MCPIP1/Regnase-1 Expression in Keratinocytes of Patients with Hidradenitis Suppurativa: Preliminary Results
Abstract
1. Introduction
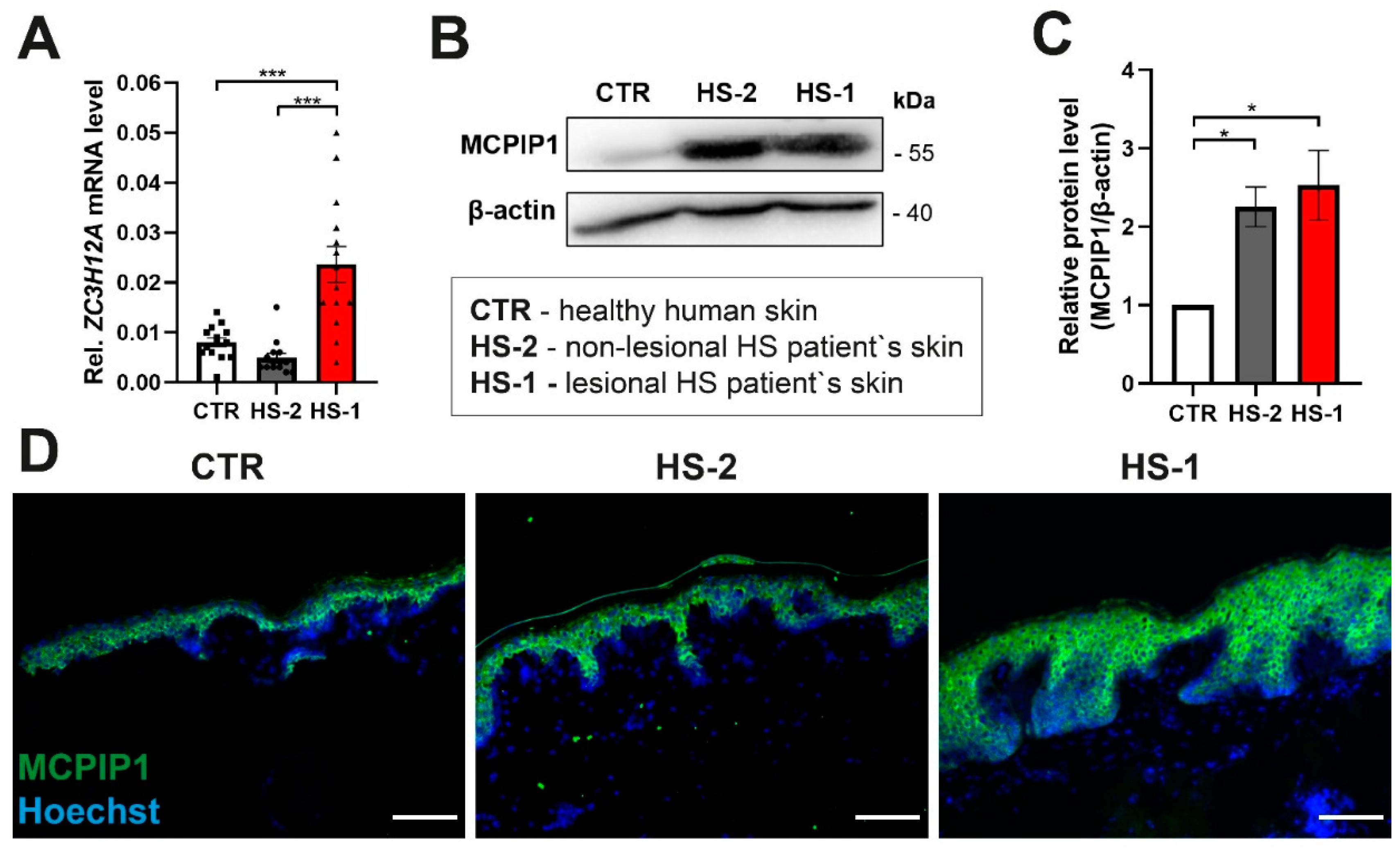
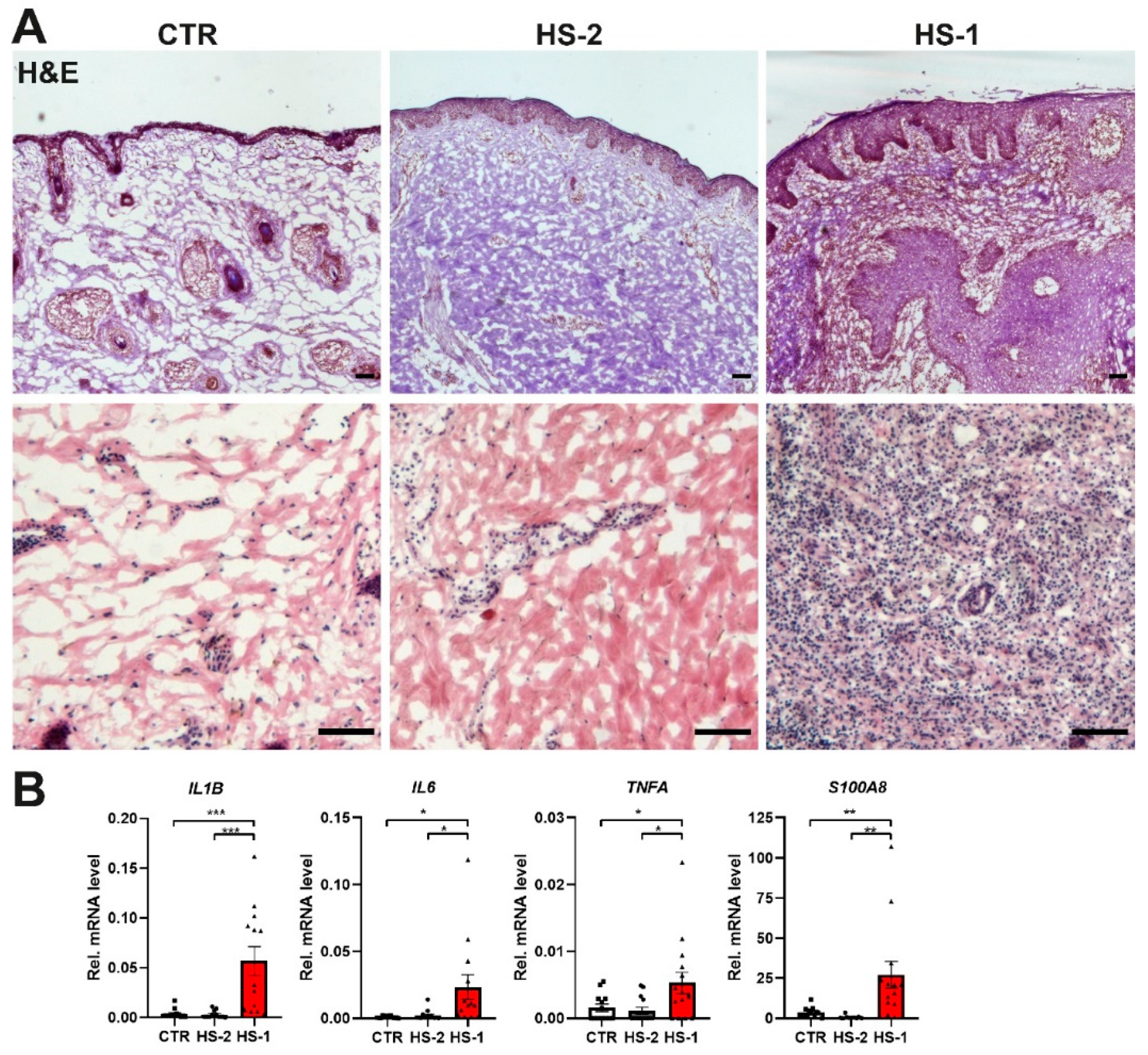
2. Results
MCPIP1 Is Aberrantly Expressed in Hidradenitis Suppurativa
3. Discussion
4. Materials and Methods
4.1. Biopsy
4.2. RNA Isolation and Quantitative Real-Time PCR
4.3. Western Blot Analysis
4.4. Immunofluorescence Staining
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Medzhitov, R. Inflammation 2010: New adventures of an old flame. Cell 2010, 140, 771–776. [Google Scholar] [CrossRef]
- Nathan, C. Points of control in inflammation. Nature 2002, 420, 846–852. [Google Scholar] [CrossRef]
- Ozen, S. What’s new in autoinflammation? Pediatr. Nephrol. 2019, 34, 2449–2456. [Google Scholar] [CrossRef]
- Seelaender, M.; Neto, J.C.; Pimentel, G.D.; Goldszmid, R.S.; Lira, F.S. Inflammation in the disease: Mechanism and therapies 2014. Mediat. Inflamm. 2015, 2015, 169852. [Google Scholar] [CrossRef]
- Pasparakis, M.; Haase, I.; Nestle, F.O. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 2014, 14, 289–301. [Google Scholar] [CrossRef]
- Vekic, D.A.; Frew, J.; Cains, G.D. Hidradenitis suppurativa, a review of pathogenesis, associations and management. Australas. J. Dermatol. 2018, 59 Pt 1, 267–277. [Google Scholar] [CrossRef]
- Prens, E.; Deckers, I. Pathophysiology of hidradenitis suppurativa: An update. J. Am. Acad. Dermatol. 2015, 73 (Suppl. 1), S8–S11. [Google Scholar] [CrossRef]
- Wlodarek, K.; Ponikowska, M.; Matusiak, L.; Szepietowski, J.C. Biologics for hidradenitis suppurativa: An update. Immunotherapy 2019, 11, 45–59. [Google Scholar] [CrossRef]
- Miller, I.; Lynggaard, C.D.; Lophaven, S.; Zachariae, C.; Dufour, D.N.; Jemec, G.B. A double-blind placebo-controlled randomized trial of adalimumab in the treatment of hidradenitis suppurativa. Br. J. Dermatol. 2011, 165, 391–398. [Google Scholar] [CrossRef]
- Grant, A.; Gonzalez, T.; Montgomery, M.O.; Cardenas, V.; Kerdel, F.A. Infliximab therapy for patients with moderate to severe hidradenitis suppurativa: A randomized, double-blind, placebo-controlled crossover trial. J. Am. Acad. Dermatol. 2010, 62, 205–217. [Google Scholar] [CrossRef]
- Matsushita, K.; Takeuchi, O.; Standley, D.M.; Kumagai, Y.; Kawagoe, T.; Miyake, T.; Satoh, T.; Kato, H.; Tsujimura, T.; Nakamura, H.; et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 2009, 458, 1185–1190. [Google Scholar] [CrossRef]
- Mizgalska, D.; Wegrzyn, P.; Murzyn, K.; Kasza, A.; Koj, A.; Jura, J.; Jarząb, B.; Jura, J. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. FEBS J. 2009, 276, 7386–7399. [Google Scholar] [CrossRef]
- Uehata, T.; Akira, S. mRNA degradation by the endoribonuclease Regnase-1/ZC3H12a/MCPIP-1. Biochim. Biophys. Acta 2013, 1829, 708–713. [Google Scholar] [CrossRef]
- Fu, M.; Blackshear, P.J. RNA-binding proteins in immune regulation: A focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 2017, 17, 130–143. [Google Scholar] [CrossRef]
- Jura, J.; Skalniak, L.; Koj, A. Monocyte chemotactic protein-1-induced protein-1 (MCPIP1) is a novel multifunctional modulator of inflammatory reactions. Biochim. Biophys. Acta 2012, 1823, 1905–1913. [Google Scholar] [CrossRef]
- Zhou, Z.; Miao, R.; Huang, S.; Elder, B.; Quinn, T.; Papasian, C.J.; Zhang, J.; Fan, D.; Chen, Y.E.; Fu, M. MCPIP1 deficiency in mice results in severe anemia related to autoimmune mechanisms. PLoS ONE 2013, 8, e82542. [Google Scholar] [CrossRef]
- Liang, J.; Saad, Y.; Lei, T.; Wang, J.; Qi, D.; Yang, Q.; Kolattukudy, P.E.; Fu, M. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J. Exp. Med. 2010, 207, 2959–2973. [Google Scholar] [CrossRef]
- Miao, R.; Huang, S.; Zhou, Z.; Quinn, T.; Van Treeck, B.; Nayyar, T. Targeted disruption of MCPIP1/Zc3h12a results in fatal inflammatory disease. Immunol. Cell Biol. 2013, 91, 368–376. [Google Scholar] [CrossRef]
- Garg, A.V.; Amatya, N.; Chen, K.; Cruz, J.A.; Grover, P.; Whibley, N. MCPIP1 Endoribonuclease Activity Negatively Regulates Interleukin-17-Mediated Signaling and Inflammation. Immunity 2015, 43, 475–487. [Google Scholar] [CrossRef]
- Ruiz-Romeu, E.; Ferran, M.; Gimenez-Arnau, A.; Bugara, B.; Lipert, B.; Jura, J. MCPIP1 RNase Is Aberrantly Distributed in Psoriatic Epidermis and Rapidly Induced by IL-17A. J. Investig. Dermatol. 2016, 136, 1599–1607. [Google Scholar] [CrossRef][Green Version]
- Takaishi, M.; Satoh, T.; Akira, S.; Sano, S. Regnase-1, an Immunomodulator, Limits the IL-36/IL-36R Autostimulatory Loop in Keratinocytes to Suppress Skin Inflammation. J. Investig. Dermatol. 2018, 138, 1439–1442. [Google Scholar] [CrossRef]
- Konieczny, P.; Lichawska-Cieslar, A.; Kwiecinska, P.; Cichy, J.; Pietrzycka, R.; Szukala, W. Keratinocyte-specific ablation of Mcpip1 impairs skin integrity and promotes local and systemic inflammation. J. Mol. Med. 2019, 97, 1669–1684. [Google Scholar] [CrossRef] [PubMed]
- Monin, L.; Gudjonsson, J.E.; Childs, E.E.; Amatya, N.; Xing, X.; Verma, A.H. MCPIP1/Regnase-1 Restricts IL-17A- and IL-17C-Dependent Skin Inflammation. J. Immunol. 2017, 198, 767–775. [Google Scholar] [CrossRef]
- Lichawska-Cieslar, A.; Konieczny, P.; Szukala, W.; Declercq, W.; Fu, M.; Jura, J. Loss of keratinocyte Mcpip1 abruptly activates the IL-23/Th17 and Stat3 pathways in skin inflammation. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118866. [Google Scholar] [CrossRef]
- Kridin, K.; Shani, M.; Schonmann, Y.; Fisher, S.; Shalom, G.; Comaneshter, D.; Batat, E.; Cohen, A.D. Psoriasis and Hidradenitis Suppurativa: A Large-scale Population-based Study. J. Am. Acad. Dermatol. 2018. [Google Scholar] [CrossRef]
- Iwasaki, H.; Takeuchi, O.; Teraguchi, S.; Matsushita, K.; Uehata, T.; Kuniyoshi, K.; Satoh, T.; Saitoh, T.; Matsushita, M.; Standley, D.M.; et al. The IkappaB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat. Immunol. 2011, 12, 1167–1175. [Google Scholar] [CrossRef]
- Deckers, I.E.; van der Zee, H.H.; Prens, E.P. Epidemiology of Hidradenitis Suppurativa: Prevalence, Pathogenesis, and Factors Associated with the Development of HS. Curr. Dermatol. Rep. 2013, 3, 54–60. [Google Scholar] [CrossRef]
- Lima, A.L.; Karl, I.; Giner, T.; Poppe, H.; Schmidt, M.; Presser, D.; Goebeler, M.; Bauer, B. Keratinocytes and neutrophils are important sources of proinflammatory molecules in hidradenitis suppurativa. Br. J. Dermatol. 2016, 174, 514–521. [Google Scholar] [CrossRef]
- van der Zee, H.H.; de Ruiter, L.; van den Broecke, D.G.; Dik, W.A.; Laman, J.D.; Prens, E.P. Elevated levels of tumour necrosis factor (TNF)-alpha, interleukin (IL)-1beta and IL-10 in hidradenitis suppurativa skin: A rationale for targeting TNF-alpha and IL-1beta. Br. J. Dermatol. 2011, 164, 1292–1298. [Google Scholar] [CrossRef]
- Matusiak, L.; Bieniek, A.; Szepietowski, J.C. Increased serum tumour necrosis factor-alpha in hidradenitis suppurativa patients: Is there a basis for treatment with anti-tumour necrosis factor-alpha agents? Acta Derm. Venereol. 2009, 89, 601–603. [Google Scholar] [CrossRef]
- Mozeika, E.; Pilmane, M.; Nurnberg, B.M.; Jemec, G.B. Tumour necrosis factor-alpha and matrix metalloproteinase-2 are expressed strongly in hidradenitis suppurativa. Acta Derm. Venereol. 2013, 93, 301–304. [Google Scholar] [CrossRef]
- Emelianov, V.U.; Bechara, F.G.; Glaser, R.; Langan, E.A.; Taungjaruwinai, W.M.; Schroder, J.M.; Meyer, K.C.; Paus, R. Immunohistological pointers to a possible role for excessive cathelicidin (LL-37) expression by apocrine sweat glands in the pathogenesis of hidradenitis suppurativa/acne inversa. Br. J. Dermatol. 2012, 166, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Lister, M.F.; Sharkey, J.; Sawatzky, D.A.; Hodgkiss, J.P.; Davidson, D.J.; Rossi, A.G.; Finlayson, K. The role of the purinergic P2X7 receptor in inflammation. J. Inflamm. 2007, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Salzer, S.; Kresse, S.; Hirai, Y.; Koglin, S.; Reinholz, M.; Ruzicka, T.; Schauber, J. Cathelicidin peptide LL-37 increases UVB-triggered inflammasome activation: Possible implications for rosacea. J. Dermatol. Sci. 2014, 76, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Manfredini, M.; Giuliani, A.L.; Ruina, G.; Gafa, R.; Bosi, C.; Zoppas, E.; Di Virgilio, F.; Bettoli, V. The P2X7 Receptor Is Overexpressed in the Lesional Skin of Subjects Affected by Hidradenitis Suppurativa: A Preliminary Study. Dermatology 2021, 237, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Killeen, M.E.; Ferris, L.; Kupetsky, E.A.; Falo, L., Jr.; Mathers, A.R. Signaling through purinergic receptors for ATP induces human cutaneous innate and adaptive Th17 responses: Implications in the pathogenesis of psoriasis. J. Immunol. 2013, 190, 4324–4336. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Del Marmol, V.; Mrowietz, U.; Prens, E.P.; Tzellos, T.; Jemec, G.B. Hidradenitis Suppurativa/Acne Inversa: Criteria for Diagnosis, Severity Assessment, Classification and Disease Evaluation. Dermatology 2015, 231, 184–190. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Tzellos, T.; Kyrgidis, A.; Jemec, G.B.E.; Bechara, F.G.; Giamarellos-Bourboulis, E.J.; Ingram, J.R.; Kanni, T.; Karagiannidis, I.; Martorell, A.; et al. Development and validation of the International Hidradenitis Suppurativa Severity Score System (IHS4), a novel dynamic scoring system to assess HS severity. Br. J. Dermatol. 2017, 177, 1401–1409. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krajewski, P.K.; Szukała, W.; Lichawska-Cieślar, A.; Matusiak, Ł.; Jura, J.; Szepietowski, J.C. MCPIP1/Regnase-1 Expression in Keratinocytes of Patients with Hidradenitis Suppurativa: Preliminary Results. Int. J. Mol. Sci. 2021, 22, 7241. https://doi.org/10.3390/ijms22147241
Krajewski PK, Szukała W, Lichawska-Cieślar A, Matusiak Ł, Jura J, Szepietowski JC. MCPIP1/Regnase-1 Expression in Keratinocytes of Patients with Hidradenitis Suppurativa: Preliminary Results. International Journal of Molecular Sciences. 2021; 22(14):7241. https://doi.org/10.3390/ijms22147241
Chicago/Turabian StyleKrajewski, Piotr K., Weronika Szukała, Agata Lichawska-Cieślar, Łukasz Matusiak, Jolanta Jura, and Jacek C. Szepietowski. 2021. "MCPIP1/Regnase-1 Expression in Keratinocytes of Patients with Hidradenitis Suppurativa: Preliminary Results" International Journal of Molecular Sciences 22, no. 14: 7241. https://doi.org/10.3390/ijms22147241
APA StyleKrajewski, P. K., Szukała, W., Lichawska-Cieślar, A., Matusiak, Ł., Jura, J., & Szepietowski, J. C. (2021). MCPIP1/Regnase-1 Expression in Keratinocytes of Patients with Hidradenitis Suppurativa: Preliminary Results. International Journal of Molecular Sciences, 22(14), 7241. https://doi.org/10.3390/ijms22147241






