Early Transcriptional Liver Signatures in Experimental Visceral Leishmaniasis
Abstract
1. Introduction
2. Results
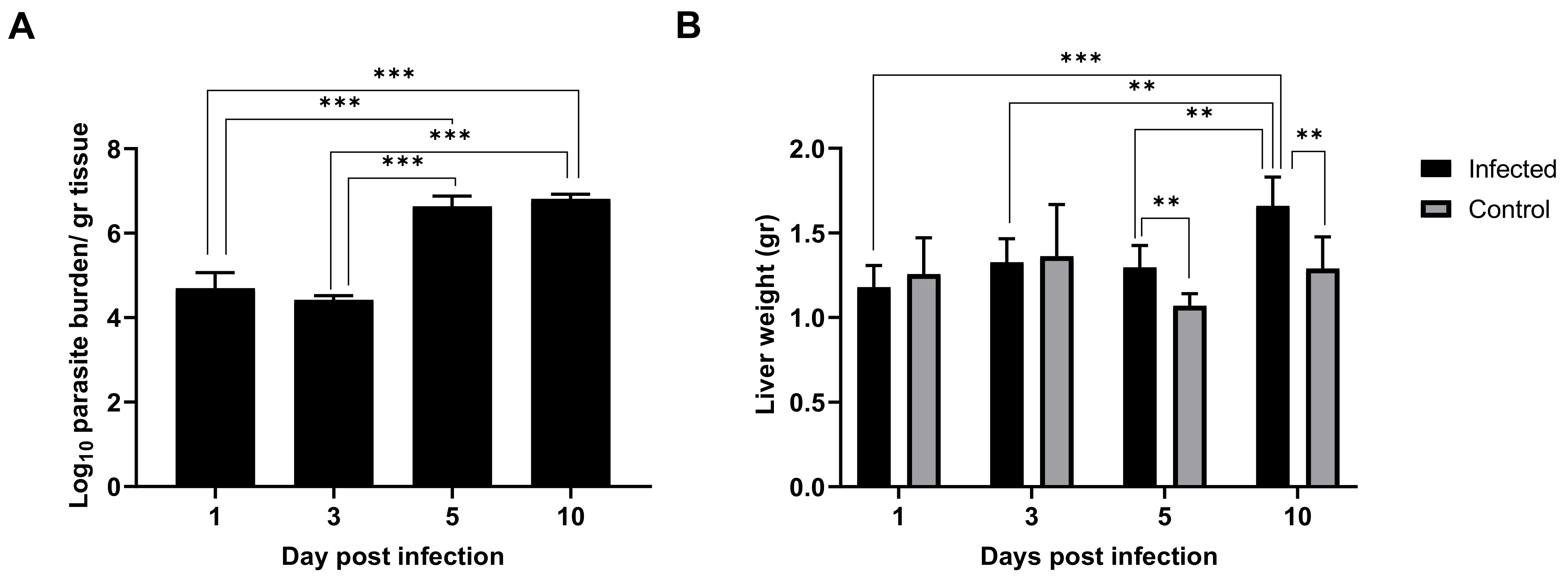
2.1. Leishmania Infantum Infection Is Established in Liver 24 h Post Inoculation
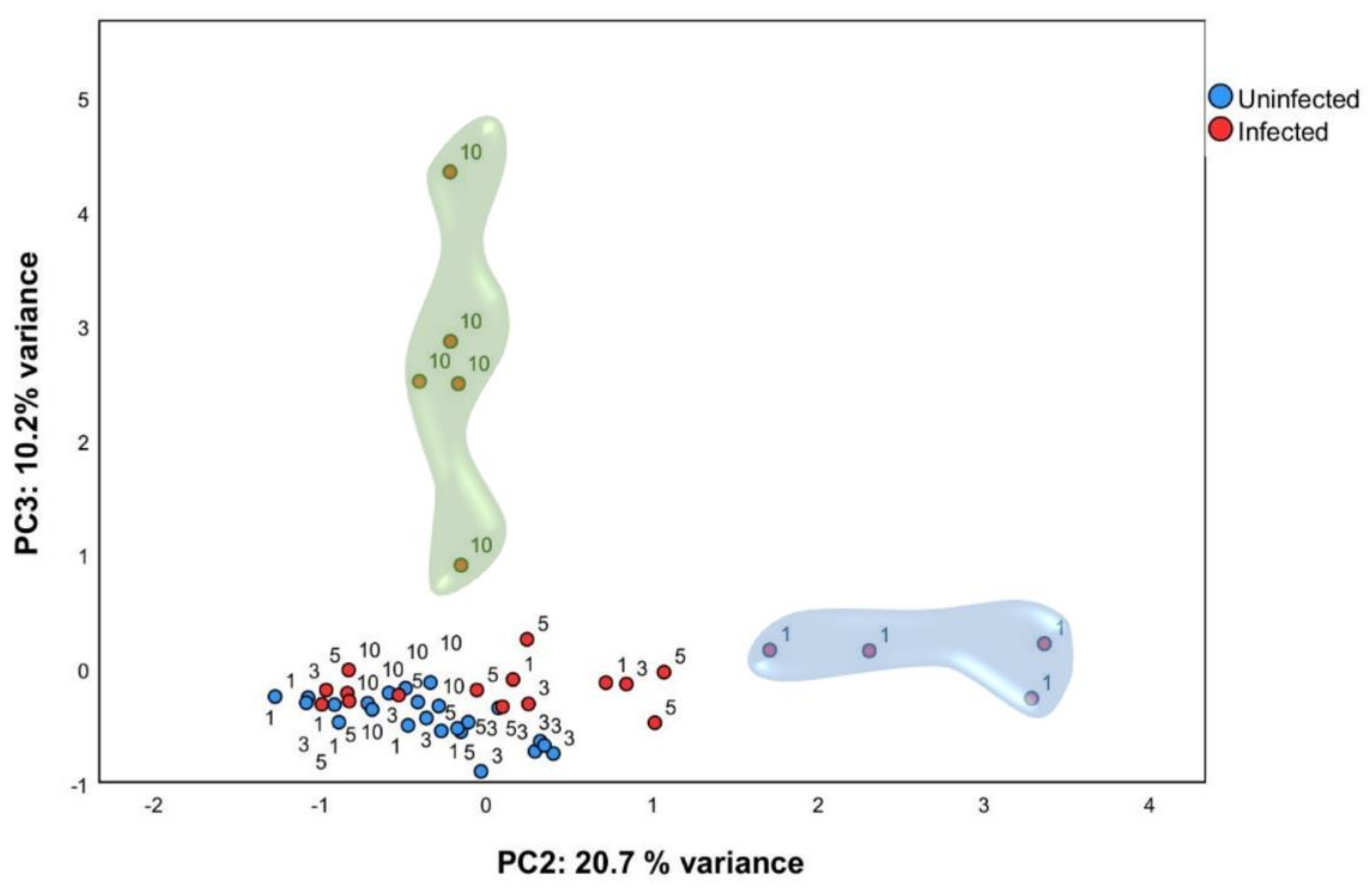
2.2. Multivariate Analysis Identified Particular Gene Expression Profiles at 1- and 10-Days Post Infection
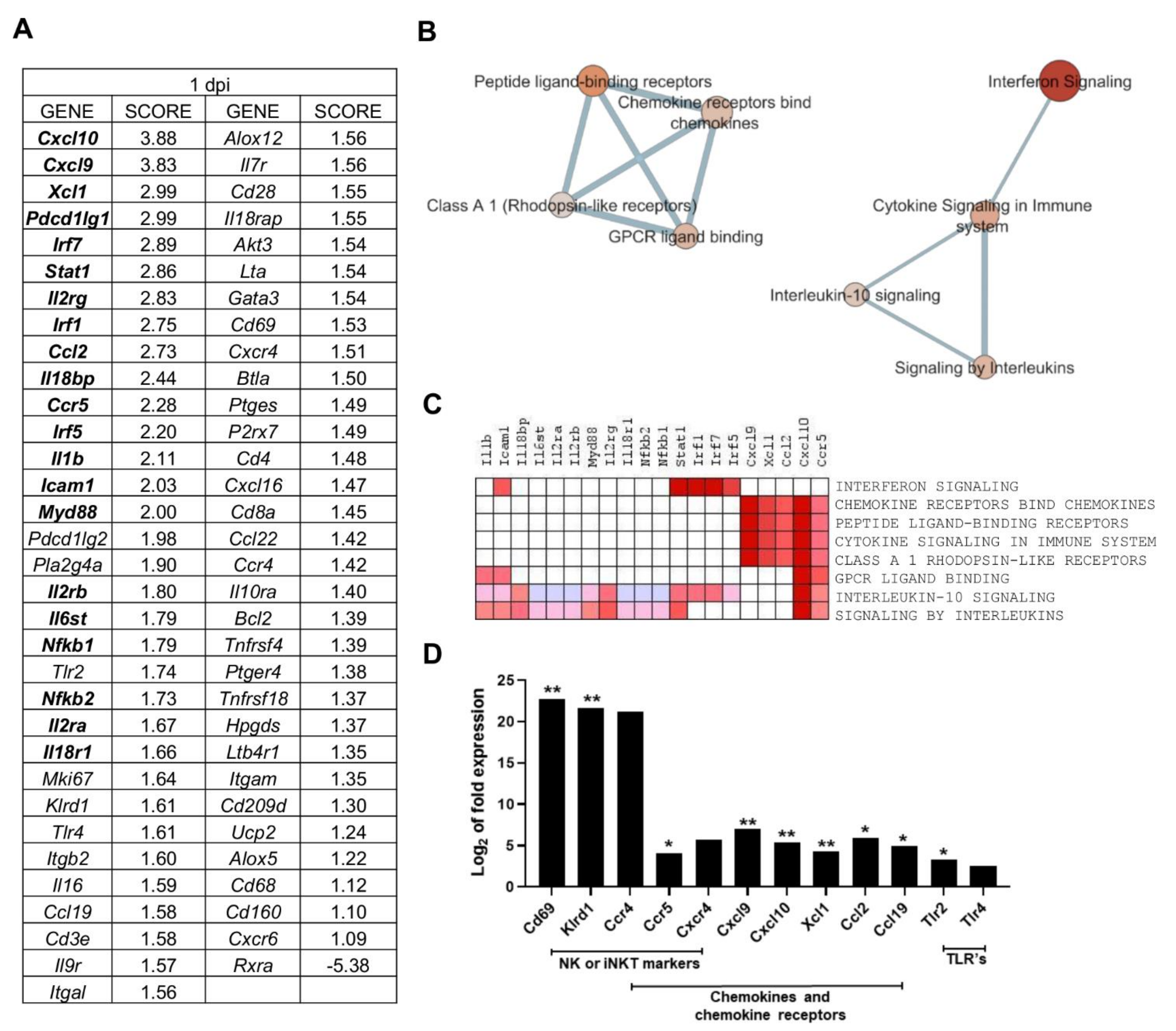
2.3. Multiple Processes Are Activated in L. infantum Infection in Livers of BALB/c Mice at 1-Day Post Inoculation
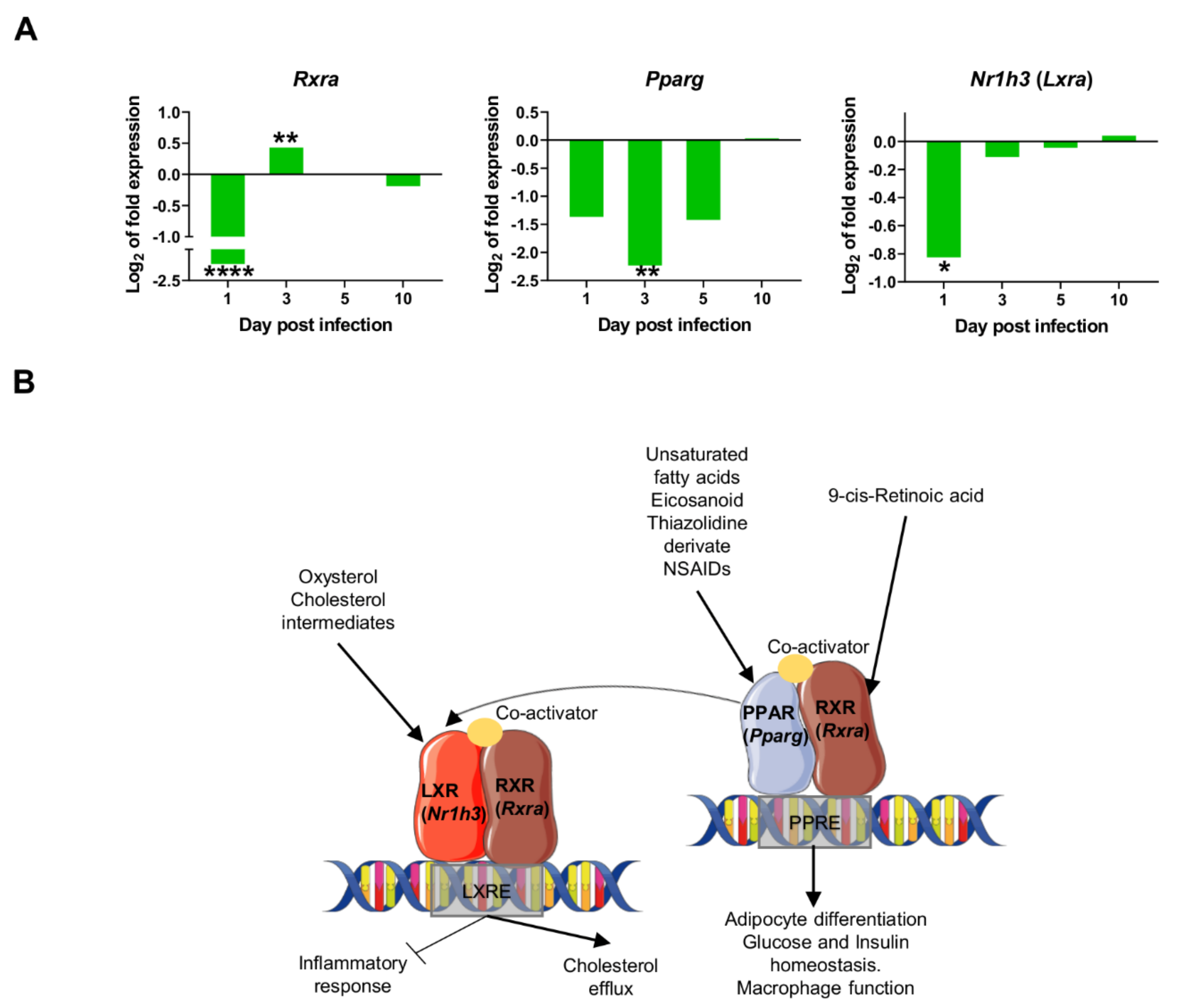
2.4. Downregulation of Transcriptional Factors Involved in Lipid Metabolism and Inflammation
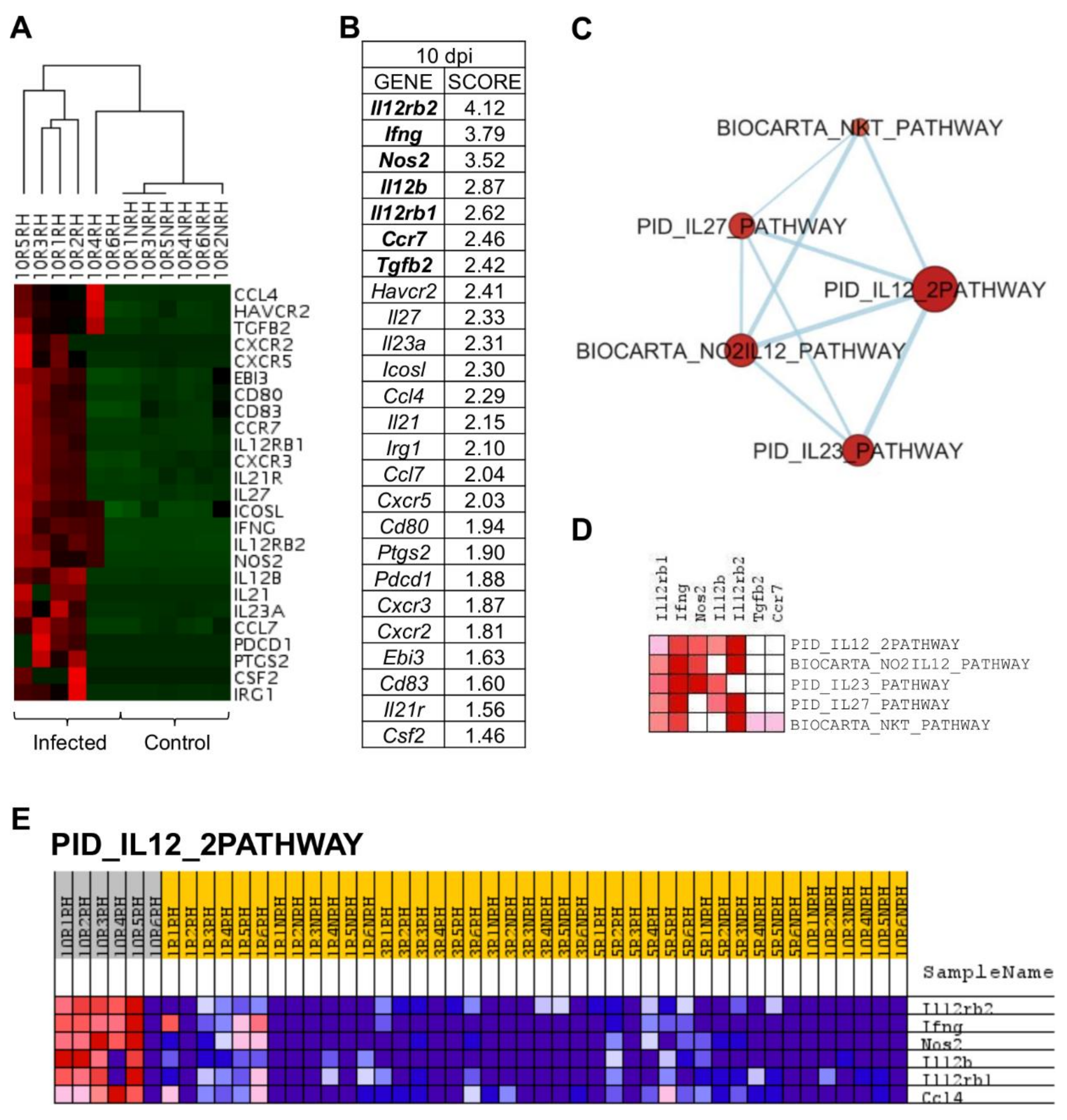
2.5. L. infantum Infection Induces Th1 Responses through Interleukin-12 Signaling at 10 Days Post Infection
3. Discussion
4. Materials and Methods
4.1. Biological Samples
4.2. RNA Isolation and Quantification
4.3. Retrotranscription and High-Throughput Real-Time Quantitative PCR (RT-qPCR)
4.4. Data Pre-Processing and Processing
4.5. Statistical Analysis
4.6. Enrichment Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loria-Cervera, E.N.; Andrade-Narvaez, F.J. Animal models for the study of leishmaniasis immunology. Rev. Inst. Med. Trop. Sao Paulo 2014, 56, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Melby, P.C.; Tryon, V.V.; Chandrasekar, B.; Freeman, G.L. Cloning of Syrian Hamster (Mesocricetus auratus) Cytokine cDNAs and Analysis of Cytokine mRNA Expression in Experimental Visceral Leishmaniasis. Infect. Immun. 1998, 66, 2135–2142. [Google Scholar] [CrossRef] [PubMed]
- Sacks, D.L.; Melby, P.C. Animal Models for the Analysis of Immune Responses to Leishmaniasis. Curr. Protoc. Immunol. 2015, 108, 19.2.1–19.2.24. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.; Santos-Mateus, D.; Alexandre-Pires, G.; Valério-Bolas, A.; Rafael-Fernandes, M.; Pereira, M.A.; Ligeiro, D.; de Jesus, J.; Alves-Azevedo, R.; Lopes-Ventura, S.; et al. Leishmania infantum exerts immunomodulation in canine Kupffer cells reverted by meglumine antimoniate. Comp. Immunol. Microbiol. Infect. Dis. 2017, 55, 42–52. [Google Scholar] [CrossRef]
- Abdullah, Z.; Knolle, P.A. Liver macrophages in healthy and diseased liver. Pflügers Arch. Eur. J. Physiol. 2017, 469, 553–560. [Google Scholar] [CrossRef]
- Kong, F.; Saldarriaga, O.A.; Spratt, H.; Osorio, E.Y.; Travi, B.L.; Luxon, B.A.; Melby, P.C. Transcriptional Profiling in Experimental Visceral Leishmaniasis Reveals a Broad Splenic Inflammatory Environment that Conditions Macrophages toward a Disease-Promoting Phenotype. PLoS Pathog. 2017, 13, e1006165. [Google Scholar] [CrossRef]
- Medina-Colorado, A.A.; Osorio, E.Y.; Saldarriaga, O.A.; Travi, B.L.; Kong, F.; Spratt, H.; Soong, L.; Melby, P.C. Splenic CD4+ T Cells in Progressive Visceral Leishmaniasis Show a Mixed Effector-Regulatory Phenotype and Impair Macrophage Effector Function through Inhibitory Receptor Expression. PLoS ONE 2017, 12, e0169496. [Google Scholar] [CrossRef]
- Ontoria, E.; Hernández-Santana, Y.E.; González-García, A.C.; López, M.C.; Valladares, B.; Carmelo, E. Transcriptional Profiling of Immune-Related Genes in Leishmania infantum-Infected Mice: Identification of Potential Biomarkers of Infection and Progression of Disease. Front. Cell. Infect. Microbiol. 2018, 8, 197. [Google Scholar] [CrossRef]
- Ashwin, H.; Seifert, K.; Forrester, S.; Brown, N.; MacDonald, S.; James, S.; Lagos, D.; Timmis, J.; Mottram, J.C.; Croft, S.L.; et al. Tissue and host species-specific transcriptional changes in models of experimental visceral leishmaniasis [version 2; referees: 4 approved]. Wellcome Open Res. 2019. [Google Scholar] [CrossRef]
- Rodrigues, V.; Cordeiro-da-Silva, A.; Laforge, M.; Silvestre, R.; Estaquier, J. Regulation of immunity during visceral Leishmania infection. Parasit. Vectors 2016, 9, 118. [Google Scholar] [CrossRef]
- Agallou, M.; Athanasiou, E.; Kammona, O.; Tastsoglou, S.; Hatzigeorgiou, A.G.; Kiparissides, C.; Karagouni, E. Transcriptome Analysis Identifies Immune Markers Related to Visceral Leishmaniasis Establishment in the Experimental Model of BALB/c Mice. Front. Immunol. 2019, 10, 2749. [Google Scholar] [CrossRef]
- Gautam, S.; Kumar, R.; Singh, N.; Singh, A.K.; Rai, M.; Sacks, D.; Sundar, S.; Nylén, S. CD8 T Cell Exhaustion in Human Visceral Leishmaniasis. J. Infect. Dis. 2014, 209, 290–299. [Google Scholar] [CrossRef]
- Kumar, R.; Bunn, P.T.; Singh, S.S.; Ng, S.S.; Montes de Oca, M.; De Labastida Rivera, F.; Chauhan, S.B.; Singh, N.; Faleiro, R.J.; Edwards, C.L.; et al. Type I Interferons Suppress Anti-parasitic Immunity and Can Be Targeted to Improve Treatment of Visceral Leishmaniasis. Cell Rep. 2020, 30, 2512–2525.e9. [Google Scholar] [CrossRef]
- Cezário, G.A.G.; de Oliveira, L.R.C.; Peresi, E.; Nicolete, V.C.; Polettini, J.; de Lima, C.R.G.; Gatto, M.; Calvi, S.A. Analysis of the expression of toll-like receptors 2 and 4 and cytokine production during experimental Leishmania chagasi infection. Mem. Inst. Oswaldo Cruz 2011, 106, 573–583. [Google Scholar] [CrossRef]
- Buscher, K.; Ehinger, E.; Gupta, P.; Pramod, A.B.; Wolf, D.; Tweet, G.; Pan, C.; Mills, C.D.; Lusis, A.J.; Ley, K. Natural variation of macrophage activation as disease-relevant phenotype predictive of inflammation and cancer survival. Nat. Commun. 2017, 8, 16041. [Google Scholar] [CrossRef]
- Wang, N.; Liang, H.; Zen, K. Molecular Mechanisms That Influence the Macrophage M1–M2 Polarization Balance. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef]
- Zhong, Y.; Yi, C. MicroRNA-720 suppresses M2 macrophage polarization by targeting GATA3. Biosci. Rep. 2016, 36. [Google Scholar] [CrossRef]
- Chiba, Y.; Mizoguchi, I.; Furusawa, J.; Hasegawa, H.; Ohashi, M.; Xu, M.; Owaki, T.; Yoshimoto, T. Interleukin-27 Exerts Its Antitumor Effects by Promoting Differentiation of Hematopoietic Stem Cells to M1 Macrophages. Cancer Res. 2018, 78, 182–194. [Google Scholar] [CrossRef]
- Lawrence, T.; Natoli, G. Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat. Rev. Immunol. 2011, 11, 750–761. [Google Scholar] [CrossRef]
- Jablonski, K.A.; Amici, S.A.; Webb, L.M.; Ruiz-Rosado, J.D.D.; Popovich, P.G.; Partida-Sanchez, S.; Guerau-de-Arellano, M. Novel Markers to Delineate Murine M1 and M2 Macrophages. PLoS ONE 2015, 10, e0145342. [Google Scholar] [CrossRef]
- Lefèvre, L.; Lugo-Villarino, G.; Meunier, E.; Valentin, A.; Olagnier, D.; Authier, H.; Duval, C.; Dardenne, C.; Bernad, J.; Lemesre, J.L.; et al. The C-type Lectin Receptors Dectin-1, MR, and SIGNR3 Contribute Both Positively and Negatively to the Macrophage Response to Leishmania infantum. Immunity 2013, 38, 1038–1049. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Canetti, C.; Coutinho-Silva, R. Crosstalk between purinergic receptors and lipid mediators in leishmaniasis. Parasit. Vectors 2016, 9, 489. [Google Scholar] [CrossRef] [PubMed]
- Rabhi, I.; Rabhi, S.; Ben-Othman, R.; Rasche, A.; Consortium, S.; Daskalaki, A.; Trentin, B.; Piquemal, D.; Regnault, B.; Descoteaux, A.; et al. Transcriptomic Signature of Leishmania Infected Mice Macrophages: A Metabolic Point of View. PLoS Negl. Trop. Dis. 2012, 6, e1763. [Google Scholar] [CrossRef] [PubMed]
- Rabhi, S.; Rabhi, I.; Trentin, B.; Piquemal, D.; Regnault, B.; Goyard, S.; Lang, T.; Descoteaux, A.; Enninga, J.; Guizani-Tabbane, L. Lipid Droplet Formation, Their Localization and Dynamics during Leishmania major Macrophage Infection. PLoS ONE 2016, 11, e0148640. [Google Scholar] [CrossRef]
- Beattie, L.; d’El-Rei Hermida, M.; Moore, J.W.J.; Maroof, A.; Brown, N.; Lagos, D.; Kaye, P.M. A Transcriptomic Network Identified in Uninfected Macrophages Responding to Inflammation Controls Intracellular Pathogen Survival. Cell Host Microbe 2013, 14, 357–368. [Google Scholar] [CrossRef][Green Version]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Rodríguez-Perea, A.L.; Arcia, E.D.; Rueda, C.M.; Velilla, P.A. Phenotypical characterization of regulatory T cells in humans and rodents. Clin. Exp. Immunol. 2016, 185, 281–291. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Svensson, M.; Zubairi, S.; Maroof, A.; Kazi, F.; Taniguchi, M.; Kaye, P.M. Invariant NKT Cells Are Essential for the Regulation of Hepatic CXCL10 Gene Expression during Leishmania donovani Infection. Infect. Immun. 2005, 73, 7541–7547. [Google Scholar] [CrossRef]
- Racoosin, E.L.; Beverley, S.M. Leishmania major:Promastigotes Induce Expression of a Subset of Chemokine Genes in Murine Macrophages. Exp. Parasitol. 1997, 85, 283–295. [Google Scholar] [CrossRef]
- Matte, C.; Olivier, M. Leishmania-Induced Cellular Recruitment during the Early Inflammatory Response: Modulation of Proinflammatory Mediators. J. Infect. Dis. 2002, 185, 673–681. [Google Scholar] [CrossRef]
- Zaph, C.; Scott, P. Interleukin-12 Regulates Chemokine Gene Expression during the Early Immune Response to Leishmania major. Infect. Immun. 2003, 71, 1587–1589. [Google Scholar] [CrossRef]
- Cotterell, S.E.J.; Engwerda, C.R.; Kaye, P.M. Leishmania donovani infection initiates T cell-independent chemokine responses, which are subsequently amplified in a T cell-dependent manner. Eur. J. Immunol. 1999, 29, 203–214. [Google Scholar] [CrossRef]
- Lamichane, S.; Dahal Lamichane, B.; Kwon, S.-M. Pivotal Roles of Peroxisome Proliferator-Activated Receptors (PPARs) and Their Signal Cascade for Cellular and Whole-Body Energy Homeostasis. Int. J. Mol. Sci. 2018, 19, 949. [Google Scholar] [CrossRef]
- Castrillo, A.; Tontonoz, P. Nuclear Receptors in Macrophage Biology: At the Crossroads of Lipid Metabolism and Inflammation. Annu. Rev. Cell Dev. Biol. 2004, 20, 455–480. [Google Scholar] [CrossRef]
- Joseph, S.B.; Castrillo, A.; Laffitte, B.A.; Mangelsdorf, D.J.; Tontonoz, P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat. Med. 2003, 9, 213–219. [Google Scholar] [CrossRef]
- Joseph, S.B.; Bradley, M.N.; Castrillo, A.; Bruhn, K.W.; Mak, P.A.; Pei, L.; Hogenesch, J.; O’Connell, R.M.; Cheng, G.; Saez, E.; et al. LXR-Dependent Gene Expression Is Important for Macrophage Survival and the Innate Immune Response. Cell 2004, 119, 299–309. [Google Scholar] [CrossRef]
- Castrillo, A.; Joseph, S.B.; Marathe, C.; Mangelsdorf, D.J.; Tontonoz, P. Liver X Receptor-dependent Repression of Matrix Metalloproteinase-9 Expression in Macrophages. J. Biol. Chem. 2003, 278, 10443–10449. [Google Scholar] [CrossRef]
- Bilotta, M.T.; Petillo, S.; Santoni, A.; Cippitelli, M. Liver X Receptors: Regulators of Cholesterol Metabolism, Inflammation, Autoimmunity, and Cancer. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Janowski, B.A.; Grogan, M.J.; Jones, S.A.; Wisely, G.B.; Kliewer, S.A.; Corey, E.J.; Mangelsdorf, D.J. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc. Natl. Acad. Sci. USA 1999, 96, 266–271. [Google Scholar] [CrossRef]
- Lima, J.B.; Araújo-Santos, T.; Lázaro-Souza, M.; Carneiro, A.B.; Ibraim, I.C.; Jesus-Santos, F.H.; Luz, N.F.; Pontes, S.D.M.; Entringer, P.F.; Descoteaux, A.; et al. Leishmania infantum lipophosphoglycan induced-Prostaglandin E2 production in association with PPAR-γ expression via activation of Toll like receptors-1 and 2. Sci. Rep. 2017, 7, 14321. [Google Scholar] [CrossRef]
- Chan, M.M.; Adapala, N.; Chen, C. Peroxisome Proliferator-Activated Receptor-γ-Mediated Polarization of Macrophages in Leishmania Infection. PPAR Res. 2012, 2012, 1–11. [Google Scholar] [CrossRef]
- Gallardo-Soler, A.; Gómez-Nieto, C.; Campo, M.L.; Marathe, C.; Tontonoz, P.; Castrillo, A.; Corraliza, I. Arginase I Induction by Modified Lipoproteins in Macrophages: A Peroxisome Proliferator-Activated Receptor-γ/δ-Mediated Effect that Links Lipid Metabolism and Immunity. Mol. Endocrinol. 2008, 22, 1394–1402. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Ricardo-Gonzalez, R.R.; Goforth, M.H.; Morel, C.R.; Subramanian, V.; Mukundan, L.; Eagle, A.R.; Vats, D.; Brombacher, F.; Ferrante, A.W.; et al. Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 2007, 447, 1116–1120. [Google Scholar] [CrossRef]
- Saunders, E.C.; McConville, M.J. Immunometabolism of Leishmania granulomas. Immunol. Cell Biol. 2020, 98, 832–844. [Google Scholar] [CrossRef]
- Kidani, Y.; Bensinger, S.J. LXR and PPAR as integrators of lipid homeostasis and immunity. J. Immunol. Rev. 2014, 249, 72–83. [Google Scholar] [CrossRef]
- Li, A.C.; Glass, C.K. PPAR- and LXR-dependent pathways controlling lipid metabolism and the development of atherosclerosis. J. Lipid Res. 2004, 45, 2161–2173. [Google Scholar] [CrossRef]
- Fernandes, M.C.; Dillon, L.A.L.; Belew, A.T.; Bravo, H.C.; Mosser, D.M.; El-Sayed, N.M. Dual Transcriptome Profiling of Leishmania -Infected Human Macrophages Reveals Distinct Reprogramming Signatures. MBio 2016, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Dillon, L.A.L.; Suresh, R.; Okrah, K.; Corrada Bravo, H.; Mosser, D.M.; El-Sayed, N.M. Simultaneous transcriptional profiling of Leishmania major and its murine macrophage host cell reveals insights into host-pathogen interactions. BMC Genomics 2015, 16, 1108. [Google Scholar] [CrossRef]
- Veras, P.S.T.; Ramos, P.I.P.; de Menezes, J.P.B. In Search of Biomarkers for Pathogenesis and Control of Leishmaniasis by Global Analyses of Leishmania-Infected Macrophages. Front. Cell. Infect. Microbiol. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Oghumu, S.; Lezama-Dávila, C.M.; Isaac-Márquez, A.P.; Satoskar, A.R. Role of chemokines in regulation of immunity against leishmaniasis. Exp. Parasitol. 2010, 126, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Eufrásio de Figueiredo, W.M.; Heredia, F.F.; Santos, A.S.; da Rocha Braga, R.; Marciano Fonseca, F.R.; Lúcia de Castro Rodrigues, N.; Abreu, T.M.; Maria de Lima Pompeu, M.; Barbosa, H.S.; Teixeira, M.J. CXCL10 treatment promotes reduction of IL-10+ regulatory T (Foxp3+ and Tr1) cells in the spleen of BALB/c mice infected by Leishmania infantum. Exp. Parasitol. 2019, 207, 107789. [Google Scholar] [CrossRef]
- Figueiredo, W.M.E.; de Viana, S.M.; Alves, D.T.; Guerra, P.V.; Coêlho, Z.C.B.; Barbosa, H.S.; Teixeira, M.J. Protection mediated by chemokine CXCL10 in BALB/c mice infected by Leishmania infantum. Mem. Inst. Oswaldo Cruz 2017, 112, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Robert-Gangneux, F.; Drogoul, A.-S.; Rostan, O.; Piquet-Pellorce, C.; Cayon, J.; Lisbonne, M.; Herbelin, A.; Gascan, H.; Guiguen, C.; Samson, M.; et al. Invariant NKT Cells Drive Hepatic Cytokinic Microenvironment Favoring Efficient Granuloma Formation and Early Control of Leishmania donovani Infection. PLoS ONE 2012, 7, e33413. [Google Scholar] [CrossRef]
- Khan, I.A.; Thomas, S.Y.; Moretto, M.M.; Lee, F.S.; Islam, S.A.; Combe, C.; Schwartzman, J.D.; Luster, A.D. CCR5 Is Essential for NK Cell Trafficking and Host Survival following Toxoplasma gondii Infection. PLoS Pathog. 2006, 2, e49. [Google Scholar] [CrossRef]
- Fouillet, A.; Mawson, J.; Suliman, O.; Sharrack, B.; Romero, I.A.; Woodroofe, M.N. CCL2 binding is CCR2 independent in primary adult human astrocytes. Brain Res. 2012, 1437, 115–126. [Google Scholar] [CrossRef]
- Murray, H.W. Tissue granuloma structure-function in experimental visceral leishmaniasis. Int. J. Exp. Pathol. 2001, 82, 249–267. [Google Scholar] [CrossRef]
- Liese, J.; Schleicher, U.; Bogdan, C. TLR9 signaling is essential for the innate NK cell response in murine cutaneous leishmaniasis. Eur. J. Immunol. 2007, 37, 3424–3434. [Google Scholar] [CrossRef]
- Tsutsui, H.; Nishiguchi, S. Importance of Kupffer Cells in the Development of Acute Liver Injuries in Mice. Int. J. Mol. Sci. 2014, 15, 7711–7730. [Google Scholar] [CrossRef]
- Murray, H.W.; Zhang, Y.; Zhang, Y.; Raman, V.S.; Reed, S.G.; Ma, X. Regulatory Actions of Toll-Like Receptor 2 (TLR2) and TLR4 in Leishmania donovani Infection in the Liver. Infect. Immun. 2013, 81, 2318–2326. [Google Scholar] [CrossRef]
- Kavoosi, G.; Ardestani, S.K.; Kariminia, A. The involvement of TLR2 in cytokine and reactive oxygen species (ROS) production by PBMCs in response to Leishmania major phosphoglycans (PGs). Parasitology 2009, 136, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Kavoosi, G.; Ardestani, S.K.; Kariminia, A.; Alimohammadian, M.H. Leishmania major lipophosphoglycan: Discrepancy in toll-like receptor signaling. Exp. Parasitol. 2010, 124, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Puddu, P.; Fantuzzi, L.; Borghi, P.; Varano, B.; Rainaldi, G.; Guillemard, E.; Malorni, W.; Nicaise, P.; Wolf, S.F.; Belardelli, F.; et al. IL-12 induces IFN-gamma expression and secretion in mouse peritoneal macrophages. J. Immunol. 1997, 159, 3490–3497. [Google Scholar] [PubMed]
- Murray, H.W.; Squires, K.E.; Miralles, C.D.; Stoeckle, M.Y.; Granger, A.M.; Granelli-Piperno, A.; Bogdan, C. Acquired resistance and granuloma formation in experimental visceral leishmaniasis. Differential T cell and lymphokine roles in initial versus established immunity. J. Immunol. 1992, 148, 1858–1863. [Google Scholar]
- O’Garra, A.; McEvoy, L.M.; Zlotnik, A. T-cell subsets: Chemokine receptors guide the way. Curr. Biol. 1998, 8, R646–R649. [Google Scholar] [CrossRef]
- Kaye, P.M.; Svensson, M.; Ato, M.; Maroof, A.; Polley, R.; Stager, S.; Zubairi, S.; Engwerda, C.R. The immunopathology of experimental visceral leishmaniasis. Immunol. Rev. 2004, 201, 239–253. [Google Scholar] [CrossRef]
- Yoshida, H.; Hamano, S.; Senaldi, G.; Covey, T.; Faggioni, R.; Mu, S.; Xia, M.; Wakeham, A.C.; Nishina, H.; Potter, J.; et al. WSX-1 Is Required for the Initiation of Th1 Responses and Resistance to L. major Infection. Immunity 2001, 15, 569–578. [Google Scholar] [CrossRef]
- Chen, Q.; Ghilardi, N.; Wang, H.; Baker, T.; Xie, M.-H.; Gurney, A.; Grewal, I.S.; de Sauvage, F.J. Development of Th1-type immune responses requires the type I cytokine receptor TCCR. Nature 2000, 407, 916–920. [Google Scholar] [CrossRef]
- Bruhn, K.W.; Marathe, C.; Maretti-Mira, A.C.; Nguyen, H.; Haskell, J.; Tran, T.A.; Vanchinathan, V.; Gaur, U.; Wilson, M.E.; Tontonoz, P.; et al. LXR Deficiency Confers Increased Protection against Visceral Leishmania Infection in Mice. PLoS Negl. Trop. Dis. 2010, 4, e886. [Google Scholar] [CrossRef]
- Gogulamudi, V.R.; Dubey, M.L.; Kaul, D.; Hubert, D.J.; Kandimalla, R.; Sehgal, R. Vitamins (A&D) and Isoprenoid (Chenodeoxycholic acid) molecules are accompanied by Th1 immunostimulatory response and therapeutic cure in vivo: Possible antileishmanial drugs. Sci. Rep. 2019, 9, 8531. [Google Scholar] [CrossRef]
- Greenwald, R.J.; Freeman, G.J.; Sharpe, A.H. The B7 Family Revisited. Annu. Rev. Immunol. 2005, 23, 515–548. [Google Scholar] [CrossRef]
- Pauken, K.E.; Wherry, E.J. Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 2015, 36, 265–276. [Google Scholar] [CrossRef]
- Agata, Y.; Kawasaki, A.; Nishimura, H.; Ishida, Y.; Tsubat, T.; Yagita, H.; Honjo, T. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int. Immunol. 1996, 8, 765–772. [Google Scholar] [CrossRef]
- Nishimura, H.; Honjo, T.; Minato, N. Facilitation of β Selection and Modification of Positive Selection in the Thymus of Pd-1–Deficient Mice. J. Exp. Med. 2000, 191, 891–898. [Google Scholar] [CrossRef]
- Gannavaram, S.; Bhattacharya, P.; Ismail, N.; Kaul, A.; Singh, R.; Nakhasi, H.L. Modulation of Innate Immune Mechanisms to Enhance Leishmania Vaccine-Induced Immunity: Role of Coinhibitory Molecules. Front. Immunol. 2016, 7. [Google Scholar] [CrossRef]
- Wu, W.; Shi, Y.; Li, S.; Zhang, Y.; Liu, Y.; Wu, Y.; Chen, Z. Blockade of Tim-3 signaling restores the virus-specific CD8 + T-cell response in patients with chronic hepatitis B. Eur. J. Immunol. 2012, 42, 1180–1191. [Google Scholar] [CrossRef]
- Schietinger, A.; Greenberg, P.D. Tolerance and exhaustion: Defining mechanisms of T cell dysfunction. Trends Immunol. 2014, 35, 51–60. [Google Scholar] [CrossRef]
- Dyck, L.; Mills, K.H.G. Immune checkpoints and their inhibition in cancer and infectious diseases. Eur. J. Immunol. 2017, 47, 765–779. [Google Scholar] [CrossRef]
- Da Silva, I.P.; Gallois, A.; Jimenez-Baranda, S.; Khan, S.; Anderson, A.C.; Kuchroo, V.K.; Osman, I.; Bhardwaj, N. Reversal of NK-Cell Exhaustion in Advanced Melanoma by Tim-3 Blockade. Cancer Immunol. Res. 2014, 2, 410–422. [Google Scholar] [CrossRef]
- Habib, S.; El Andaloussi, A.; Elmasry, K.; Handoussa, A.; Azab, M.; Elsawey, A.; Al-Hendy, A.; Ismail, N. PDL-1 Blockade Prevents T Cell Exhaustion, Inhibits Autophagy, and Promotes Clearance of Leishmania donovani. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef]
- Da Fonseca-Martins, A.M.; Ramos, T.D.; Pratti, J.E.S.; Firmino-Cruz, L.; Gomes, D.C.O.; Soong, L.; Saraiva, E.M.; de Matos Guedes, H.L. Immunotherapy using anti-PD-1 and anti-PD-L1 in Leishmania amazonensis-infected BALB/c mice reduce parasite load. Sci. Rep. 2019, 9, 20275. [Google Scholar] [CrossRef]
- Hsu, J.; Hodgins, J.J.; Marathe, M.; Nicolai, C.J.; Bourgeois-Daigneault, M.-C.; Trevino, T.N.; Azimi, C.S.; Scheer, A.K.; Randolph, H.E.; Thompson, T.W.; et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J. Clin. Investig. 2018, 128, 4654–4668. [Google Scholar] [CrossRef]
- Kaye, P.M.; Cruz, I.; Picado, A.; Van Bocxlaer, K.; Croft, S.L. Leishmaniasis immunopathology—impact on design and use of vaccines, diagnostics and drugs. Semin. Immunopathol. 2020, 42, 247–264. [Google Scholar] [CrossRef]
- Akbari, M.; Oryan, A.; Hatam, G. Immunotherapy in treatment of leishmaniasis. Immunol. Lett. 2021, 233, 80–86. [Google Scholar] [CrossRef]
- Leopold Wager, C.M.; Arnett, E.; Schlesinger, L.S. Macrophage nuclear receptors: Emerging key players in infectious diseases. PLoS Pathog. 2019, 15, e1007585. [Google Scholar] [CrossRef]
- Buffet, P.A.; Sulahian, A.; Garin, Y.J.; Nassar, N.; Derouin, F. Culture microtitration: A sensitive method for quantifying Leishmania infantum in tissues of infected mice. Antimicrob. Agents Chemother. 1995, 39, 2167–2168. [Google Scholar] [CrossRef]
- Hernandez-Santana, Y.E.; Ontoria, E.; Gonzalez-García, A.C.; Quispe-Ricalde, M.A.; Larraga, V.; Valladares, B.; Carmelo, E. The Challenge of Stability in High-Throughput Gene Expression Analysis: Comprehensive Selection and Evaluation of Reference Genes for BALB/c Mice Spleen Samples in the Leishmania infantum Infection Model. PLoS ONE 2016, 11, e0163219. [Google Scholar] [CrossRef]
- Shannon, P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Morris, J.H.; Apeltsin, L.; Newman, A.M.; Baumbach, J.; Wittkop, T.; Su, G.; Bader, G.D.; Ferrin, T.E. clusterMaker: A multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 2011, 12, 436. [Google Scholar] [CrossRef]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 2010. [Google Scholar] [CrossRef]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]

| PC2 Genes | |||||
| Gene | Factor Loading | Gene | Factor Loading | Gene | Factor Loading |
| Il18r1 | 0.742 | Cd8a | 0.683 | Tlr2 | 0.594 |
| Pdcd1lg2 (Pd-l2)e | 0.738 | Cd4 | 0.682 | Cd160 | 0.592 |
| P2rx7 (P2x7)a | 0.733 | Ptgesa | 0.679 | Irf1 | 0.591 |
| Bcl2 | 0.723 | Il2rg | 0.678 | Myd88 | 0.585 |
| Gata3 | 0.72 | Ptger4a | 0.677 | Pla2g4ab | 0.582 |
| Il16 | 0.719 | Tnfrsf18 (Gitr)f | 0.668 | Stat1 | 0.58 |
| Cd3e | 0.718 | Il2rb | 0.653 | Ccl2 | 0.58 |
| Il2ra (Cd25)f | 0.717 | Tnfrsf4 (Ox40)f | 0.652 | Il18bp | 0.577 |
| Lta | 0.713 | Klrd1 (Cd94)c | 0.639 | Irf5 | 0.573 |
| Il10ra | 0.713 | Cd209dc | 0.634 | Tlr4 | 0.572 |
| Cd69 | 0.712 | Itgam | 0.633 | Il6st | 0.572 |
| Akt3d | 0.707 | Icam1 | 0.632 | Ltb4r1 (Blt1)a | 0.571 |
| Cd28 | 0.706 | Alox12a | 0.632 | Il1b | 0.569 |
| Il7r (Cd127) | 0.704 | Xcl1 | 0.622 | Nfkb1 | 0.567 |
| Hpgds (Ptgds2)a | 0.704 | Ucp2 | 0.621 | Itgal | 0.567 |
| Mki67 (Ki67) | 0.702 | Pdcd1lg1 (Pd-l1)e | 0.619 | Cxcl9 | 0.567 |
| Ccr4 | 0.701 | Cxcl10 | 0.614 | Nfkb2 | 0.566 |
| Il9r | 0.697 | Ccl22 | 0.614 | Cxcl16 | 0.562 |
| Cxcr4 | 0.693 | Ccr5 | 0.607 | Cxcr6 | 0.552 |
| Ccl19 | 0.689 | Irf7 | 0.605 | Alox5a | 0.544 |
| Btla | 0.685 | Itgb2 | 0.603 | Rxrab | −0.569 |
| Il18rap | 0.684 | Cd68 | 0.6 | ||
| PC3 Genes | |||||
| Gene | Factor Loading | Gene | Factor Loading | Gene | Factor Loading |
| Il12rb1 | 0.943 | Cxcr2 | 0.825 | Cd83 | 0.63 |
| Il12rb2 | 0.941 | Il27 | 0.803 | Csf2 | 0.619 |
| Nos2 | 0.928 | Cd80 | 0.782 | Ccl4 | 0.614 |
| Ccr7 | 0.914 | Irg1 (acod1) | 0.768 | Havcr2 (Tim-3)e,f | 0.591 |
| Il12b | 0.884 | Tgfb2 | 0.693 | Icosl | 0.574 |
| Ifng | 0.867 | Cxcr3 | 0.663 | Ebi3 (Il27b) | 0.562 |
| Cxcr5 | 0.864 | Ccl7 | 0.657 | Il21r | 0.558 |
| Il23a | 0.842 | Pdcd1 (Pd-1)e,f | 0.643 | ||
| Il21 | 0.839 | Ptgs2a | 0.64 | ||
| Nº | GENE SET NAME | RATIO | NES | NOM p-Value | FDR q-Value |
|---|---|---|---|---|---|
| 1 | INTERFERON SIGNALING (REACTOME R-HSA-913531,2) | 5/149 | 2.050 | 0.000 | 0.039 |
| 2 | CHEMOKINE RECEPTORS BIND CHEMOKINES (REACTOME DATABASE ID RELEASE 75 380108) | 10/49 | 1.782 | 0.013 | 0.199 |
| 3 | PEPTIDE LIGAND-BINDING RECEPTORS (REACTOME R-HSA-375276,5) | 10/182 | 1.781 | 0.013 | 0.133 |
| 4 | CYTOKINE SIGNALING IN IMMUNE SYSTEM (REACTOME DATABASE ID RELEASE 75 1280215) | 32/608 | 1.708 | 0.024 | 0.170 |
| 5 | CLASS A 1 RHODOPSIN-LIKE RECEPTORS (REACTOME R-HSA-373076,8) | 12/307 | 1.629 | 0.036 | 0.227 |
| 6 | GPCR LIGAND BINDING (REACTOME R-HSA-500792,3) | 12/420 | 1.629 | 0.035 | 0.190 |
| 7 | INTERLEUKIN-10 SIGNALING (REACTOME R-HSA-6783783,3) | 6/42 | 1.584 | 0.040 | 0.212 |
| 8 | SIGNALING BY INTERLEUKINS (REACTOME DATABASE ID RELEASE 75 449147) | 25/417 | 1.580 | 0.047 | 0.189 |
| Nº | GENE SET NAME | RATIO | NES | NOM p-Value | FDR q-Value |
|---|---|---|---|---|---|
| 1 | PID_IL12_2PATHWAY (MSIGDB_C2 PID_IL12_2PATHWAY) | 6/59 | 2.188 | 0.000 | 0.003 |
| 2 | BIOCARTA_NO2IL12_PATHWAY (MSIGDB_C2 BIOCARTA_NO2IL12_PATHWAY) | 5/15 | 1.965 | 0.003 | 0.013 |
| 3 | PID_IL23_PATHWAY (MSIGDB_C2 PID_IL23_PATHWAY) | 5/36 | 1.879 | 0.006 | 0.016 |
| 4 | PID_IL27_PATHWAY (MSIGDB_C2 PID_IL27_PATHWAY) | 6/25 | 1.788 | 0.012 | 0.024 |
| 5 | BIOCARTA_NKT_PATHWAY (MSIGDB_C2 BIOCARTA_NKT_PATHWAY) | 8/25 | 1.642 | 0.035 | 0.052 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palacios, G.; Diaz-Solano, R.; Valladares, B.; Dorta-Guerra, R.; Carmelo, E. Early Transcriptional Liver Signatures in Experimental Visceral Leishmaniasis. Int. J. Mol. Sci. 2021, 22, 7161. https://doi.org/10.3390/ijms22137161
Palacios G, Diaz-Solano R, Valladares B, Dorta-Guerra R, Carmelo E. Early Transcriptional Liver Signatures in Experimental Visceral Leishmaniasis. International Journal of Molecular Sciences. 2021; 22(13):7161. https://doi.org/10.3390/ijms22137161
Chicago/Turabian StylePalacios, Génesis, Raquel Diaz-Solano, Basilio Valladares, Roberto Dorta-Guerra, and Emma Carmelo. 2021. "Early Transcriptional Liver Signatures in Experimental Visceral Leishmaniasis" International Journal of Molecular Sciences 22, no. 13: 7161. https://doi.org/10.3390/ijms22137161
APA StylePalacios, G., Diaz-Solano, R., Valladares, B., Dorta-Guerra, R., & Carmelo, E. (2021). Early Transcriptional Liver Signatures in Experimental Visceral Leishmaniasis. International Journal of Molecular Sciences, 22(13), 7161. https://doi.org/10.3390/ijms22137161







