Inherited Retinal Diseases Due to RPE65 Variants: From Genetic Diagnostic Management to Therapy
Abstract
1. Introduction
2. Epidemiology of RPE65-IRDs
3. Molecular Biology of RPE65
4. Sequence Variants in the RPE65 Gene: An Overview
4.1. Variants
4.2. Variants of Uncertain Significance Assessment
4.3. Challenging RPE65-IRD Cases
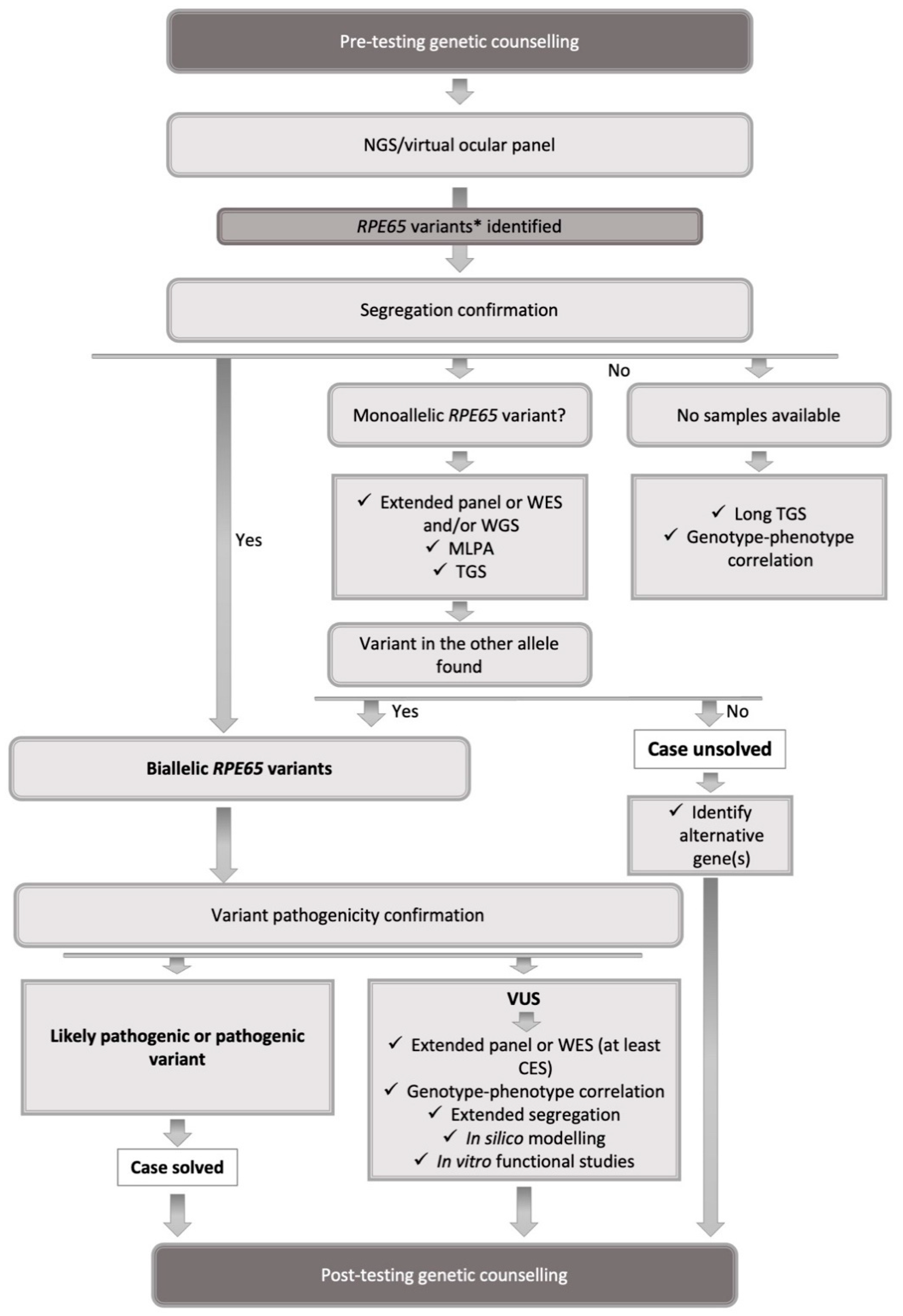
5. Testing Strategy
6. Role of Genetic Counseling
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rivolta, C.; Sharon, D.; DeAngelis, M.M.; Dryja, T.P. Retinitis pigmentosa and allied diseases: Numerous diseases, genes, and inheritance patterns. Hum. Mol. Genet. 2002, 11, 1219–1227. [Google Scholar] [CrossRef]
- Royal Australian and New Zealand college of Ophthalmologists (RANZCO). Guidelines for the Assessment and Management of Patients with Inherited Retinal Diseases (IRD). Available online: https://ranzco.edu/wp-content/uploads/2020/05/RANZCO-Guidelines-for-the-assessment-and-management-of-patients-with-inherited-retinal-diseases-IRD.pdf (accessed on 22 December 2020).
- Verbakel, S.K.; van Huet, R.A.C.; Boon, C.J.F.; den Hollander, A.I.; Collin, R.W.J.; Klaver, C.C.W.; Hoyng, C.B.; Roepman, R.; Klevering, B.J. Non-syndromic retinitis pigmentosa. Prog. Retin. Eye Res. 2018, 66, 157–186. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.F.; Joo, K.; Kemp, J.A.; Fialho, S.L.; da Silva Cunha, A., Jr.; Woo, S.J.; Kwon, Y.J. Molecular genetics and emerging therapies for retinitis pigmentosa: Basic research and clinical perspectives. Prog. Retin. Eye Res. 2018, 63, 107–131. [Google Scholar] [CrossRef]
- Hamel, C. Retinitis pigmentosa. Orphanet J. Rare Dis. 2006, 1, 40. [Google Scholar] [CrossRef] [PubMed]
- Hanein, S.; Perrault, I.; Gerber, S.; Tanguy, G.; Barbet, F.; Ducroq, D.; Calvas, P.; Dollfus, H.; Hamel, C.; Lopponen, T.; et al. Leber congenital amaurosis: Comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum. Mutat. 2004, 23, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, N.; Moore, A.T.; Weleber, R.G.; Michaelides, M. Leber congenital amaurosis/early-onset severe retinal dystrophy: Clinical features, molecular genetics and therapeutic interventions. Br. J. Ophthalmol. 2017, 101, 1147–1154. [Google Scholar] [CrossRef]
- Retinal Information Network. RetNet. Available online: https://sph.uth.edu/retnet/ (accessed on 8 April 2021).
- Tsang, S.H.; Sharma, T. Retinitis Pigmentosa (Non-syndromic). In Atlas of Inherited Retinal Diseases; Tsang, S.H., Sharma, T., Eds.; Springer International Publishing: Cham, Swizerland, 2018; pp. 125–130. [Google Scholar]
- Tsang, S.H.; Sharma, T. Leber Congenital Amaurosis. Adv. Exp. Med. Biol. 2018, 1085, 131–137. [Google Scholar] [CrossRef]
- Chao, D.L.; Burr, A.; Pennesi, M. RPE65-Related Leber Congenital Amaurosi /Early-Onset Severe Retinal Dystrophy. In GeneReviews((R)); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Mirzaa, G., Amemiya, A., Eds.; University of Washington: Seattle WA, USA, 1993–2021.
- Chung, D.C.; Bertelsen, M.; Lorenz, B.; Pennesi, M.E.; Leroy, B.P.; Hamel, C.P.; Pierce, E.; Sallum, J.; Larsen, M.; Stieger, K.; et al. The Natural History of Inherited Retinal Dystrophy Due to Biallelic Mutations in the RPE65 Gene. Am. J. Ophthalmol. 2019, 199, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, B.; Wabbels, B.; Wegscheider, E.; Hamel, C.P.; Drexler, W.; Preising, M.N. Lack of fundus autofluorescence to 488 nanometers from childhood on in patients with early-onset severe retinal dystrophy associated with mutations in RPE65. Ophthalmology 2004, 111, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, S.G.; Aleman, T.S.; Cideciyan, A.V.; Roman, A.J.; Sumaroka, A.; Windsor, E.A.; Schwartz, S.B.; Heon, E.; Stone, E.M. Defining the residual vision in leber congenital amaurosis caused by RPE65 mutations. Invest. Ophthalmol. Vis. Sci. 2009, 50, 2368–2375. [Google Scholar] [CrossRef] [PubMed]
- Hull, S.; Holder, G.E.; Robson, A.G.; Mukherjee, R.; Michaelides, M.; Webster, A.R.; Moore, A.T. Preserved visual function in retinal dystrophy due to hypomorphic RPE65 mutations. Br. J. Ophthalmol. 2016, 100, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yu, S.; Duncan, T.; Li, Y.; Liu, P.; Gene, E.; Cortes-Pena, Y.; Qian, H.; Dong, L.; Redmond, T.M. Mouse model of human RPE65 P25L hypomorph resembles wild type under normal light rearing but is fully resistant to acute light damage. Hum. Mol. Genet. 2015, 24, 4417–4428. [Google Scholar] [CrossRef][Green Version]
- Lorenz, B.; Poliakov, E.; Schambeck, M.; Friedburg, C.; Preising, M.N.; Redmond, T.M. A comprehensive clinical and biochemical functional study of a novel RPE65 hypomorphic mutation. Invest. Ophthalmol. Vis. Sci. 2008, 49, 5235–5242. [Google Scholar] [CrossRef]
- Samardzija, M.; von Lintig, J.; Tanimoto, N.; Oberhauser, V.; Thiersch, M.; Reme, C.E.; Seeliger, M.; Grimm, C.; Wenzel, A. R91W mutation in Rpe65 leads to milder early-onset retinal dystrophy due to the generation of low levels of 11-cis-retinal. Hum. Mol. Genet. 2008, 17, 281–292. [Google Scholar] [CrossRef]
- Li, S.; Xiao, X.; Yi, Z.; Sun, W.; Wang, P.; Zhang, Q. RPE65 mutation frequency and phenotypic variation according to exome sequencing in a tertiary centre for genetic eye diseases in China. Acta Ophthalmol. (Copenh.) 2020, 98, e181–e190. [Google Scholar] [CrossRef]
- Choi, E.H.; Suh, S.; Sander, C.L.; Hernandez, C.J.O.; Bulman, E.R.; Khadka, N.; Dong, Z.; Shi, W.; Palczewski, K.; Kiser, P.D. Insights into the pathogenesis of dominant retinitis pigmentosa associated with a D477G mutation in RPE65. Hum. Mol. Genet. 2018, 27, 2225–2243. [Google Scholar] [CrossRef]
- Hull, S.; Mukherjee, R.; Holder, G.E.; Moore, A.T.; Webster, A.R. The clinical features of retinal disease due to a dominant mutation in RPE65. Mol. Vis. 2016, 22, 626–635. [Google Scholar]
- Jauregui, R.; Park, K.S.; Tsang, S.H. Two-year progression analysis of RPE65 autosomal dominant retinitis pigmentosa. Ophthalmic Genet. 2018, 39, 544–549. [Google Scholar] [CrossRef]
- US Food and Drug Administration (FDA). Luxturna: Prescribing Information. Available online: https://www.fda.gov/media/109906/download (accessed on 4 November 2020).
- European Medicines Agency (EMA). Luxturna: EPAR—Product Information. Available online: https://www.ema.europa.eu/en/documents/product-information/luxturna-epar-product-information_en.pdf (accessed on 4 November 2020).
- Dockery, A.; Stephenson, K.; Keegan, D.; Wynne, N.; Silvestri, G.; Humphries, P.; Kenna, P.F.; Carrigan, M.; Farrar, G.J. Target 5000: Target Capture Sequencing for Inherited Retinal Degenerations. Genes 2017, 8, 304. [Google Scholar] [CrossRef] [PubMed]
- Hamel, C.P.; Tsilou, E.; Pfeffer, B.A.; Hooks, J.J.; Detrick, B.; Redmond, T.M. Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem. 1993, 268, 15751–15757. [Google Scholar] [CrossRef]
- Pagon, R.A. Retinitis pigmentosa. Surv. Ophthalmol. 1988, 33, 137–177. [Google Scholar] [CrossRef]
- Avela, K.; Salonen-Kajander, R.; Laitinen, A.; Ramsden, S.; Barton, S.; Rudanko, S.L. The genetic aetiology of retinal degeneration in children in Finland—New founder mutations identified. Acta Ophthalmol. 2019, 97, 805–814. [Google Scholar] [CrossRef]
- Holtan, J.P.; Selmer, K.K.; Heimdal, K.R.; Bragadottir, R. Inherited retinal disease in Norway—A characterisation of current clinical and genetic knowledge. Acta Ophthalmol. 2020, 98, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Koenekoop, R.K. An overview of Leber congenital amaurosis: A model to understand human retinal development. Surv. Ophthalmol. 2004, 49, 379–398. [Google Scholar] [CrossRef] [PubMed]
- Simonelli, F.; Ziviello, C.; Testa, F.; Rossi, S.; Fazzi, E.; Bianchi, P.E.; Fossarello, M.; Signorini, S.; Bertone, C.; Galantuomo, S.; et al. Clinical and molecular genetics of Leber’s congenital amaurosis: A multicenter study of Italian patients. Invest. Ophthalmol. Vis. Sci. 2007, 48, 4284–4290. [Google Scholar] [CrossRef] [PubMed]
- Stone, E.M.; Andorf, J.L.; Whitmore, S.S.; DeLuca, A.P.; Giacalone, J.C.; Streb, L.M.; Braun, T.A.; Mullins, R.F.; Scheetz, T.E.; Sheffield, V.C.; et al. Clinically Focused Molecular Investigation of 1000 Consecutive Families with Inherited Retinal Disease. Ophthalmology 2017, 124, 1314–1331. [Google Scholar] [CrossRef]
- Hanany, M.; Rivolta, C.; Sharon, D. Worldwide carrier frequency and genetic prevalence of autosomal recessive inherited retinal diseases. Proc. Natl. Acad. Sci. USA 2020, 117, 2710–2716. [Google Scholar] [CrossRef] [PubMed]
- Pontikos, N.; Arno, G.; Jurkute, N.; Schiff, E.; Ba-Abbad, R.; Malka, S.; Gimenez, A.; Georgiou, M.; Wright, G.; Armengol, M.; et al. Genetic Basis of Inherited Retinal Disease in a Molecularly Characterised Cohort of More Than 3000 Families from the United Kingdom. Ophthalmology 2020, 127, 1384–1394. [Google Scholar] [CrossRef] [PubMed]
- Sharon, D.; Ben-Yosef, T.; Goldenberg-Cohen, N.; Pras, E.; Gradstein, L.; Soudry, S.; Mezer, E.; Zur, D.; Abbasi, A.H.; Zeitz, C.; et al. A nationwide genetic analysis of inherited retinal diseases in Israel as assessed by the Israeli inherited retinal disease consortium (IIRDC). Hum. Mutat. 2020, 41, 140–149. [Google Scholar] [CrossRef]
- Whelan, L.; Dockery, A.; Wynne, N.; Zhu, J.; Stephenson, K.; Silvestri, G.; Turner, J.; O’Byrne, J.J.; Carrigan, M.; Humphries, P.; et al. Findings from a Genotyping Study of Over 1000 People with Inherited Retinal Disorders in Ireland. Genes 2020, 11, 105. [Google Scholar] [CrossRef]
- Colombo, L.; Maltese, P.E.; Castori, M.; El Shamieh, S.; Zeitz, C.; Audo, I.; Zulian, A.; Marinelli, C.; Benedetti, S.; Costantini, A.; et al. Molecular Epidemiology in 591 Italian Probands With Nonsyndromic Retinitis Pigmentosa and Usher Syndrome. Invest. Ophthalmol. Vis. Sci. 2021, 62, 13. [Google Scholar] [CrossRef] [PubMed]
- Perea-Romero, I.; Gordo, G.; Iancu, I.F.; Del Pozo-Valero, M.; Almoguera, B.; Blanco-Kelly, F.; Carreno, E.; Jimenez-Rolando, B.; Lopez-Rodriguez, R.; Lorda-Sanchez, I.; et al. Genetic landscape of 6089 inherited retinal dystrophies affected cases in Spain and their therapeutic and extended epidemiological implications. Sci. Rep. 2021, 11, 1526. [Google Scholar] [CrossRef] [PubMed]
- Alastalo, T.-P.; Kämpjärvi, K.; Guidugli, L.; Känsäkoski, J.; Wells, K.; Västinsalo, H.; Sarantaus, L.; Salmenperä, P.; Myllykangas, S.; Sankila, E.-M.K.; et al. Prevalence and genetic characteristics of RPE65-associated retinal disease. Invest. Ophthalmol. Vis. Sci. 2019, 60, 400. [Google Scholar]
- Wigman, L.; van Zelst-Stams, W.A.; Pfundt, R.; van den Born, L.I.; Klaver, C.C.; Verheij, J.B.; Hoyng, C.B.; Breuning, M.H.; Boon, C.J.; Kievit, A.J.; et al. Diagnostic exome sequencing in 266 Dutch patients with visual impairment. Eur. J. Hum. Genet. 2017, 25, 591–599. [Google Scholar] [CrossRef]
- Bernardis, I.; Chiesi, L.; Tenedini, E.; Artuso, L.; Percesepe, A.; Artusi, V.; Simone, M.L.; Manfredini, R.; Camparini, M.; Rinaldi, C.; et al. Unravelling the Complexity of Inherited Retinal Dystrophies Molecular Testing: Added Value of Targeted Next-Generation Sequencing. BioMed Res. Int. 2016, 2016, 6341870. [Google Scholar] [CrossRef]
- den Hollander, A.I.; Roepman, R.; Koenekoop, R.K.; Cremers, F.P. Leber congenital amaurosis: Genes, proteins and disease mechanisms. Prog. Retin. Eye Res. 2008, 27, 391–419. [Google Scholar] [CrossRef]
- Yzer, S.; Leroy, B.P.; De Baere, E.; de Ravel, T.J.; Zonneveld, M.N.; Voesenek, K.; Kellner, U.; Ciriano, J.P.; de Faber, J.T.; Rohrschneider, K.; et al. Microarray-based mutation detection and phenotypic characterisation of patients with Leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci. 2006, 47, 1167–1176. [Google Scholar] [CrossRef]
- Lorenz, B.; Tavares, J.; van den Born, L.I.; Marques, J.P.; Scholl, H.P.N.; EVICR.net Group. Current management of patients with RPE65 mutation-associated inherited retinal degenerations (IRDs) in Europe. Results of a multinational survey by the European Vision Institute Clinical Research Network EVICR.net. Ophthalmic Res. 2021. [Google Scholar] [CrossRef]
- Cai, X.; Conley, S.M.; Naash, M.I. RPE65: Role in the visual cycle, human retinal disease, and gene therapy. Ophthalmic Genet. 2009, 30, 57–62. [Google Scholar] [CrossRef]
- Travis, G.H.; Golczak, M.; Moise, A.R.; Palczewski, K. Diseases caused by defects in the visual cycle: Retinoids as potential therapeutic agents. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 469–512. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Yuan, Q.; Li, S.; Travis, G.H. Role of LRAT on the retinoid isomerase activity and membrane association of Rpe65. J. Biol. Chem. 2007, 282, 20915–20924. [Google Scholar] [CrossRef]
- Yang, U.; Gentleman, S.; Gai, X.; Gorin, M.B.; Borchert, M.S.; Lee, T.C.; Villanueva, A.; Koenekoop, R.; Maguire, A.M.; Bennett, J.; et al. Utility of In Vitro Mutagenesis of RPE65 Protein for Verification of Mutational Pathogenicity Before Gene Therapy. JAMA Ophthalmol. 2019, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sodi, A.; Banfi, S.; Testa, F.; Della Corte, M.; Passerini, I.; Pelo, E.; Rossi, S.; Simonelli, F. Italian IRD Working Group. RPE65-associated inherited retinal diseases: Consensus recommendations for eligibility to gene therapy. Orphanet J. Rare Dis. 2021, 16, 257. [Google Scholar] [CrossRef]
- Carr, T.H.; McEwen, R.; Dougherty, B.; Johnson, J.H.; Dry, J.R.; Lai, Z.; Ghazoui, Z.; Laing, N.M.; Hodgson, D.R.; Cruzalegui, F.; et al. Defining actionable mutations for oncology therapeutic development. Nat. Rev. Cancer 2016, 16, 319–329. [Google Scholar] [CrossRef]
- Gu, S.M.; Thompson, D.A.; Srikumari, C.R.; Lorenz, B.; Finckh, U.; Nicoletti, A.; Murthy, K.R.; Rathmann, M.; Kumaramanickavel, G.; Denton, M.J.; et al. Mutations in RPE65 cause autosomal recessive childhood-onset severe retinal dystrophy. Nat. Genet. 1997, 17, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Marlhens, F.; Bareil, C.; Griffoin, J.M.; Zrenner, E.; Amalric, P.; Eliaou, C.; Liu, S.Y.; Harris, E.; Redmond, T.M.; Arnaud, B.; et al. Mutations in RPE65 cause Leber’s congenital amaurosis. Nat. Genet. 1997, 17, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information (NCBI). ClinVar. Available online: https://www.ncbi.nlm.nih.gov/clinvar/?term=rpe65%5Bgene%5D (accessed on 5 January 2021).
- Leiden University Medical Center. LOVD. Available online: https://databases.lovd.nl/shared/variants/RPE65/unique?search_var_status=%3D%22Marked%22%7C%3D%22Public%22#object_id=VariantOnTranscriptUnique%2CVariantOnGenome&id=RPE65&order=VariantOnTranscript%2FDNA%2CASC&search_transcriptid=00018066&page_size=500&page=1 (accessed on 5 January 2021).
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Chung, D.C.; Trzupek, K.; Wellman, J.; High, K. Absence of genotype-phenotype correlations in RPE65 gene mutations associated with autosomal recessive retinal dystrophy. Invest. Ophthalmol. Vis. Sci. 2015, 56, 2891. [Google Scholar]
- Pierrache, L.H.M.; Ghafaryasl, B.; Khan, M.I.; Yzer, S.; van Genderen, M.M.; Schuil, J.; Boonstra, F.N.; Pott, J.W.R.; de Faber, J.; Tjon-Fo-Sang, M.J.H.; et al. Longitudinal Study of Rpe65-Associated Inherited Retinal Degenerations. Retina 2020, 40, 1812–1828. [Google Scholar] [CrossRef]
- Lopez-Rodriguez, R.; Lantero, E.; Blanco-Kelly, F.; Avila-Fernandez, A.; Martin Merida, I.; del Pozo-Valero, M.; Perea-Romero, I.; Zurita, O.; Jiménez-Rolando, B.; Swafiri, S.T.; et al. RPE65/-related retinal dystrophy: Mutational and phenotypic spectrum in 45 affected patients. medRxiv 2021. [Google Scholar] [CrossRef]
- Mahajan, V.; Bennett, J.; Maguire, A.M.; Haller, J.A.; Leroy, B.P.; Sohn, E.H.; Drack, A.V.; Yu, Z.-F.; Tillman, A.; Ciulla, T.A.; et al. RPE65 Mutation subtype effects on baseline visual function and treatment response in phase 3 voretigene neparvovec trial. In Proceedings of the American Academy of Ophthalmology Meeting, Chicago, IL, USA, 27–30 October 2018. [Google Scholar]
- Motta, F.L.; Martin, R.P.; Porto, F.B.O.; Wohler, E.S.; Resende, R.G.; Gomes, C.P.; Pesquero, J.B.; Sallum, J.M.F. Pathogenicity Reclasssification of RPE65 Missense Variants Related to Leber Congenital Amaurosis and Early-Onset Retinal Dystrophy. Genes 2019, 11, 24. [Google Scholar] [CrossRef]
- Ellard, S.; Baple, E.L.; Callaway, A.; Berry, I.; Forrester, N.; Turnbull, C.; Owens, M.; Eccles, D.M.; Abbs, S.; Scott, R.; et al. ACGS Best Practice Guidelines for Variant Classification in Rare Disease 2020. Available online: https://www.acgs.uk.com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf (accessed on 5 January 2021).
- Philp, A.R.; Jin, M.; Li, S.; Schindler, E.I.; Iannaccone, A.; Lam, B.L.; Weleber, R.G.; Fishman, G.A.; Jacobson, S.G.; Mullins, R.F.; et al. Predicting the pathogenicity of RPE65 mutations. Hum. Mutat. 2009, 30, 1183–1188. [Google Scholar] [CrossRef][Green Version]
- Iancu, I.-F.; Avila-Fernandez, A.; Arteche, A.; Trujillo-Tiebas, M.J.; Riveiro-Alvarez, R.; Almoguera, B.; Martin-Merida, I.; Del Pozo-Valero, M.; Perea-Romero, I.; Corton, M. Prioritizing variants of uncertain significance for reclassification using a rule-based algorithm in inherited retinal dystrophies. NPJ Genom. Med. 2021, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Redmond, T.M.; Poliakov, E.; Yu, S.; Tsai, J.Y.; Lu, Z.; Gentleman, S. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc. Natl. Acad. Sci. USA 2005, 102, 13658–13663. [Google Scholar] [CrossRef]
- Ellingford, J.M.; Horn, B.; Campbell, C.; Arno, G.; Barton, S.; Tate, C.; Bhaskar, S.; Sergouniotis, P.I.; Taylor, R.L.; Carss, K.J.; et al. Assessment of the incorporation of CNV surveillance into gene panel next-generation sequencing testing for inherited retinal diseases. J. Med. Genet. 2018, 55, 114–121. [Google Scholar] [CrossRef]
- Thompson, D.A.; McHenry, C.L.; Li, Y.; Richards, J.E.; Othman, M.I.; Schwinger, E.; Vollrath, D.; Jacobson, S.G.; Gal, A. Retinal dystrophy due to paternal isodisomy for chromosome 1 or chromosome 2, with homoallelism for mutations in RPE65 or MERTK, respectively. Am. J. Hum. Genet. 2002, 70, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Mantere, T.; Kersten, S.; Hoischen, A. Long-Read Sequencing Emerging in Medical Genetics. Front. Genet. 2019, 10, 426. [Google Scholar] [CrossRef]
- Black, G.C.; Sergouniotis, P.; Sodi, A.; Leroy, B.P.; Van Cauwenbergh, C.; Liskova, P.; Gronskov, K.; Klett, A.; Kohl, S.; Taurina, G.; et al. The need for widely available genomic testing in rare eye diseases: An ERN-EYE position statement. Orphanet J. Rare Dis. 2021, 16, 142. [Google Scholar] [CrossRef] [PubMed]
- Weisschuh, N.; Mayer, A.K.; Strom, T.M.; Kohl, S.; Glockle, N.; Schubach, M.; Andreasson, S.; Bernd, A.; Birch, D.G.; Hamel, C.P.; et al. Mutation Detection in Patients with Retinal Dystrophies Using Targeted Next Generation Sequencing. PLoS ONE 2016, 11, e0145951. [Google Scholar] [CrossRef]
- Martin-Sanchez, M.; Bravo-Gil, N.; Gonzalez-Del Pozo, M.; Mendez-Vidal, C.; Fernandez-Suarez, E.; Rodriguez-de la Rua, E.; Borrego, S.; Antinolo, G. A Multi-Strategy Sequencing Workflow in Inherited Retinal Dystrophies: Routine Diagnosis, Addressing Unsolved Cases and Candidate Genes Identification. Int. J. Mol. Sci. 2020, 21, 9355. [Google Scholar] [CrossRef] [PubMed]
- Zeitz, C.; Michiels, C.; Neuille, M.; Friedburg, C.; Condroyer, C.; Boyard, F.; Antonio, A.; Bouzidi, N.; Milicevic, D.; Veaux, R.; et al. Where are the missing gene defects in inherited retinal disorders? Intronic and synonymous variants contribute at least to 4% of CACNA1F-mediated inherited retinal disorders. Hum. Mutat. 2019, 40, 765–787. [Google Scholar] [CrossRef] [PubMed]
- Cremers, F.P.M.; Lee, W.; Collin, R.W.J.; Allikmets, R. Clinical spectrum, genetic complexity and therapeutic approaches for retinal disease caused by ABCA4 mutations. Prog. Retin. Eye Res. 2020, 79, 100861. [Google Scholar] [CrossRef]
- Lorenz, B.; Tavares, J.; van den Born, L.I.; Marques, J.P.; Scholl, H.P.N. Current management of Inherited Retinal Degenerations (IRD) patients in Europe. Results of a multinational survey by the European Vision Institute Clinical Research Network EVICR.net. Ophthalmic Res. 2021. [Google Scholar] [CrossRef]
- Jespersgaard, C.; Fang, M.; Bertelsen, M.; Dang, X.; Jensen, H.; Chen, Y.; Bech, N.; Dai, L.; Rosenberg, T.; Zhang, J.; et al. Molecular genetic analysis using targeted NGS analysis of 677 individuals with retinal dystrophy. Sci. Rep. 2019, 9, 1219. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Q.; Wang, Z. The evolution of nanopore sequencing. Front. Genet. 2014, 5, 449. [Google Scholar] [CrossRef]
- Xiao, T.; Zhou, W. The third generation sequencing: The advanced approach to genetic diseases. Transl. Ped. 2020, 9, 163–173. [Google Scholar] [CrossRef]
- Dias, R.; Torkamani, A. Artificial intelligence in clinical and genomic diagnostics. Genome Med. 2019, 11, 70. [Google Scholar] [CrossRef]
- Fujinami-Yokokawa, Y.; Pontikos, N.; Yang, L.; Tsunoda, K.; Yoshitake, K.; Iwata, T.; Miyata, H.; Fujinami, K. Japan Eye Genetics Consortium. Prediction of Causative Genes in Inherited Retinal Disorders from Spectral-Domain Optical Coherence Tomography Utilising Deep Learning Techniques. J. Ophthalmol. 2019, 2019, 1691064. [Google Scholar] [CrossRef] [PubMed]
- Resta, R.; Biesecker, B.B.; Bennett, R.L.; Blum, S.; Hahn, S.E.; Strecker, M.N.; Williams, J.L.; National Society of Genetic Counselors’ Definition Task Force. A new definition of Genetic Counseling: National Society of Genetic Counselors’ Task Force report. J. Genet. Couns. 2006, 15, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Austin, J.; Semaka, A.; Hadjipavlou, G. Conceptualising genetic counseling as psychotherapy in the era of genomic medicine. J. Genet. Couns. 2014, 23, 903–909. [Google Scholar] [CrossRef]
- Madlensky, L.; Trepanier, A.M.; Cragun, D.; Lerner, B.; Shannon, K.M.; Zierhut, H. A rapid systematic review of outcomes studies in genetic counseling. J. Genet. Couns. 2017, 26, 361–378. [Google Scholar] [CrossRef]
- Hafler, B.P. Clinical Progress in Inherited Retinal Degenerations: Gene Therapy Clinical Trials and Advances in Genetic Sequencing. Retina 2017, 37, 417–423. [Google Scholar] [CrossRef] [PubMed]
| Study | Participants * | Findings * |
|---|---|---|
| Simonelli et al. 2007 [31] | Italian patients with LCA/EORD N = 95 | RPE65 mutations in sample: n = 8 (8.4%) |
| Stone et al. 2017 [32] | Consecutive patients with IRD seen by a single physician N = 1000 | RPE65 mutations in the sample: n = 3 Estimated frequency in the US general population: 1:576667 |
| Avela et al. 2019 [28] | Finnish children with IRD N = 68 | RP: n = 14 (21%) LCA/EORD: n = 15 (22%) |
| Chung et al. 2019 [12] | Patients with IRD due to biallelic mutations in RPE65 N = 70 | LCA/EORD: n=39 (50.0%) RP: n = 6 (7.7%) |
| Hanany et al. 2020 [33] | Genotype data on six main world populations | Autosomal recessive IRD: 1:1380 RP: 23% LCA/EORD: 7% Biallelic RPE65: n = 15,620 individuals or 0.3% of all patients with IRD |
| Holtan et al. 2020 [29] | Norwegian patients with IRDN = 866 N = 685 (living in the south-east of Norway) | RP: n = 468 (54.0%) LCA/EORD: n = 45 (5.2%) RPE65-related RP and LCA/EORD: 0.6% Minimum adjusted prevalence of IRD in the south-east of Norway: 1:3856 |
| Pontikos et al. 2020 [34] | Patients with IRD Individuals: N = 4236 | RPE65: n = 51 (1.2%) LCA/EORD: 3% |
| Sharon et al. 2020 [35] | Israeli patients with IRD Individuals: N = 3413 | RP: 43% LCA/EORD: 4% RPE65: 1% of IRDs |
| Whelan et al. 2020 [36] | Irish patients with IRD N = 1004 | RP: 37.75% RPE65: 7.41% of autosomal dominant RP LCA/EORD: 2.59% RPE65: 6.25% of LCA/EORD |
| Colombo et al. (2021) [37] | Italian patients with RP N = 591 | AR-RP: 103/591 (17.5%) RPE65: 2/103 (2%) of AR-RP |
| Perea-Romero et al. (2021) [38] | Spanish patients with IRD N = 6089 individuals (4403 families) | Non-syndromic RP: 55.6% of families RPE65: 23/666 (3%) of families with AR-RP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aoun, M.; Passerini, I.; Chiurazzi, P.; Karali, M.; De Rienzo, I.; Sartor, G.; Murro, V.; Filimonova, N.; Seri, M.; Banfi, S. Inherited Retinal Diseases Due to RPE65 Variants: From Genetic Diagnostic Management to Therapy. Int. J. Mol. Sci. 2021, 22, 7207. https://doi.org/10.3390/ijms22137207
Aoun M, Passerini I, Chiurazzi P, Karali M, De Rienzo I, Sartor G, Murro V, Filimonova N, Seri M, Banfi S. Inherited Retinal Diseases Due to RPE65 Variants: From Genetic Diagnostic Management to Therapy. International Journal of Molecular Sciences. 2021; 22(13):7207. https://doi.org/10.3390/ijms22137207
Chicago/Turabian StyleAoun, Manar, Ilaria Passerini, Pietro Chiurazzi, Marianthi Karali, Irene De Rienzo, Giovanna Sartor, Vittoria Murro, Natalia Filimonova, Marco Seri, and Sandro Banfi. 2021. "Inherited Retinal Diseases Due to RPE65 Variants: From Genetic Diagnostic Management to Therapy" International Journal of Molecular Sciences 22, no. 13: 7207. https://doi.org/10.3390/ijms22137207
APA StyleAoun, M., Passerini, I., Chiurazzi, P., Karali, M., De Rienzo, I., Sartor, G., Murro, V., Filimonova, N., Seri, M., & Banfi, S. (2021). Inherited Retinal Diseases Due to RPE65 Variants: From Genetic Diagnostic Management to Therapy. International Journal of Molecular Sciences, 22(13), 7207. https://doi.org/10.3390/ijms22137207








