Feedback, Crosstalk and Competition: Ingredients for Emergent Non-Linear Behaviour in the PI3K/mTOR Signalling Network
Abstract
1. Introduction
2. Signal Transduction within the PI3K/mTOR Signalling Pathway
3. Complex Regulatory Mechanisms within the PI3K/mTOR Network
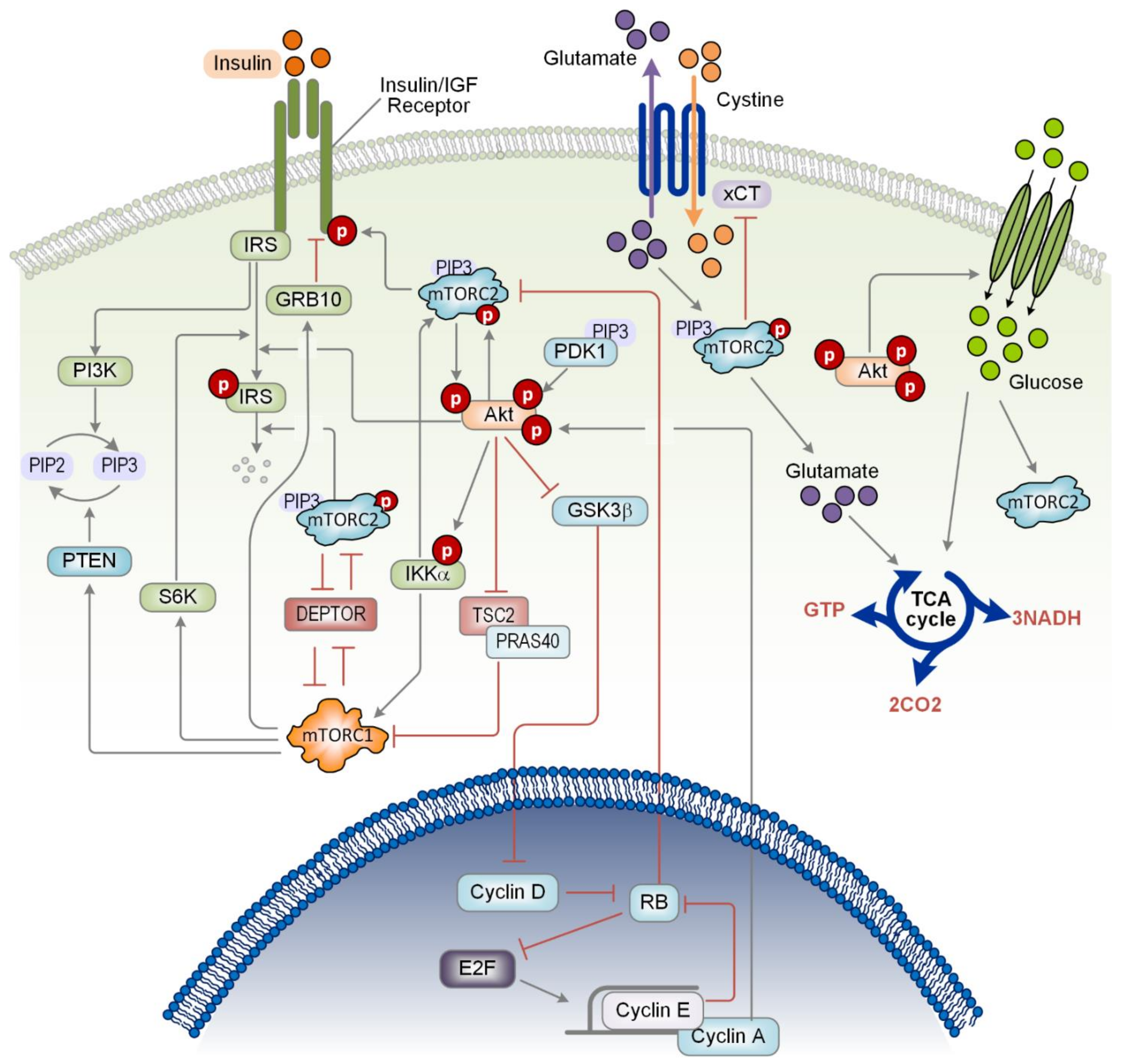
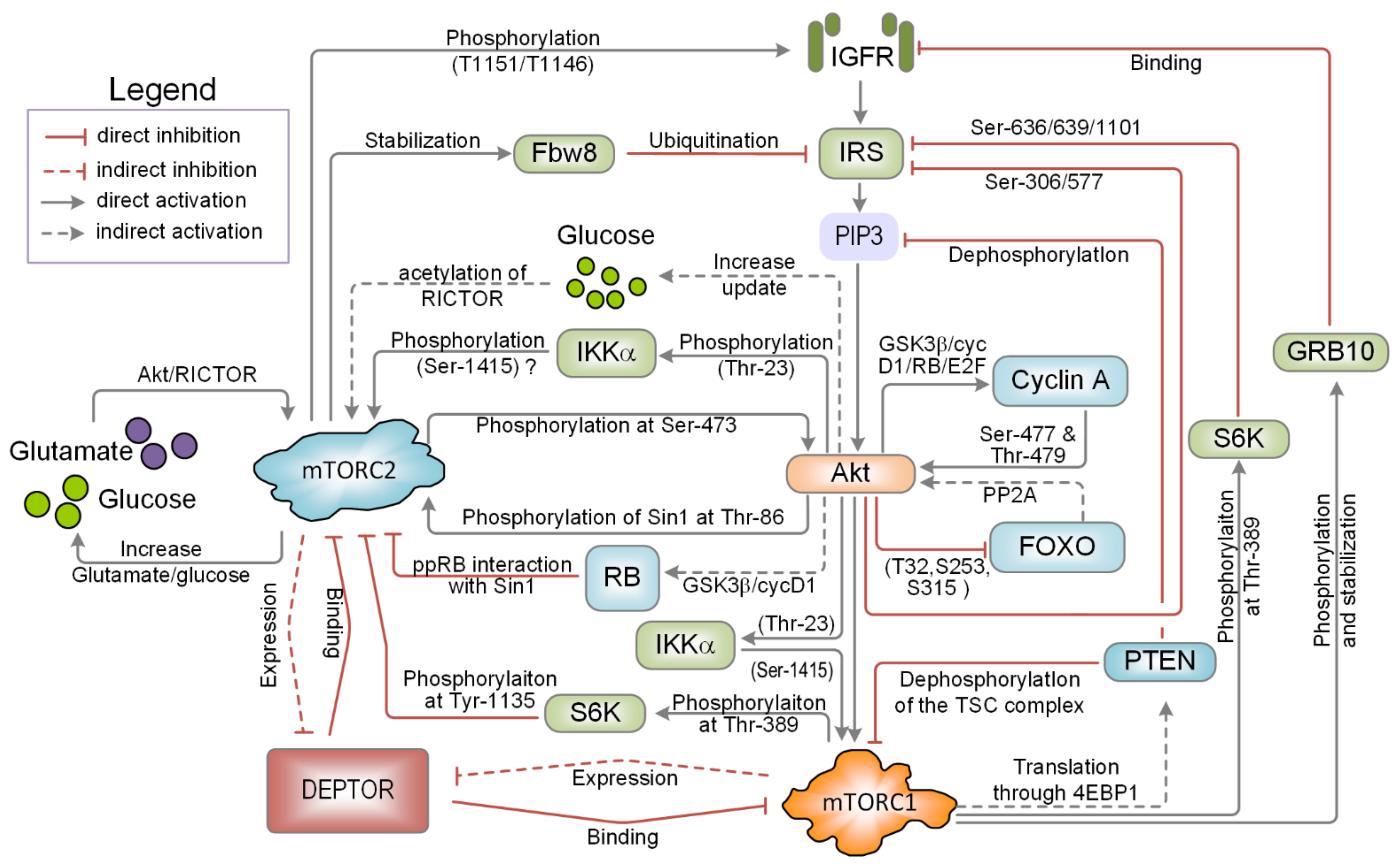
3.1. Feedback Mechanisms Mediated by mTOR
3.2. Feedback Mechanisms Mediated by Akt
3.3. Mutual Inhibition Mediated by Protein-Protein Competition
4. Crosstalk between PI3K/mTOR and Other Signalling Pathways
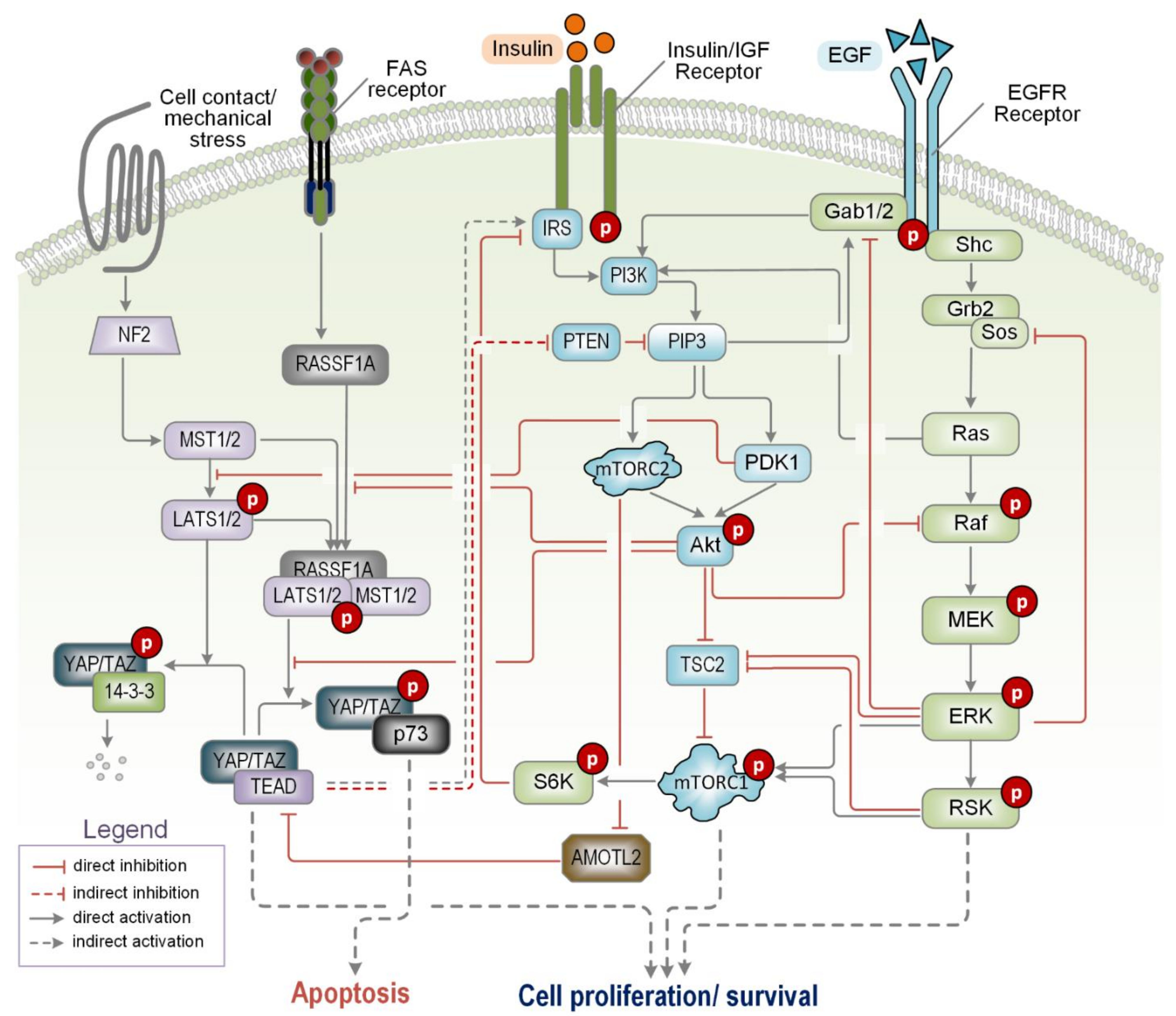
4.1. Complex Crosstalk with the Cell-Cycle Signalling Network
4.2. Crosstalk with the NF-κB Signalling Pathway
4.3. Crosstalk between mTOR and Nutrient Sources
4.4. Crosstalk with the Ras/ERK Signalling Pathway
4.5. Crosstalk with the Hippo/MST Signalling Pathway
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Xie, J.; Proud, C.G. Crosstalk between mTOR complexes. Nat. Cell Biol. 2013, 15, 1263–1265. [Google Scholar] [CrossRef] [PubMed]
- Guantes, R.; Poyatos, J.F. Multistable Decision Switches for Flexible Control of Epigenetic Differentiation. PLoS Comput. Biol. 2008, 4, e1000235. [Google Scholar] [CrossRef]
- Brandman, O.; Meyer, T. Feedback Loops Shape Cellular Signals in Space and Time. Science 2008, 322, 390–395. [Google Scholar] [CrossRef]
- Mangan, S.; Alon, U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 2003, 100, 11980–11985. [Google Scholar] [CrossRef] [PubMed]
- Goentoro, L.; Shoval, O.; Kirschner, M.W.; Alon, U. The Incoherent Feedforward Loop Can Provide Fold-Change Detection in Gene Regulation. Mol. Cell 2009, 36, 894–899. [Google Scholar] [CrossRef]
- Tian, T.; Li, X.; Zhang, J. mTOR signaling in cancer and mTOR inhibitors in solid tumor targeting therapy. Int. J. Mol. Sci. 2019, 20, 755. [Google Scholar] [CrossRef]
- Samuels, Y.; Velculescu, V. Oncogenic Mutations of PIK3CA in Human Cancers. Cell Cycle 2004, 3, 1221–1224. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 1–28. [Google Scholar] [CrossRef]
- Hanker, A.; Kaklamani, V.; Arteaga, C.L. Challenges for the Clinical Development of PI3K Inhibitors: Strategies to Improve Their Impact in Solid Tumors. Cancer Discov. 2019, 9, 482–491. [Google Scholar] [CrossRef]
- Cremers, C.G.; Nguyen, L.K. Network rewiring, adaptive resistance and combating strategies in breast cancer. Cancer Drug Resist. 2019, 2, 1106–1126. [Google Scholar] [CrossRef]
- Fayçal, C.A.; Hatat, A.-S.; Gazzeri, S.; Eymin, B. Splice Variants of the RTK Family: Their Role in Tumour Progression and Response to Targeted Therapy. Int. J. Mol. Sci. 2017, 18, 383. [Google Scholar] [CrossRef]
- Mardilovich, K.; Pankratz, S.L.; Shaw, L.M. Expression and function of the insulin receptor substrate proteins in cancer. Cell Commun. Signal. 2009, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Voliovitch, H.; Schindler, D.G.; Hadari, Y.R.; Taylor, S.I.; Accili, D.; Zick, Y. Tyrosine Phosphorylation of Insulin Receptor Substrate-1 in Vivo Depends upon the Presence of Its Pleckstrin Homology Region. J. Biol. Chem. 1995, 270, 18083–18087. [Google Scholar] [CrossRef]
- Sun, X.J.; Crimmins, D.; Myers, M.; Miralpeix, M.; White, M. Pleiotropic insulin signals are engaged by multisite phosphorylation of IRS-1. Mol. Cell. Biol. 1993, 13, 7418–7428. [Google Scholar] [CrossRef] [PubMed]
- Csolle, M.P.; Ooms, L.M.; Papa, A.; Mitchell, C.A. PTEN and Other PtdIns(3,4,5)P3 Lipid Phosphatases in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 9189. [Google Scholar] [CrossRef]
- Liu, P.; Gan, W.; Chin, Y.M.R.; Ogura, K.; Guo, J.; Zhang, J.; Wang, B.; Blenis, J.; Cantley, L.C.; Toker, A.; et al. PtdIns(3,4,5)P3-Dependent Activation of the mTORC2 Kinase Complex. Cancer Discov. 2015, 5, 1194–1209. [Google Scholar] [CrossRef]
- Gan, X.; Wang, J.; Su, B.; Wu, D. Evidence for Direct Activation of mTORC2 Kinase Activity by Phosphatidylinositol 3,4,5-Trisphosphate. J. Biol. Chem. 2011, 286, 10998–11002. [Google Scholar] [CrossRef]
- Alessi, D.; Andjelkovic, M.; Caudwell, B.; Cron, P.; Morrice, N.; Cohen, P.; Hemmings, B.A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996, 15, 6541–6551. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef]
- Job, C.; Lagnado, L. Calcium and Protein Kinase C Regulate the Actin Cytoskeleton in the Synaptic Terminal of Retinal Bipolar Cells. J. Cell Biol. 1998, 143, 1661–1672. [Google Scholar] [CrossRef]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt Promotes Cell Survival by Phosphorylating and Inhibiting a Forkhead Transcription Factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef]
- Huang, H.; Tindall, D.J.; Zhang, X.; Sejas, D.P.; Qiu, Y.; Williams, D.A.; Pang, Q. Dynamic FoxO transcription factors. J. Cell Sci. 2007, 120, 2479–2487. [Google Scholar] [CrossRef]
- Nguyen, L.K.; Kholodenko, B.N. Feedback regulation in cell signalling: Lessons for cancer therapeutics. Semin. Cell Dev. Biol. 2016, 50, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Tzatsos, A. Raptor binds the SAIN (Shc and IRS-1 NPXY binding) domain of insulin receptor substrate-1 (IRS-1) and regulates the phosphorylation of IRS-1 at Ser-636/639 by mTOR. J. Biol. Chem. 2009, 284, 22525–22534. [Google Scholar] [CrossRef]
- Shah, O.J.; Hunter, T. Turnover of the active fraction of IRS1 involves raptor-mTOR-and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Mol. Cell. Biol. 2006, 26, 6425–6434. [Google Scholar] [CrossRef]
- Harrington, L.S.; Findlay, G.M.; Gray, A.; Tolkacheva, T.; Wigfield, S.; Rebholz, H.; Barnett, J.; Leslie, N.; Cheng, S.; Shepherd, P.R.; et al. The TSC1-2 tumor suppressor controls insulin–PI3K signaling via regulation of IRS proteins. J. Cell Biol. 2004, 166, 213–223. [Google Scholar] [CrossRef]
- Dong, L.Q.; Porter, S.; Hu, D.; Liu, F. Inhibition of hGrb10 Binding to the Insulin Receptor by Functional Domain-mediated Oligomerization. J. Biol. Chem. 1998, 273, 17720–17725. [Google Scholar] [CrossRef]
- Laviola, L.; Giorgino, F.; Chow, J.C.; Baquero, J.A.; Hansen, H.; Ooi, J.; Zhu, J.; Riedel, H.; Smith, R.J. The adapter protein Grb10 associates preferentially with the insulin receptor as compared with the IGF-I receptor in mouse fibroblasts. J. Clin. Investig. 1997, 99, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Frantz, J.D.; Giorgetti-Peraldi, S.; Ottinger, E.A.; Shoelson, S.E. Human GRB-IRβ/GRB10: Splice variants of an insulin and growth factor receptor-binding protein with PH and SH2 domains. J. Biol. Chem. 2001, 272, 2659–2667. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yoon, S.-O.; Poulogiannis, G.; Yang, Q.; Ma, X.M.; Villén, J.; Kubica, N.; Hoffman, G.R.; Cantley, L.C.; Gygi, S.P.; et al. Phosphoproteomic Analysis Identifies Grb10 as an mTORC1 Substrate That Negatively Regulates Insulin Signaling. Science 2011, 332, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.P.; Kang, S.A.; Rameseder, J.; Zhang, Y.; Ottina, K.A.; Lim, D.; Peterson, T.R.; Choi, Y.; Gray, N.S.; Yaffe, M.B.; et al. The mTOR-Regulated Phosphoproteome Reveals a Mechanism of mTORC1-Mediated Inhibition of Growth Factor Signaling. Science 2011, 332, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.K. Regulation of oscillation dynamics in biochemical systems with dual negative feedback loops. J. R. Soc. Interface 2012, 9, 1998–2010. [Google Scholar] [CrossRef]
- Kim, S.J.; DeStefano, M.A.; Oh, W.J.; Wu, C.-C.; Vega-Cotto, N.M.; Finlan, M.; Liu, D.; Su, B.; Jacinto, E. mTOR Complex 2 Regulates Proper Turnover of Insulin Receptor Substrate-1 via the Ubiquitin Ligase Subunit Fbw8. Mol. Cell 2012, 48, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, R.; Vanaja, K.G.; Boyer, J.A.; Gadal, S.; Solomon, H.; Chandarlapaty, S.; Levchenko, A.; Rosen, N. Regulation of PTEN translation by PI3K signaling maintains pathway homeostasis. Mol. Cell 2021, 81, 708–723.e5. [Google Scholar] [CrossRef]
- Garami, A.; Zwartkruis, F.J.; Nobukuni, T.; Joaquin, M.; Roccio, M.; Stocker, H.; Kozma, S.C.; Hafen, E.; Bos, J.L.; Thomas, G. Insulin Activation of Rheb, a Mediator of mTOR/S6K/4E-BP Signaling, Is Inhibited by TSC1 and 2. Mol. Cell 2003, 11, 1457–1466. [Google Scholar] [CrossRef]
- Stambolic, V.; Suzuki, A.; De La Pompa, J.L.; Brothers, G.M.; Mirtsos, C.; Sasaki, T.; Ruland, J.; Penninger, J.M.; Siderovski, D.; Mak, T.W. Negative Regulation of PKB/Akt-Dependent Cell Survival by the Tumor Suppressor PTEN. Cell 1998, 95, 29–39. [Google Scholar] [CrossRef]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef]
- Inoki, K.; Zhu, T.; Guan, K.-L. TSC2 Mediates Cellular Energy Response to Control Cell Growth and Survival. Cell 2003, 115, 577–590. [Google Scholar] [CrossRef]
- Ling, N.X.Y.; Kaczmarek, A.; Hoque, A.; Davie, E.; Ngoei, K.R.W.; Morrison, K.R.; Smiles, W.J.; Forte, G.M.; Wang, T.; Lie, S.; et al. mTORC1 directly inhibits AMPK to promote cell proliferation under nutrient stress. Nat. Metab. 2020, 2, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Saqcena, M.; Chatterjee, A.; Garcia, A.; Frias, M.A.; Foster, D.A. Reciprocal Regulation of AMP-activated Protein Kinase and Phospholipase D. J. Biol. Chem. 2015, 290, 6986–6993. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Frias, M.A.; Chatterjee, A.; Yellen, P.; Foster, D.A.A. The Enigma of Rapamycin Dosage. Mol. Cancer Ther. 2016, 15, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Chatterjee, A.; Kogan, D.; Patel, D.; A Foster, D. 5-Aminoimidazole-4-carboxamide-1-β-4-ribofuranoside (AICAR) enhances the efficacy of rapamycin in human cancer cells. Cell Cycle 2015, 14, 3331–3339. [Google Scholar] [CrossRef] [PubMed]
- Molina, J.R.; Agarwal, N.K.; Morales, F.C.; Hayashi, Y.; Aldape, K.D.; Cote, G.; Georgescu, M.-M. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene 2011, 31, 1264–1274. [Google Scholar] [CrossRef]
- Liu, J.; Stevens, P.D.; Li, X.; Schmidt, M.D.; Gao, T. PHLPP-Mediated Dephosphorylation of S6K1 Inhibits Protein Translation and Cell Growth. Mol. Cell. Biol. 2011, 31, 4917–4927. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Brognard, J.; Newton, A.C. The Phosphatase PHLPP Controls the Cellular Levels of Protein Kinase C. J. Biol. Chem. 2008, 283, 6300–6311. [Google Scholar] [CrossRef]
- Gao, T.; Furnari, F.; Newton, A.C. PHLPP: A Phosphatase that Directly Dephosphorylates Akt, Promotes Apoptosis, and Suppresses Tumor Growth. Mol. Cell 2005, 18, 13–24. [Google Scholar] [CrossRef]
- Brognard, J.; Sierecki, E.; Gao, T.; Newton, A.C. PHLPP and a Second Isoform, PHLPP2, Differentially Attenuate the Amplitude of Akt Signaling by Regulating Distinct Akt Isoforms. Mol. Cell 2007, 25, 917–931. [Google Scholar] [CrossRef]
- Liu, J.; Stevens, P.D.; Gao, T. mTOR-dependent regulation of PHLPP expression controls the rapamycin sensitivity in cancer cells. J. Biol. Chem. 2011, 286, 6510–6520. [Google Scholar] [CrossRef]
- Li, X.; Liu, J.; Gao, T. β-TrCP-mediated ubiquitination and degradation of PHLPP1 are negatively regulated by Akt. Mol. Cell. Biol. 2009, 29, 6192–6205. [Google Scholar] [CrossRef]
- Cross, D.A.E.; Alessi, D.; Cohen, P.; Andjelkovich, M.; Hemmings, B.A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nat. Cell Biol. 1995, 378, 785–789. [Google Scholar] [CrossRef]
- Yin, Y.; Hua, H.; Li, M.; Liu, S.; Kong, Q.; Shao, T.; Wang, J.; Luo, Y.; Wang, Q.; Luo, T.; et al. mTORC2 promotes type I insulin-like growth factor receptor and insulin receptor activation through the tyrosine kinase activity of mTOR. Cell Res. 2015, 26, 46–65. [Google Scholar] [CrossRef]
- Liu, P.; Gan, W.; Inuzuka, H.; Lazorchak, A.S.; Gao, D.; Arojo, O.; Liu, D.; Wan, L.; Zhai, B.; Yu, Y.; et al. Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signalling to suppress tumorigenesis. Nat. Cell Biol. 2013, 15, 1340–1350. [Google Scholar] [CrossRef] [PubMed]
- Julien, L.-A.; Carriere, A.; Moreau, J.; Roux, P.P. mTORC1-Activated S6K1 Phosphorylates Rictor on Threonine 1135 and Regulates mTORC2 Signaling. Mol. Cell. Biol. 2010, 30, 908–921. [Google Scholar] [CrossRef]
- Yang, G.; Murashige, D.S.; Humphrey, S.J.; James, D.E. A Positive Feedback Loop between Akt and mTORC2 via SIN1 Phosphorylation. Cell Rep. 2015, 12, 937–943. [Google Scholar] [CrossRef]
- Treins, C.; Warne, P.; Magnuson, M.; Pende, M.; Downward, J. Rictor is a novel target of p70 S6 kinase-1. Oncogene 2010, 29, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Xu, W.; Li, G.; Cui, W. Weighing In on mTOR Complex 2 Signaling: The Expanding Role in Cell Metabolism. Oxidative Med. Cell. Longev. 2018, 2018, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Angeli, D.; Ferrell, J.E.; Sontag, E.D. Detection of multistability, bifurcations, and hysteresis in a large class of biological positive-feedback systems. Proc. Natl. Acad. Sci. USA 2004, 101, 1822–1827. [Google Scholar] [CrossRef] [PubMed]
- Chu, N.; Salguero, A.L.; Liu, A.Z.; Chen, Z.; Dempsey, D.R.; Ficarro, S.B.; Alexander, W.M.; Marto, J.A.; Li, Y.; Amzel, L.M.; et al. Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis. Cell 2018, 174, 897–907.e14. [Google Scholar] [CrossRef] [PubMed]
- Guertin, D.A.; Stevens, D.M.; Thoreen, C.C.; Burds, A.A.; Kalaany, N.Y.; Moffat, J.; Brown, M.; Fitzgerald, K.J.; Sabatini, D.M. Ablation in Mice of the mTORC Components raptor, rictor, or mLST8 Reveals that mTORC2 Is Required for Signaling to Akt-FOXO and PKCα, but Not S6K1. Dev. Cell 2006, 11, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Jacinto, E.; Facchinetti, V.; Liu, D.; Soto, N.; Wei, S.; Jung, S.Y.; Huang, Q.; Qin, J.; Su, B. SIN1/MIP1 Maintains rictor-mTOR Complex Integrity and Regulates Akt Phosphorylation and Substrate Specificity. Cell 2006, 127, 125–137. [Google Scholar] [CrossRef]
- Kearney, A.L.; Cooke, K.C.; Norris, D.M.; Zadoorian, A.; Krycer, J.R.; Fazakerley, D.J.; Burchfield, J.G.; James, D.E. Serine 474 phosphorylation is essential for maximal Akt2 kinase activity in adipocytes. J. Biol. Chem. 2019, 294, 16729–16739. [Google Scholar] [CrossRef]
- Kearney, A.L.N.; Ghomlaghi, M.; Wong, M.; Humphrey, M.; Carroll, L.; Yang, G.; Cooke, K.; Yang, P.; Geddes, T.; Shin, S.F.; et al. Akt phosphorylates insulin receptor substrate (IRS) to limit PI3K-mediated PI(3,4,5)P3 synthesis. eLife 2021, in press. [Google Scholar]
- Nitulescu, G.M.; Van De Venter, M.; Nitulescu, G.; Ungurianu, A.; Juzenas, P.; Peng, Q.; Olaru, O.T.; Grădinaru, D.; Tsatsakis, A.; Tsoukalas, D.; et al. The Akt pathway in oncology therapy and beyond (Review). Int. J. Oncol. 2018, 53, 2319–2331. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.G.; Wang, N.; Cao, D.J.; Sachan, N.; Morris, D.J.; Gerard, R.D.; Kuro-O, M.; Rothermel, B.A.; Hill, J.A. FoxO transcription factors activate Akt and attenuate insulin signaling in heart by inhibiting protein phosphatases. Proc. Natl. Acad. Sci. USA 2007, 104, 20517–20522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tang, N.; Hadden, T.J.; Rishi, A.K. Akt, FoxO and regulation of apoptosis. Biochim. Biophys. Acta (BBA) Bioenerg. 2011, 1813, 1978–1986. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, J.-M.; Sotnikova, T.D.; Marion, S.; Lefkowitz, R.J.; Gainetdinov, R.; Caron, M.G. An Akt/β-Arrestin 2/PP2A Signaling Complex Mediates Dopaminergic Neurotransmission and Behavior. Cell 2005, 122, 261–273. [Google Scholar] [CrossRef]
- Tzivion, G.; Dobson, M.; Ramakrishnan, G. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim. Biophys. Acta (BBA) Bioenerg. 2011, 1813, 1938–1945. [Google Scholar] [CrossRef]
- Dobson, M.; Ramakrishnan, G.; Ma, S.; Kaplun, L.; Balan, V.; Fridman, R.; Tzivion, G. Bimodal regulation of FoxO3 by AKT and 14-3-3. Biochim. Biophys. Acta (BBA) Bioenerg. 2011, 1813, 1453–1464. [Google Scholar] [CrossRef]
- Hui, R.C.-Y.; Gomes, A.R.; Constantinidou, D.; Costa, J.R.; Karadedou, C.T.; de Mattos, S.F.; Wymann, M.P.; Brosens, J.J.; Schulze, A.; Lam, E.W.-F. The Forkhead Transcription Factor FOXO3a Increases Phosphoinositide-3 Kinase/Akt Activity in Drug-Resistant Leukemic Cells through Induction of PIK3CA Expression. Mol. Cell. Biol. 2008, 28, 5886–5898. [Google Scholar] [CrossRef] [PubMed]
- Puig, O.; Tjian, R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 2005, 19, 2435–2446. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Jie, Z.; Joo, D.; Ordureau, A.; Liu, P.; Gan, W.; Guo, J.; Zhang, J.; North, B.J.; Dai, X.; et al. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nat. Cell Biol. 2017, 545, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Romano, D.; Nguyen, L.K.; Matallanas, D.; Halasz, M.; Doherty, C.; Kholodenko, B.N.; Kolch, W. Protein interaction switches coordinate Raf-1 and MST2/Hippo signalling. Nat. Cell Biol. 2014, 16, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Varusai, T.M.; Kolch, W.; Kholodenko, B.N.; Nguyen, L.K. Protein–protein interactions generate hidden feedback and feed-forward loops to trigger bistable switches, oscillations and biphasic dose–responses. Mol. BioSyst. 2015, 11, 2750–2762. [Google Scholar] [CrossRef] [PubMed]
- Ghomlaghi, M.; Shin, S.; Yang, G.; James, D.E.; Nguyen, L.K. Dynamic modelling of the PI3K/mTOR signalling network uncovers biphasic dependence of mTORC1 activation on the mTORC2 subunit Sin1. bioRxiv 2020. [Google Scholar] [CrossRef]
- Peterson, T.R.; Laplante, M.; Thoreen, C.C.; Sancak, Y.; Kang, S.A.; Kuehl, W.M.; Gray, N.S.; Sabatini, D.M. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009, 137, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xiong, X.; Sun, Y. DEPTOR, an mTOR Inhibitor, is a Physiological Substrate of SCFβTrCP E3 Ubiquitin Ligase and Regulates Survival and Autophagy. Mol. Cell 2011, 44, 304–316. [Google Scholar] [CrossRef]
- Varusai, T.M.; Nguyen, L.K. Dynamic modelling of the mTOR signalling network reveals complex emergent behaviours conferred by DEPTOR. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Caron, A.; Briscoe, D.M.; Richard, D.; Laplante, M. DEPTOR at the Nexus of Cancer, Metabolism, and Immunity. Physiol. Rev. 2018, 98, 1765–1803. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.K.; Matallanas, D.; Romano, D.; Kholodenko, B.N.; Kolch, W. Competing to coordinate cell fate decisions: The MST2-Raf-1 signaling device. Cell Cycle 2015, 14, 189–199. [Google Scholar] [CrossRef]
- Shin, S.-Y.; Nguyen, L.K. Unveiling Hidden Dynamics of Hippo Signalling: A Systems Analysis. Genes 2016, 7, 44. [Google Scholar] [CrossRef]
- Weinberg, R.A. The retinoblastoma protein and cell cycle control. Cell 1995, 81, 323–330. [Google Scholar] [CrossRef]
- Dowdy, S.F.; Hinds, P.W.; Louie, K.; Reed, S.I.; Arnold, A.; Weinberg, R.A. Physical interaction of the retinoblastoma protein with human D cyclins. Cell 1993, 73, 499–511. [Google Scholar] [CrossRef]
- Giacinti, C.; Giordano, A. RB and cell cycle progression. Oncogene 2006, 25, 5220–5227. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, K.; Liu, P.; Geng, Y.; Wang, B.; Gan, W.; Guo, J.; Wu, F.; Chin, Y.M.R.; Berrios, C.; et al. Inhibition of Rb Phosphorylation Leads to mTORC2-Mediated Activation of Akt. Mol. Cell 2016, 62, 929–942. [Google Scholar] [CrossRef]
- Burkhart, D.L.; Sage, J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 2008, 8, 671–682. [Google Scholar] [CrossRef]
- Choi, Y.J.; Li, X.; Hydbring, P.; Sanda, T.; Stefano, J.; Christie, A.L.; Signoretti, S.; Look, A.T.; Kung, A.L.; von Boehmer, H.; et al. The Requirement for Cyclin D Function in Tumor Maintenance. Cancer Cell 2012, 22, 438–451. [Google Scholar] [CrossRef]
- Liu, P.; Begley, M.J.; Michowski, W.; Inuzuka, H.; Ginzberg, M.; Gao, D.; Tsou, P.; Gan, W.; Papa, A.; Kim, B.M.; et al. Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nat. Cell Biol. 2014, 508, 541–545. [Google Scholar] [CrossRef]
- Lawrence, T. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harbor Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef]
- Bai, D.; Ueno, L.; Vogt, P.K. Akt-mediated regulation of NFκB and the essentialness of NFκB for the oncogenicity of PI3K and Akt. Int. J. Cancer 2009, 125, 2863–2870. [Google Scholar] [CrossRef]
- Dan, H.C.; Ebbs, A.; Pasparakis, M.; Van Dyke, T.; Basseres, D.S.; Baldwin, A.S. Akt-dependent activation of mTORC1 Involves phosphorylation of mTOR by IKKalpha. J. Biol. Chem. 2014, 289, 25227–25240. [Google Scholar] [CrossRef]
- Dan, H.C.; Antonia, R.; Baldwin, A.S. PI3K/Akt promotes feedforward mTORC2 activation through IKKα. Oncotarget 2016, 7, 21064–21075. [Google Scholar] [CrossRef]
- Pavlova, N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Koppula, P.; Zhang, Y.; Shi, J.; Li, W.; Gan, B. The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. J. Biol. Chem. 2017, 292, 14240–14249. [Google Scholar] [CrossRef]
- Gu, Y.; Albuquerque, C.P.; Braas, D.; Zhang, W.; Villa, G.R.; Bi, J.; Ikegami, S.; Masui, K.; Gini, B.; Yang, H.; et al. mTORC2 Regulates Amino Acid Metabolism in Cancer by Phosphorylation of the Cystine-Glutamate Antiporter xCT. Mol. Cell 2017, 67, 128–138.e7. [Google Scholar] [CrossRef]
- Vincent, E.; Sergushichev, A.; Griss, T.; Gingras, M.-C.; Samborska, B.; Ntimbane, T.; Coelho, P.P.; Blagih, J.; Raissi, T.C.; Choinière, L.; et al. Mitochondrial Phosphoenolpyruvate Carboxykinase Regulates Metabolic Adaptation and Enables Glucose-Independent Tumor Growth. Mol. Cell 2015, 60, 195–207. [Google Scholar] [CrossRef]
- Shin, C.-S.; Mishra, P.; Watrous, J.D.; Carelli, V.; D’Aurelio, M.; Jain, M.; Chan, D.C. The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR Signaling in Growth Control and Disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef]
- Pezze, P.D.; Ruf, S.; Sonntag, A.G.; Langelaar-Makkinje, M.; Hall, P.; Heberle, A.M.; Navas, P.R.; Van Eunen, K.; Tölle, R.C.; Schwarz, J.J. A systems study reveals concurrent activation of AMPK and mTOR by amino acids. Nat. Commun. 2016, 7, 1–19. [Google Scholar]
- Beg, M.; Abdullah, N.; Thowfeik, F.S.; Altorki, N.K.; McGraw, T.E. Distinct Akt phosphorylation states are required for insulin regulated Glut4 and Glut1-mediated glucose uptake. eLife 2017, 6, e26896. [Google Scholar] [CrossRef]
- Kleinert, M.; Parker, B.L.; Fritzen, A.M.; Knudsen, J.R.; Jensen, T.E.; Kjøbsted, R.; Sylow, L.; Ruegg, M.; James, D.E.; Richter, E.A. Mammalian target of rapamycin complex 2 regulates muscle glucose uptake during exercise in mice. J. Physiol. 2017, 595, 4845–4855. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, B.K.; Lamming, D.W. The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging. Cell Metab. 2016, 23, 990–1003. [Google Scholar] [CrossRef]
- Lamming, D.; Ye, L.; Katajisto, P.; Goncalves, M.; Saitoh, M.; Stevens, D.M.; Davis, J.G.; Salmon, A.B.; Richardson, A.; Ahima, R.S.; et al. Rapamycin-Induced Insulin Resistance Is Mediated by mTORC2 Loss and Uncoupled from Longevity. Science 2012, 335, 1638–1643. [Google Scholar] [CrossRef]
- Masui, K.; Tanaka, K.; Ikegami, S.; Villa, G.R.; Yang, H.; Yong, W.H.; Cloughesy, T.F.; Yamagata, K.; Arai, N.; Cavenee, W.K.; et al. Glucose-dependent acetylation of Rictor promotes targeted cancer therapy resistance. Proc. Natl. Acad. Sci. USA 2015, 112, 9406–9411. [Google Scholar] [CrossRef]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011, 36, 320–328. [Google Scholar] [CrossRef]
- Rodriguez-Viciana, P.; Warne, P.H.; Dhand, R.; Vanhaesebroeck, B.; Gout, I.; Fry, M.; Waterfield, M.D.; Downward, J. Phosphatidylinositol-3-OH kinase direct target of Ras. Nat. Cell Biol. 1994, 370, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Kodaki, T.; Woscholski, R.; Hallberg, B.; Downward, P.R.-V.J.; Parker, P.J. The activation of phosphatidylinositol 3-kinase by Ras. Curr. Biol. 1994, 4, 798–806. [Google Scholar] [CrossRef]
- Gupta, S.; Ramjaun, A.R.; Haiko, P.; Wang, Y.; Warne, P.H.; Nicke, B.; Nye, E.; Stamp, G.; Alitalo, K.; Downward, J. Binding of Ras to Phosphoinositide 3-Kinase p110α Is Required for Ras- Driven Tumorigenesis in Mice. Cell 2007, 129, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Heiden, M.G.V.; McCormick, F. The metabolic landscape of RAS-driven cancers from biology to therapy. Nat. Rev. Cancer 2021, 2, 271–283. [Google Scholar] [CrossRef]
- Zimmermann, S.; Moelling, K. Phosphorylation and regulation of Raf by Akt (protein kinase B). Science 1999, 286, 1741–1744. [Google Scholar] [CrossRef]
- Dhillon, A.S.; Meikle, S.; Yazici, Z.A.; Eulitz, M.; Kolch, W. Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J. 2002, 21, 64–71. [Google Scholar] [CrossRef]
- Guan, K.-L.; Figueroa, C.; Brtva, T.R.; Zhu, T.; Taylor, J.; Barber, T.D.; Vojtek, A.B. Negative Regulation of the Serine/Threonine Kinase B-Raf by Akt. J. Biol. Chem. 2000, 275, 27354–27359. [Google Scholar] [CrossRef]
- Carriere, A.; Romeo, Y.; Acosta-Jaquez, H.A.; Moreau, J.; Bonneil, E.; Thibault, P.; Fingar, D.C.; Roux, P.P. ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1). J. Biol. Chem. 2011, 286, 567–577. [Google Scholar] [CrossRef]
- Foster, K.G.; Acosta-Jaquez, H.A.; Romeo, Y.; Ekim, B.; Soliman, G.; Carriere, A.; Roux, P.; Ballif, B.A.; Fingar, D.C. Regulation of mTOR Complex 1 (mTORC1) by Raptor Ser863 and Multisite Phosphorylation*. J. Biol. Chem. 2010, 285, 80–94. [Google Scholar] [CrossRef]
- Rodrigues, G.A.; Falasca, M.; Zhang, Z.; Ong, S.H.; Schlessinger, J. A Novel Positive Feedback Loop Mediated by the Docking Protein Gab1 and Phosphatidylinositol 3-Kinase in Epidermal Growth Factor Receptor Signaling. Mol. Cell. Biol. 2000, 20, 1448–1459. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Schlessinger, J. Cell Signaling by Receptor Tyrosine Kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Wöhrle, F.U.; Daly, R.J.; Brummer, T. Function, regulation and pathological roles of the Gab/DOS docking proteins. Cell Commun. Signal. 2009, 7, 1–28. [Google Scholar] [CrossRef]
- Lehr, S.; Kotzka, J.; Avci, H.; Sickmann, A.; Meyer, H.E.; Herkner, A.A.; Muller-Wieland, D. Identification of Major ERK-Related Phosphorylation Sites in Gab1. Biochemistry 2004, 43, 12133–12140. [Google Scholar] [CrossRef] [PubMed]
- Tyson, J.J.; Chen, K.C.; Novak, B. Sniffers, buzzers, toggles and blinkers: Dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 2003, 15, 221–231. [Google Scholar] [CrossRef]
- Frame, S.; Cohen, P. The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2001, 2, 769–776. [Google Scholar]
- Sato, S.; Fujita, N.; Tsuruo, T. Involvement of 3-Phosphoinositide-dependent Protein Kinase-1 in the MEK/MAPK Signal Transduction Pathway. J. Biol. Chem. 2004, 279, 33759–33767. [Google Scholar] [CrossRef]
- Mora, A.; Komander, D.; van Aalten, D.M.; Alessi, D.R. PDK1, the master regulator of AGC kinase signal transduction. Semin. Cell Dev. Biol. 2004, 15, 161–170. [Google Scholar] [CrossRef]
- King, C.C.; Gardiner, E.M.M.; Zenke, F.T.; Bohl, B.P.; Newton, A.C.; Hemmings, B.A.; Bokoch, G.M. p21-activated Kinase (PAK1) Is Phosphorylated and Activated by 3-Phosphoinositide-dependent Kinase-1 (PDK1). J. Biol. Chem. 2000, 275, 41201–41209. [Google Scholar] [CrossRef]
- Harvey, K.F.; Zhang, X.; Thomas, D. The Hippo pathway and human cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef]
- Ehmer, U.; Sage, J. Control of Proliferation and Cancer Growth by the Hippo Signaling Pathway. Mol. Cancer Res. 2016, 14, 127–140. [Google Scholar] [CrossRef]
- Fallahi, E.; O’Driscoll, N.A.; Matallanas, D. The MST/Hippo Pathway and Cell Death: A Non-Canonical Affair. Genes 2016, 7, 28. [Google Scholar] [CrossRef]
- Romano, D.; Matallanas, D.; Frederick, D.T.; Flaherty, K.T.; Kolch, W. One Hippo and many masters: Differential regulation of the Hippo pathway in cancer. Biochem. Soc. Trans. 2014, 42, 816–821. [Google Scholar] [CrossRef]
- Romano, D.; Matallanas, D.; Weitsman, G.; Preisinger, C.; Ng, T.; Kolch, W. Proapoptotic Kinase MST2 Coordinates Signaling Crosstalk between RASSF1A, Raf-1, and Akt. Cancer Res. 2010, 70, 1195–1203. [Google Scholar] [CrossRef]
- Kim, N.; Shu, S.; Coppola, M.D.; Kaneko, S.; Yuan, Z.-Q.; Cheng, J.Q. Regulation of Proapoptotic Mammalian ste20–Like Kinase MST2 by the IGF1-Akt Pathway. PLoS ONE 2010, 5, e9616. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Kim, N.-G.; Gumbiner, B.M. Regulation of Hippo pathway by mitogenic growth factors via phosphoinositide 3-kinase and phosphoinositide-dependent kinase-1. Proc. Natl. Acad. Sci. USA 2013, 110, 2569–2574. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.; Irvine, K.D. Regulation of Hippo Signaling by EGFR-MAPK Signaling through Ajuba Family Proteins. Dev. Cell 2013, 24, 459–471. [Google Scholar] [CrossRef]
- Basu, S.; Totty, N.F.; Irwin, M.S.; Sudol, M.; Downward, J. Akt Phosphorylates the Yes-Associated Protein, YAP, to Induce Interaction with 14-3-3 and Attenuation of p73-Mediated Apoptosis. Mol. Cell 2003, 11, 11–23. [Google Scholar] [CrossRef]
- Artinian, N.; Cloninger, C.; Holmes, B.; Benavides-Serrato, A.; Bashir, T.; Gera, J. Phosphorylation of the Hippo Pathway Component AMOTL2 by the mTORC2 Kinase Promotes YAP Signaling, Resulting in Enhanced Glioblastoma Growth and Invasiveness. J. Biol. Chem. 2015, 290, 19387–19401. [Google Scholar] [CrossRef]
- Wu, S.-H.; Bi, J.-F.; Cloughesy, T.; Cavenee, W.K.; Mischel, P.S. Emerging function of mTORC2 as a core regulator in glioblastoma: Metabolic reprogramming and drug resistance. Cancer Biol. Med. 2014, 11, 255–263. [Google Scholar] [CrossRef]
- Read, R.D.; Cavenee, W.K.; Furnari, F.; Thomas, J.B. A Drosophila Model for EGFR-Ras and PI3K-Dependent Human Glioma. PLoS Genet. 2009, 5, e1000374. [Google Scholar] [CrossRef]
- Bashir, T.; Cloninger, C.; Artinian, N.; Anderson, L.; Bernath, A.; Holmes, B.; Benavides-Serrato, A.; Sabha, N.; Nishimura, R.N.; Guha, A.; et al. Conditional Astroglial Rictor Overexpression Induces Malignant Glioma in Mice. PLoS ONE 2012, 7, e47741. [Google Scholar] [CrossRef]
- Jhanwar-Uniyal, M.; Gulati, N.; Karsy, M.; Albert, L.; Murali, R. Involvement of mTORC1 and mTORC2 in regulation of glioblastoma multiforme growth and motility. Int. J. Oncol. 2009, 35, 731–740. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.; Pan, P.; Wang, Z.; Zhang, Y.; Xie, P.; Geng, D.; Jiang, Y.; Yu, R.; Zhou, X. β-catenin-mediated YAP signaling promotes human glioma growth. J. Exp. Clin. Cancer Res. 2017, 36, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cinar, B.; Fang, P.-K.; Lutchman, M.; Di Vizio, D.; Adam, R.M.; Pavlova, N.; Rubin, M.A.; Yelick, P.C.; Freeman, M.R. The pro-apoptotic kinase Mst1 and its caspase cleavage products are direct inhibitors of Akt1. EMBO J. 2007, 26, 4523–4534. [Google Scholar] [CrossRef]
- Tumaneng, K.; Schlegelmilch, K.; Russell, R.C.; Yimlamai, D.; Basnet, H.; Mahadevan, N.R.; Fitamant, J.; Bardeesy, N.; Camargo, F.D.; Guan, K.-L. YAP mediates crosstalk between the Hippo and PI(3)K–TOR pathways by suppressing PTEN via miR-29. Nat. Cell Biol. 2012, 14, 1322–1329. [Google Scholar] [CrossRef]
- Jeong, S.-H.; Kim, H.-B.; Kim, M.-C.; Lee, J.-m.; Lee, J.H.; Kim, J.-H.; Kim, J.-W.; Park, W.-Y.; Kim, S.-Y.; Kim, J.B. Hippo-mediated suppression of IRS2/AKT signaling prevents hepatic steatosis and liver cancer. J. Clin. Investig. 2018, 128, 1010–1025. [Google Scholar] [CrossRef]
- Dey, A.; Varelas, X.; Guan, K.-L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 2020, 19, 480–494. [Google Scholar] [CrossRef]
- Ozbudak, E.M.; Thattai, M.; Lim, H.N.; Shraiman, B.I.; Van Oudenaarden, A. Multistability in the lactose utilization network of Escherichia coli. Nat. Cell Biol. 2004, 427, 737–740. [Google Scholar] [CrossRef] [PubMed]
- Francis, V.I.; Porter, S.L. Multikinase Networks: Two-Component Signaling Networks Integrating Multiple Stimuli. Annu. Rev. Microbiol. 2019, 73, 199–223. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.N.; Jefferson, L.S.; Kimball, S.R. ERK and Akt signaling pathways function through parallel mechanisms to promote mTORC1 signaling. Am. J. Physiol. Physiol. 2011, 300, C1172–C1180. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.-Y.; Müller, A.-K.; Verma, N.; Lev, S.; Nguyen, L.K. Systems modelling of the EGFR-PYK2-c-Met interaction network predicts and prioritizes synergistic drug combinations for triple-negative breast cancer. PLoS Comput. Biol. 2018, 14, e1006192. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghomlaghi, M.; Hart, A.; Hoang, N.; Shin, S.; Nguyen, L.K. Feedback, Crosstalk and Competition: Ingredients for Emergent Non-Linear Behaviour in the PI3K/mTOR Signalling Network. Int. J. Mol. Sci. 2021, 22, 6944. https://doi.org/10.3390/ijms22136944
Ghomlaghi M, Hart A, Hoang N, Shin S, Nguyen LK. Feedback, Crosstalk and Competition: Ingredients for Emergent Non-Linear Behaviour in the PI3K/mTOR Signalling Network. International Journal of Molecular Sciences. 2021; 22(13):6944. https://doi.org/10.3390/ijms22136944
Chicago/Turabian StyleGhomlaghi, Milad, Anthony Hart, Nhan Hoang, Sungyoung Shin, and Lan K. Nguyen. 2021. "Feedback, Crosstalk and Competition: Ingredients for Emergent Non-Linear Behaviour in the PI3K/mTOR Signalling Network" International Journal of Molecular Sciences 22, no. 13: 6944. https://doi.org/10.3390/ijms22136944
APA StyleGhomlaghi, M., Hart, A., Hoang, N., Shin, S., & Nguyen, L. K. (2021). Feedback, Crosstalk and Competition: Ingredients for Emergent Non-Linear Behaviour in the PI3K/mTOR Signalling Network. International Journal of Molecular Sciences, 22(13), 6944. https://doi.org/10.3390/ijms22136944





