In Silico Target Prediction of Overexpressed microRNAs from LPS-Challenged Zebrafish (Danio rerio) Treated with the Novel Anti-Inflammatory Peptide TnP
Abstract
:1. Introduction
2. Results
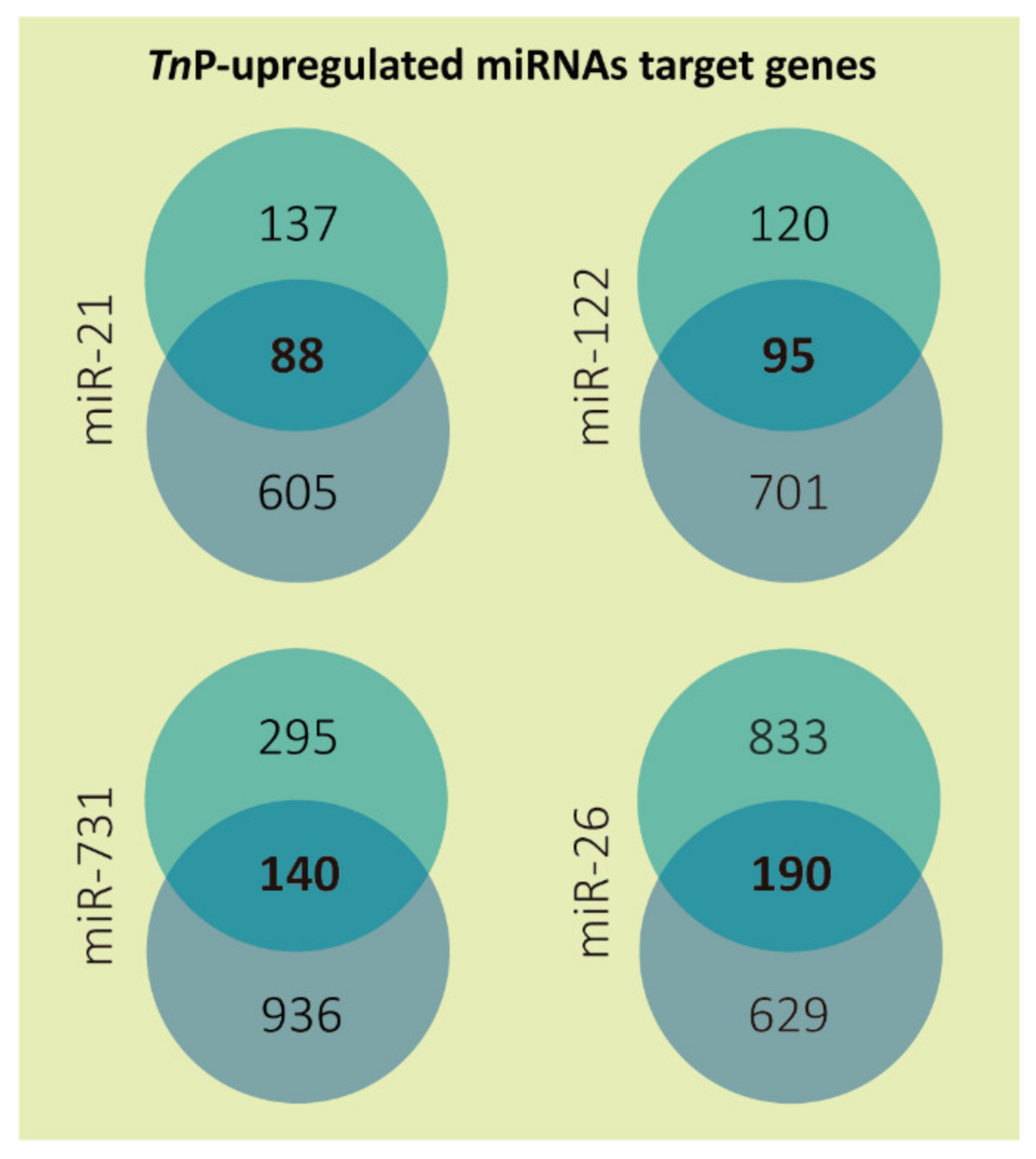
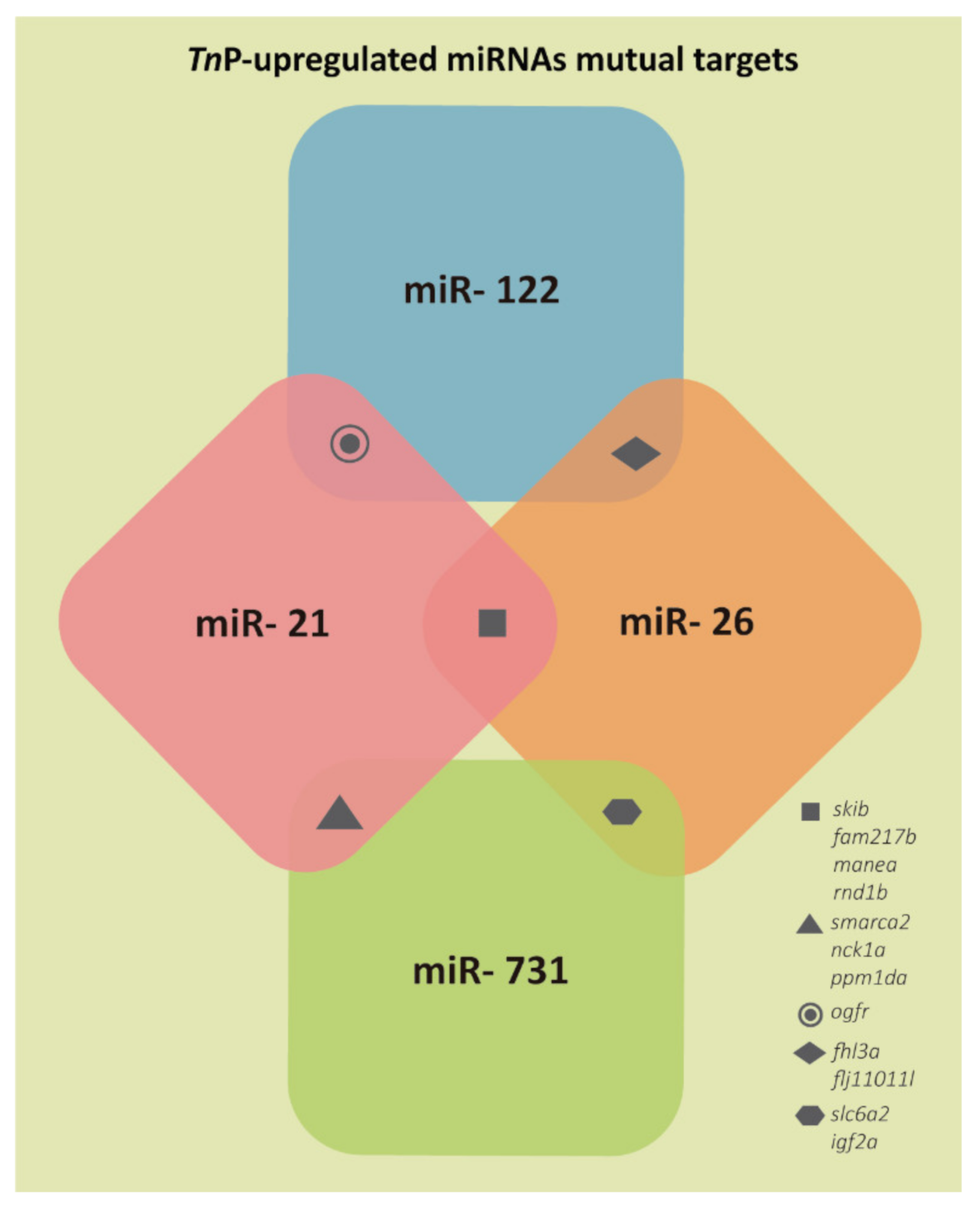
2.1. miRNAs Putative Target Genes
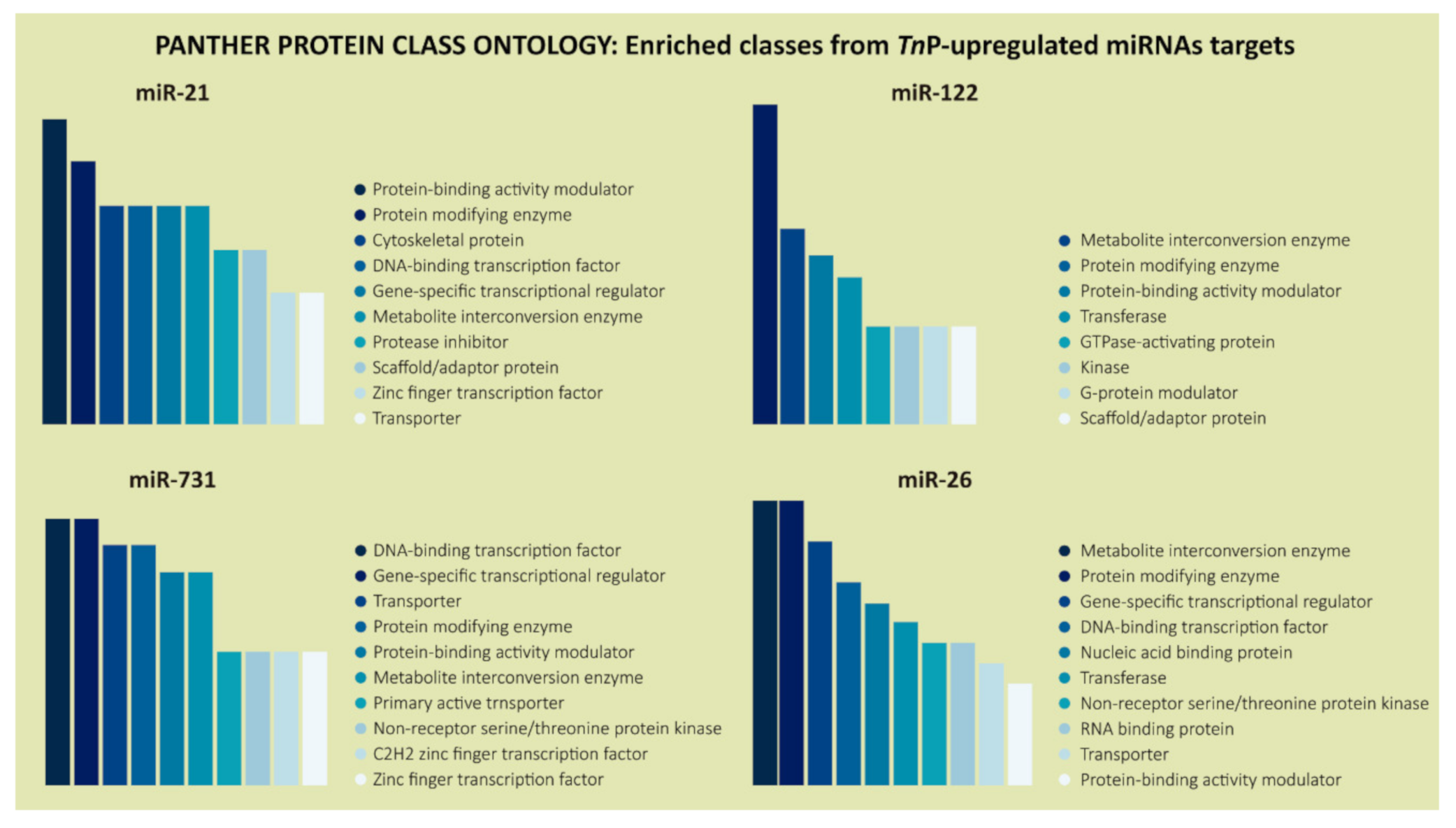
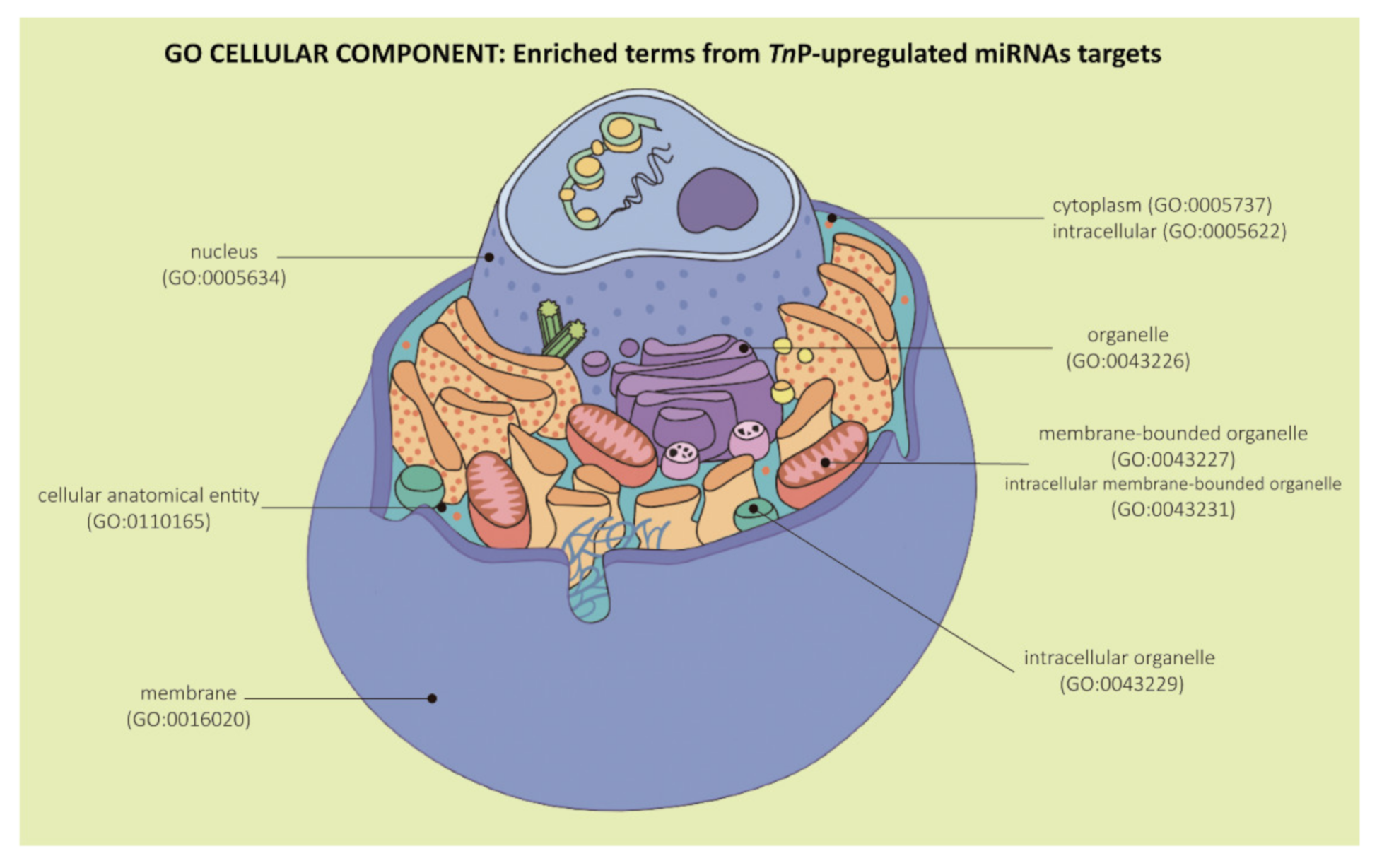
2.2. Biological Significance of Identified Putative Target Genes
3. Discussion
4. Methods
4.1. LPS Challenge and TnP Treatment
4.2. Selection of dre-miRNAs for the In Silico Target Prediction Analysis
4.3. Data Sources
4.4. In Silico Target Prediction Analysis
4.5. Gene Ontology Enrichment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Jirtle, R.L.; Skinner, M.K. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007, 8, 253–262. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, Biogenesis, and Their Evolving Role in Animal Development and Disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slota, J.A.; Booth, A.S. MicroRNAs in Neuroinflammation: Implications in Disease Pathogenesis, Biomarker Discovery and Therapeutic Applications. Noncoding RNA 2019, 5, 35. [Google Scholar] [CrossRef] [Green Version]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Thompson, F.; de Oliveira, B.C.; Cordeiro, M.C.; Masi, B.P.; Rangel, T.P.; Paz, P.; Freitas, T.; Lopes, G.; Silva, B.S.; Cabral, A.; et al. Severe impacts of the Brumadinho dam failure (Minas Gerais, Brazil) on the water quality of the Paraopeba River. Sci. Total Environ. 2020, 705, 135914. [Google Scholar] [CrossRef]
- Lopes-Ferreira, M.; Falcão, M.A.P.; Disner, G.R.; Lima, C. O modelo Zebrafish e sua contribuição ao meio ambiente. In Recurso Água: Tecnologias e Pesquisas Para o uso e a Conservação de Ecossistemas Aquáticos, 1st ed.; Silva, C.V.R.D., Queiroz, L.G., Gomes, L.E.T., Eds.; Cubo Editora: São Carlos, Brazil, 2021; pp. 188–219. [Google Scholar]
- Disner, G.R.; Falcao, M.A.P.; Andrade-Barros, A.I.; Leite-Santos, N.V.; Soares, A.B.S.; Marcolino-Souza, M.; Gomes, K.S.; Lima, C.; Lopes-Ferreira, M. The toxic effects of glyphosate, chlorpyrifos, abamectin, and 2,4-D on animal models: A systematic review of Brazilian studies. Integr. Environ. Assess. Manag. 2020. [Google Scholar] [CrossRef]
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug. Discov. 2015, 14, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Garcia, G.R.; Noyes, P.D.; Tanguay, R.L. Advancements in zebrafish applications for 21st century toxicology. Pharmacol. Ther. 2016, 161, 11–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakaria, Z.Z.; Benslimane, F.M.; Nasrallah, G.K.; Shurbaji, S.; Younes, N.N.; Mraiche, F.; Da’as, S.I.; Yalcin, H.C. Using Zebrafish for Investigating the Molecular Mechanisms of Drug-Induced Cardiotoxicity. Biomed. Res. Int. 2018, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Batista-Filho, J.; Falcão, M.A.P.; Maleski, A.L.A.; Soares, A.B.S.; Balan-Lima, L.; Disner, G.R.; Lima, C.; Lopes-Ferreira, M. Early preclinical screening using zebrafish (Danio rerio) reveals the safety of the candidate anti-inflammatory therapeutic agent TnP. Toxicol. Rep. 2021, 8, 13–22. [Google Scholar] [CrossRef]
- Bizuayehu, T.T.; Babiak, I. MicroRNA in teleost fish. Genome Biol. Evol. 2014, 6, 1911–1937. [Google Scholar] [CrossRef] [Green Version]
- Desvignes, T.; Beam, M.J.; Batzel, P.; Sydes, J.; Postlethwait, J.H. Expanding the annotation of zebrafish microRNAs based on small RNA sequencing. Gene 2014, 546, 386–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin Tai, J.K.A.; Freeman, J.L. Zebrafish as an integrative vertebrate model to identify miRNA mechanisms regulating toxicity. Toxicol. Rep. 2020, 7, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Gurol, T.; Zhou, W.; Deng, Q. MicroRNAs in neutrophils: Potential next generation therapeutics for inflammatory ailments. Immunol. Rev. 2016, 273, 29–47. [Google Scholar] [CrossRef] [PubMed]
- Hsu, A.Y.; Wang, D.; Gurol, T.; Zhou, W.; Zhu, X.; Lu, H.; Deng, Q. Overexpression of microRNA-722 fine-tunes neutrophilic inflammation by inhibiting Rac2 in zebrafish. Dis. Model. Mech. 2017, 10, 1323–1332. [Google Scholar] [CrossRef] [Green Version]
- Hsu, A.Y.; Wang, D.; Liu, S.; Lu, J.; Syahirah, R.; Bennin, D.A.; Huttenlocher, A.; Umulis, D.M.; Wan, J.; Deng, Q. Phenotypical microRNA screen reveals a noncanonical role of CDK2 in regulating neutrophil migration. Proc. Natl. Acad. Sci. USA 2019, 116, 18561–18570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, C.; Guo, X.; Ren, J.; Zu, Y.; Li, W.; Zhang, Q. Transcriptomic analysis of microRNAs-mRNAs regulating innate immune response of zebrafish larvae against Vibrio parahaemolyticus infection. Fish Shellfish Immunol. 2019, 91, 333–342. [Google Scholar] [CrossRef]
- Lopes-Ferreira, M.V.A.; Lima, C.; Pimenta, D.C.; Conceição, K.; Demasi, M.; Portaro, F.C.V. Peptídeos Cíclicos Anti-Inflamatórios e Anti-Alérgicos. PI0602885-3A2, 2019. [Google Scholar]
- Komegae, E.N.; Souza, T.A.; Grund, L.Z.; Lima, C.; Lopes-Ferreira, M. Multiple functional therapeutic effects of TnP: A small stable synthetic peptide derived from fish venom in a mouse model of multiple sclerosis. PLoS ONE 2017, 12, e0171796. [Google Scholar] [CrossRef]
- Lopes-Ferreira, M.; Lima, C. Personal Communication, Butantan Institute: São Paulo, Brazil, 2021.
- Peterson, S.M.; Thompson, J.A.; Ufkin, M.L.; Sathyanarayana, P.; Liaw, L.; Congdon, C.B. Common features of microRNA target prediction tools. Front. Genet. 2014, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Riffo-Campos, Á.L.; Riquelme, I.; Brebi-Mieville, P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? Int. J. Mol. Sci. 2016, 17, 1987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.Y.; Lin, Y.C.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajewsky, N. microRNA target predictions in animals. Nat. Genet. 2006, 38, S8–S13. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef] [Green Version]
- Forn-Cuní, G.; Varela, M.; Pereiro, P.; Novoa, B.; Figueras, A. Conserved gene regulation during acute inflammation between zebrafish and mammals. Sci. Rep. 2017, 7, 41905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harvey, A.; Edrada-Ebel, R.; Quinn, R. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akondi, K.B.; Muttenthaler, M.; Dutertre, S.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Discovery, synthesis, and structure-activity relationships of conotoxins. Chem. Rev. 2014, 114, 5815–5847. [Google Scholar] [CrossRef]
- Mahadevappa, R.; Ma, R.; Kwok, H.F. Venom Peptides: Improving Specificity in Cancer Therapy. Trends Cancer 2017, 3, 611–614. [Google Scholar] [CrossRef]
- Pope, J.E.; Deer, T.R. Ziconotide: A clinical update and pharmacologic review. Exp. Opin. Pharmacother. 2013, 14, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Furman, B.L. The development of Byetta (exenatide) from the venom of the Gila monster as an anti-diabetic agent. Toxicon 2012, 59, 464–471. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucl. Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [Green Version]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P. Prediction of mammalian MicroRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [Green Version]
- UniProt Knowledgebase. The UniProt Consortium: European Bioinformatics Institute (EMBL-EBI), SIB Swiss Institute of Bioinformatics, and Protein Information Resource (PIR). Available online: https://www.uniprot.org/ (accessed on 10 January 2021).
- Thomas, P.D. The Gene Ontology and the Meaning of Biological Function. In The Gene Ontology Handbook. Methods in Molecular Biology; Dessimoz, C., Škunca, N., Eds.; Humana Press: New York, NY, USA, 2017; p. 1446. [Google Scholar]
- Vauti, F.; Stegemann, L.A.; Vögele, V.; Köster, R.W. All-age whole mount in situ hybridization to reveal larval and juvenile expression patterns in zebrafish. PLoS ONE 2020, 15, e0237167. [Google Scholar] [CrossRef]
- Zebrafish Information Network (ZFIN). University of Oregon, Eugene, OR 97403-5274. Available online: http://zfin.org/ (accessed on 10 January 2021).
- Nohra, R.; Beyeen, A.D.; Guo, J.P.; Khademi, M.; Sundqvist, E.; Hedreul, M.T.; Sellebjerg, F.; Smestad, C.; Oturai, A.B.; Harbo, H.F.; et al. RGMA and IL21R show association with experimental inflammation and multiple sclerosis. Genes Immun. 2010, 11, 279–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreassen, R.; Høyheim, B. miRNAs associated with immune response in teleost fish. Dev. Comp. Immunol. 2017, 75, 77–85. [Google Scholar] [CrossRef]
- Zanetti, M.; Castiglioni, P.; Schoenberger, S.; Gerloni, M. The role of relB in regulating the adaptive immune response. Ann. N.Y. Acad. Sci. 2003, 987, 249–257. [Google Scholar] [CrossRef]
- Pradhan, A.; Olsson, P.E. Juvenile Ovary to Testis Transition in Zebrafish Involves Inhibition of Ptges. Biology Reprod. 2014, 91, 1–15. [Google Scholar] [CrossRef]
- Bethune, J.; Artus-Revel, C.G.; Filipowicz, W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012, 13, 716–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez-Vázquez, C.; Quintana, F.J. Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity 2018, 48, 19–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanieh, H.; Mohafez, O.; Hairul-Islam, V.I.; Alzahrani, A.; Bani Ismail, M.; Thirugnanasambantham, K. Novel Aryl Hydrocarbon Receptor Agonist Suppresses Migration and Invasion of Breast Cancer Cells. PLoS ONE 2016, 11, e0167650. [Google Scholar] [CrossRef] [Green Version]
- Lv, Q.; Shi, C.; Qiao, S.; Cao, N.; Guan, C.; Dai, Y.; Wei, Z. Alpinetin exerts anti-colitis efficacy by activating AhR, regulating miR-302/DNMT-1/CREB signals, and therefore promoting Treg differentiation. Cell Death Dis. 2018, 9, 890. [Google Scholar] [CrossRef]
- Liu, C.C.; Xia, M.; Zhang, Y.J.; Jin, P.; Zhao, L.; Zhang, J.; Li, T.; Zhou, X.M.; Tu, Y.Y.; Kong, F.; et al. Micro124-mediated AHR expression regulates the inflammatory response of chronic rhinosinusitis (CRS) with nasal polyps. Biochem. Biophys. Res. Commun. 2018, 500, 145–151. [Google Scholar] [CrossRef]
- Rogers, S.; Souza, A.R.; Zago, M.; Iu, M.; Guerrina, N.; Gomez, A.; Matthews, J.; Baglole, C.J. Aryl hydrocarbon receptor (AhR)-dependent regulation of pulmonary miRNA by chronic cigarette smoke exposure. Sci. Rep. 2017, 7, 40539. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.-C.; Chang, H.-Y.; Chen, C.-Y.; Wu, P.-Y.; Lee, H.; Liao, Y.-F.; Hsu, W.-M.; Huang, H.-C.; Juan, H.-F. Silencing of miR-124 induces neuroblastoma SK-N-SH cell differentiation, cell cycle arrest and apoptosis through promoting AHR. FEBS Lett. 2011, 585, 3582–3586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Ma, T.; Chen, W.; Chen, Y.; Li, M.; Ren, L.; Chen, J.; Cao, R.; Feng, Y.; Zhang, H.; et al. MicroRNA-124 Promotes Intestinal Inflammation by Targeting Aryl Hydrocarbon Receptor in Crohn’s Disease. J. Crohns. Colitis 2016, 10, 703–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wienholds, E.; Kloosterman, W.P.; Miska, E.; Alvarez-Saavedra, E.; Berezikov, E.; de Bruijn, E.; Horvitz, H.R.; Kauppinen, S.; Plasterk, R.H. MicroRNA expression in zebrafish embryonic development. Science 2005, 309, 310–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jopling, C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012, 9, 137–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasqualotto, A.; Ayres, R.; Longo, L.; Lima, D.D.; Oliveira, D.L.; Alvares-da-Silva, M.R.; Silveira, T.R.; Uribe-Cruz, C. Chronic exposure to ethanol alters the expression of miR-155, miR-122 and miR-217 in alcoholic liver disease in an adult zebrafish model. Biomarkers 2021, 1–18. [Google Scholar] [CrossRef]
- Wu, T.-H.; Pan, C.-Y.; Lin, M.-C.; Hsieh, J.-C.; Hui, C.-F.; Chen, J.-Y. In vivo screening of zebrafish microRNA responses to bacterial infection and their possible roles in regulating immune response genes after lipopolysaccharide stimulation. Fish Physiol. Biochem. 2012, 38, 1299–1310. [Google Scholar] [CrossRef]
- Tao, L.; Xu, X.; Fang, Y.; Wang, A.; Zhou, F.; Shen, Y.; Li, J. miR-21 targets jnk and ccr7 to modulate the inflammatory response of grass carp following bacterial infection. Fish Shellfish Immunol. 2019, 94, 258–263. [Google Scholar] [CrossRef]
- Wu, Y.; Lou, Q.Y.; Ge, F.; Xiong, Q. Quantitative Proteomics Analysis Reveals Novel Targets of miR-21 in Zebrafish Embryos. Sci. Rep. 2017, 7, 4022. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Huang, C.-X.; Wang, W.-F.; Liu, H.; Wang, H.-L. Zebrafish miR-462-731 is required for digestive organ development. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 34, 100679. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, J.; Gong, Y.; Cai, J.; Zhang, Z. Role of miR-731 and miR-2188-3p in mediating chlorpyrifos induced head kidney injury in common carp via targeting TLR and apoptosis pathways. Aquat. Toxicol. 2019, 215, 105286. [Google Scholar] [CrossRef]
- Icli, B.; Wara, A.K.; Moslehi, J.; Sun, X.; Plovie, E.; Cahill, M.; Marchini, J.F.; Schissler, A.; Padera, R.F.; Shi, J.; et al. MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ. Res. 2013, 113, 1231–1241. [Google Scholar] [CrossRef] [Green Version]
- Acharya, A.; Berry, D.C.; Zhang, H.; Jiang, Y.; Jones, B.T.; Hammer, R.E.; Graff, J.M.; Mendell, J.T. miR-26 suppresses adipocyte progenitor differentiation and fat production by targeting Fbxl19. Genes Dev. 2019, 33, 1367–1380. [Google Scholar] [CrossRef] [Green Version]
- Falcao, M.A.P.; Walker, C.I.B.; Disner, G.R.; Batista-Filho, J.; Soares, A.B.S.; Balan-Lima, L.; Lima, C.; Lopes-Ferreira, M. Knockdown of miR-26a in Zebrafish Leads to Impairment of the Anti-Inflammatory Function of TnP in the Control of Neutrophilia. Fish Shellfish Immunol. under review.
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucl. Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Reczko, M.; Maragkakis, M.; Alexiou, P.; Grosse, I.; Hatzigeorgiou, A.G. Functional microRNA targets in protein coding sequences. Bioinformatics 2012, 28, 771–776. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Disner, G.R.; Falcão, M.A.P.; Lima, C.; Lopes-Ferreira, M. In Silico Target Prediction of Overexpressed microRNAs from LPS-Challenged Zebrafish (Danio rerio) Treated with the Novel Anti-Inflammatory Peptide TnP. Int. J. Mol. Sci. 2021, 22, 7117. https://doi.org/10.3390/ijms22137117
Disner GR, Falcão MAP, Lima C, Lopes-Ferreira M. In Silico Target Prediction of Overexpressed microRNAs from LPS-Challenged Zebrafish (Danio rerio) Treated with the Novel Anti-Inflammatory Peptide TnP. International Journal of Molecular Sciences. 2021; 22(13):7117. https://doi.org/10.3390/ijms22137117
Chicago/Turabian StyleDisner, Geonildo R., Maria A. P. Falcão, Carla Lima, and Monica Lopes-Ferreira. 2021. "In Silico Target Prediction of Overexpressed microRNAs from LPS-Challenged Zebrafish (Danio rerio) Treated with the Novel Anti-Inflammatory Peptide TnP" International Journal of Molecular Sciences 22, no. 13: 7117. https://doi.org/10.3390/ijms22137117
APA StyleDisner, G. R., Falcão, M. A. P., Lima, C., & Lopes-Ferreira, M. (2021). In Silico Target Prediction of Overexpressed microRNAs from LPS-Challenged Zebrafish (Danio rerio) Treated with the Novel Anti-Inflammatory Peptide TnP. International Journal of Molecular Sciences, 22(13), 7117. https://doi.org/10.3390/ijms22137117






