou, H.; Malissen, B.; Tajbakhsh, S.; Galy, A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Dev. Camb. Engl. 2011, 138, 3647–3656. [Google Scholar]
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef] [Green Version]
- Atwood, B.K.; Lopez, J.; Wager-Miller, J.; Mackie, K.; Straiker, A. Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genom. 2011, 12, 14. [Google Scholar] [CrossRef] [Green Version]
- Ziegelstein, R.C.; Xiong, Y.; He, C.; Hu, Q. Expression of a functional extracellular calcium-sensing receptor in human aortic endothelial cells. Biochem. Biophys. Res. Commun. 2006, 342, 153–163. [Google Scholar] [CrossRef]
- Tu, C.L.; Chang, W.; Bikle, D.D. The extracellular calcium-sensing receptor is required for calcium-induced differentiation in human keratinocytes. J. Biol. Chem. 2001, 276, 41079–41085. [Google Scholar] [CrossRef] [Green Version]
- Smith, K.A.; Ayon, R.J.; Tang, H.; Makino, A.; Yuan, J.X.-J. Calcium-Sensing Receptor Regulates Cytosolic [Ca2+] and Plays a Major Role in the Development of Pulmonary Hypertension. Front. Physiol. 2016, 7, 517. [Google Scholar] [CrossRef] [Green Version]
- Pi, M.; Faber, P.; Ekema, G.; Jackson, P.D.; Ting, A.; Wang, N.; Fontilla-Poole, M.; Mays, R.W.; Brunden, K.R.; Harrington, J.J.; et al. Identification of a novel extracellular cation-sensing G-protein-coupled receptor. J. Biol. Chem. 2005, 280, 40201–40209. [Google Scholar] [CrossRef] [Green Version]
- Faure, H.; Gorojankina, T.; Rice, N.; Dauban, P.; Dodd, R.H.; Bräuner-Osborne, H.; Rognan, D.; Ruat, M. Molecular determinants of non-competitive antagonist binding to the mouse GPRC6A receptor. Cell Calcium 2009, 46, 323–332. [Google Scholar] [CrossRef]
- Jacobsen, S.E.; Nørskov-Lauritsen, L.; Thomsen, A.R.B.; Smajilovic, S.; Wellendorph, P.; Larsson, N.H.P.; Lehmann, A.; Bhatia, V.K.; Bräuner-Osborne, H. Delineation of the GPRC6A receptor signaling pathways using a mammalian cell line stably expressing the receptor. J. Pharmacol. Exp. Ther. 2013, 347, 298–309. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef] [Green Version]
- Snijders, T.; Verdijk, L.B.; van Loon, L.J.C. The impact of sarcopenia and exercise training on skeletal muscle satellite cells. Ageing Res. Rev. 2009, 8, 328–338. [Google Scholar] [CrossRef]
- Roth, S.M.; Martel, G.F.; Ivey, F.M.; Lemmer, J.T.; Tracy, B.L.; Metter, E.J.; Hurley, B.F.; Rogers, M.A. Skeletal muscle satellite cell characteristics in young and older men and women after heavy resistance strength training. J. Gerontol. A Biol. Sci. Med. Sci. 2001, 56, B240–B247. [Google Scholar] [CrossRef] [Green Version]
- Jean-Baptiste, G.; Yang, Z.; Khoury, C.; Gaudio, S.; Greenwood, M.T. Peptide and non-peptide G-protein coupled receptors (GPCRs) in skeletal muscle. Peptides 2005, 26, 1528–1536. [Google Scholar] [CrossRef]
- Wang, R.; Xu, C.; Zhao, W.; Zhang, J.; Cao, K.; Yang, B.; Wu, L. Calcium and polyamine regulated calcium-sensing receptors in cardiac tissues. Eur. J. Biochem. 2003, 270, 2680–2688. [Google Scholar] [CrossRef] [Green Version]
- Hendy, G.N.; Canaff, L. Calcium-Sensing Receptor Gene: Regulation of Expression. Front. Physiol. 2016, 7, 394. [Google Scholar] [CrossRef] [Green Version]
- Tfelt-Hansen, J.; Hansen, J.L.; Smajilovic, S.; Terwilliger, E.F.; Haunso, S.; Sheikh, S.P. Calcium receptor is functionally expressed in rat neonatal ventricular cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2006, 290, H1165–H1171. [Google Scholar] [CrossRef] [Green Version]
- Smajilovic, S.; Hansen, J.L.; Christoffersen, T.E.H.; Lewin, E.; Sheikh, S.P.; Terwilliger, E.F.; Brown, E.M.; Haunso, S.; Tfelt-Hansen, J. Extracellular calcium sensing in rat aortic vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2006, 348, 1215–1223. [Google Scholar] [CrossRef]
- Ortiz-Capisano, M.C.; Ortiz, P.A.; Garvin, J.L.; Harding, P.; Beierwaltes, W.H. Expression and function of the calcium-sensing receptor in juxtaglomerular cells. Hypertension 2007, 50, 737–743. [Google Scholar] [CrossRef] [Green Version]
- Hinson, T.K.; Damodaran, T.V.; Chen, J.; Zhang, X.; Qumsiyeh, M.B.; Seldin, M.F.; Quarles, L.D. Identification of putative transmembrane receptor sequences homologous to the calcium-sensing G-protein-coupled receptor. Genomics 1997, 45, 279–289. [Google Scholar] [CrossRef]
- Quarles, L.D.; Hartle, J.E.; Siddhanti, S.R.; Guo, R.; Hinson, T.K. A distinct cation-sensing mechanism in MC3T3-E1 osteoblasts functionally related to the calcium receptor. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 1997, 12, 393–402. [Google Scholar] [CrossRef]
- Zaidi, M.; Kerby, J.; Huang, C.L.; Alam, T.; Rathod, H.; Chambers, T.J.; Moonga, B.S. Divalent cations mimic the inhibitory effect of extracellular ionised calcium on bone resorption by isolated rat osteoclasts: Further evidence for a “calcium receptor”. J. Cell. Physiol. 1991, 149, 422–427. [Google Scholar] [CrossRef]
- Wellendorph, P.; Bräuner-Osborne, H. Molecular cloning, expression, and sequence analysis of GPRC6A, a novel family C G-protein-coupled receptor. Gene 2004, 335, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, S.; Zonefrati, R.; Galli, G.; Gronchi, G.; Perigli, G.; Borrelli, A.; Brandi, M.L. In Vitro Control of Genes Critical for Parathyroid Embryogenesis by Extracellular Calcium. J. Endocr. Soc. 2020, 4, bvaa058. [Google Scholar] [CrossRef] [PubMed]
- Mos, I.; Jacobsen, S.E.; Foster, S.R.; Bräuner-Osborne, H. Calcium-Sensing Receptor Internalization Is β-Arrestin-Dependent and Modulated by Allosteric Ligands. Mol. Pharmacol. 2019, 96, 463–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerbino, A.; Colella, M. The Different Facets of Extracellular Calcium Sensors: Old and New Concepts in Calcium-Sensing Receptor Signalling and Pharmacology. Int. J. Mol. Sci. 2018, 19, 999. [Google Scholar] [CrossRef] [Green Version]
| MRFs Genes | T0 HSCs1 | T0 HSCs2 | T0 HSCs3 | 7 Days hSCs1 | 7 Days hSCs2 | 7 Days hSCs3 |
|---|---|---|---|---|---|---|
| Myf5 | + | + | + | + | + | + |
| MyoD1 | + | + | + | + | + | + |
| Myogenin | -- | -- | -- | + | + | + |
| MHC | -- | -- | -- | + | + | + |
| Name of Gene | Primer Sequence | Tm (°C) | Amplicon Size (bp) |
|---|---|---|---|
| β-actin | Forward 5′-AGCCTCGCCTTTGCCGA-3′ | 60 °C | 174 |
| Reverse 5′-CTGGTGCCTGGGGCG-3′ | |||
| Pax7 | Forward 5′-GGTACCGAGAATGATGCGG-3′ | 55 °C | 124 |
| Reverse 5′-CCCATTGATGAAGACCCCTC-3′ | |||
| Myf5 | Forward 5′-ATGCCATCCGCTACATCG-3′ | 55 °C | 145 |
| Reverse 5′-ACAGGACTGTTACATTCGGC-3′ | |||
| MyoD1 | Forward 5′-GACGTGCCTTCTGAGTCG-3′ | 55 °C | 148 |
| Reverse 5′-CTCAGAGCACCTGGTATATCG-3′ | |||
| Myogenin | Forward 5′-AGCGAATGCAGCTCTCAC-3′ | 55 °C | 150 |
| Reverse 5′-TGTGATGCTGTCCACGATG-3′ | |||
| MHC | Forward 5′-GAGTCCTTTGTGAAAGCAACAG-3′ | 55 °C | 143 |
| Reverse 5′-GCCATGTCCTCGATCTTGTC-3′ | |||
| CaSR exons 2/3 | Forward 5′-GATCAAGATCTCAAATCAAG-3′ | 57 °C | 139 |
| Reverse 5′-CCAGCGTCAAGTTGGGAAGA-3′ | |||
| CaSR exons 4/5 | Forward 5′-CTGAGAGGTCACGAAGAAAGTG-3′ | 58 °C | 367 |
| Reverse 5′-GGTGCCAGTTGATGATGGAATA-3′ | |||
| CaSR exons 6/7 | Forward 5′-CTGCTGCTTTGAGTGTGTGG-3′ | 60 °C | 124 |
| Reverse 5′-CTTGGCAATGCAGGAGGTGT-3′ | |||
| GPRC6A | Forward 5′-CAGGAGTGTGTTGGCTTTGA-3′ | 58 °C | 238 |
| Reverse 5′-CTCTTGGCATGTAGCTGGAA-3′ |
| Name of Gene | Probe and Primer Sequences | Tm (°C) | Amplicon Size (bp) |
|---|---|---|---|
| RPS18 | Forward 5′-GATGGCAAAGGCTATTTTCCG-3′ | 58 °C | 132 |
| Reverse 5′-TCTTCCACAGGAGGCCTAC-3′ | |||
| Probe 5′-/56-FAM/TTCAGGGAT/ZEN/CACTAG AGACATGGCTGC/31ABkFQ/-3′ | |||
| CaSR exons 6/7 | Forward 5′-TGCTTTGAGTGTGTGGAGTG-3′ | 58 °C | 100 |
| Reverse 5′-GGTTCTCATTGGACCAGAAGTC-3′ | |||
| Probe 5′-/56-FAM/AGGCACTGG/ZEN/CATCTGTC | |||
| TCATCAC/31ABkFQ/-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romagnoli, C.; Sharma, P.; Zonefrati, R.; Palmini, G.; Lucattelli, E.; Ward, D.T.; Ellinger, I.; Innocenti, M.; Brandi, M.L. Study of the Expression and Function of Calcium-Sensing Receptor in Human Skeletal Muscle. Int. J. Mol. Sci. 2021, 22, 7282. https://doi.org/10.3390/ijms22147282
Romagnoli C, Sharma P, Zonefrati R, Palmini G, Lucattelli E, Ward DT, Ellinger I, Innocenti M, Brandi ML. Study of the Expression and Function of Calcium-Sensing Receptor in Human Skeletal Muscle. International Journal of Molecular Sciences. 2021; 22(14):7282. https://doi.org/10.3390/ijms22147282
Chicago/Turabian StyleRomagnoli, Cecilia, Preeti Sharma, Roberto Zonefrati, Gaia Palmini, Elena Lucattelli, Donald T. Ward, Isabella Ellinger, Marco Innocenti, and Maria Luisa Brandi. 2021. "Study of the Expression and Function of Calcium-Sensing Receptor in Human Skeletal Muscle" International Journal of Molecular Sciences 22, no. 14: 7282. https://doi.org/10.3390/ijms22147282
APA StyleRomagnoli, C., Sharma, P., Zonefrati, R., Palmini, G., Lucattelli, E., Ward, D. T., Ellinger, I., Innocenti, M., & Brandi, M. L. (2021). Study of the Expression and Function of Calcium-Sensing Receptor in Human Skeletal Muscle. International Journal of Molecular Sciences, 22(14), 7282. https://doi.org/10.3390/ijms22147282








