Insulin-Like Growth Factor 2 mRNA-Binding Protein 1 (IGF2BP1) Is a Prognostic Biomarker and Associated with Chemotherapy Responsiveness in Colorectal Cancer
Abstract
:1. Introduction
2. Results
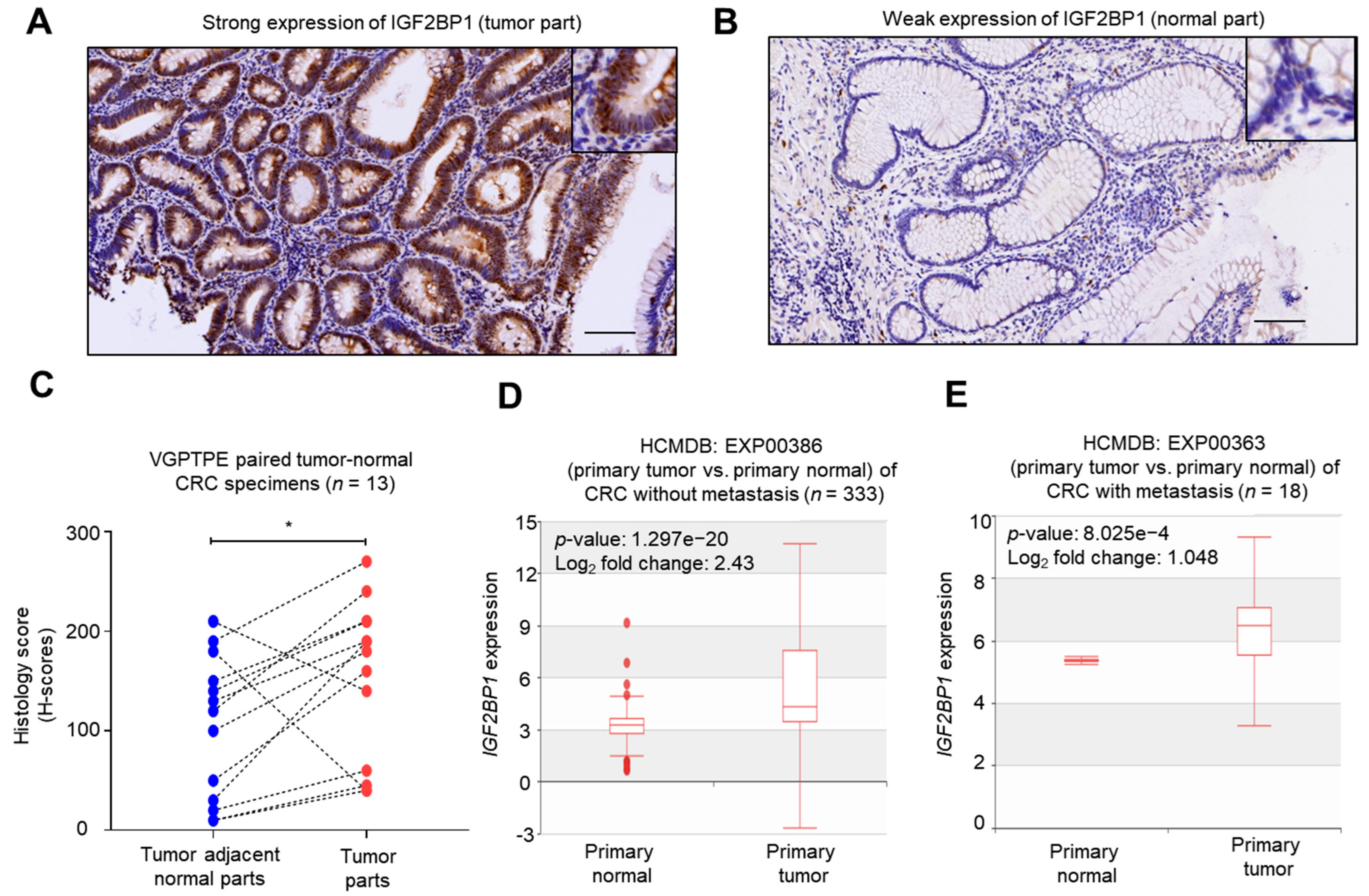
2.1. IGF2BP1 Was Overexpressed in CRC Tumors Comparing to Normal Counterparts
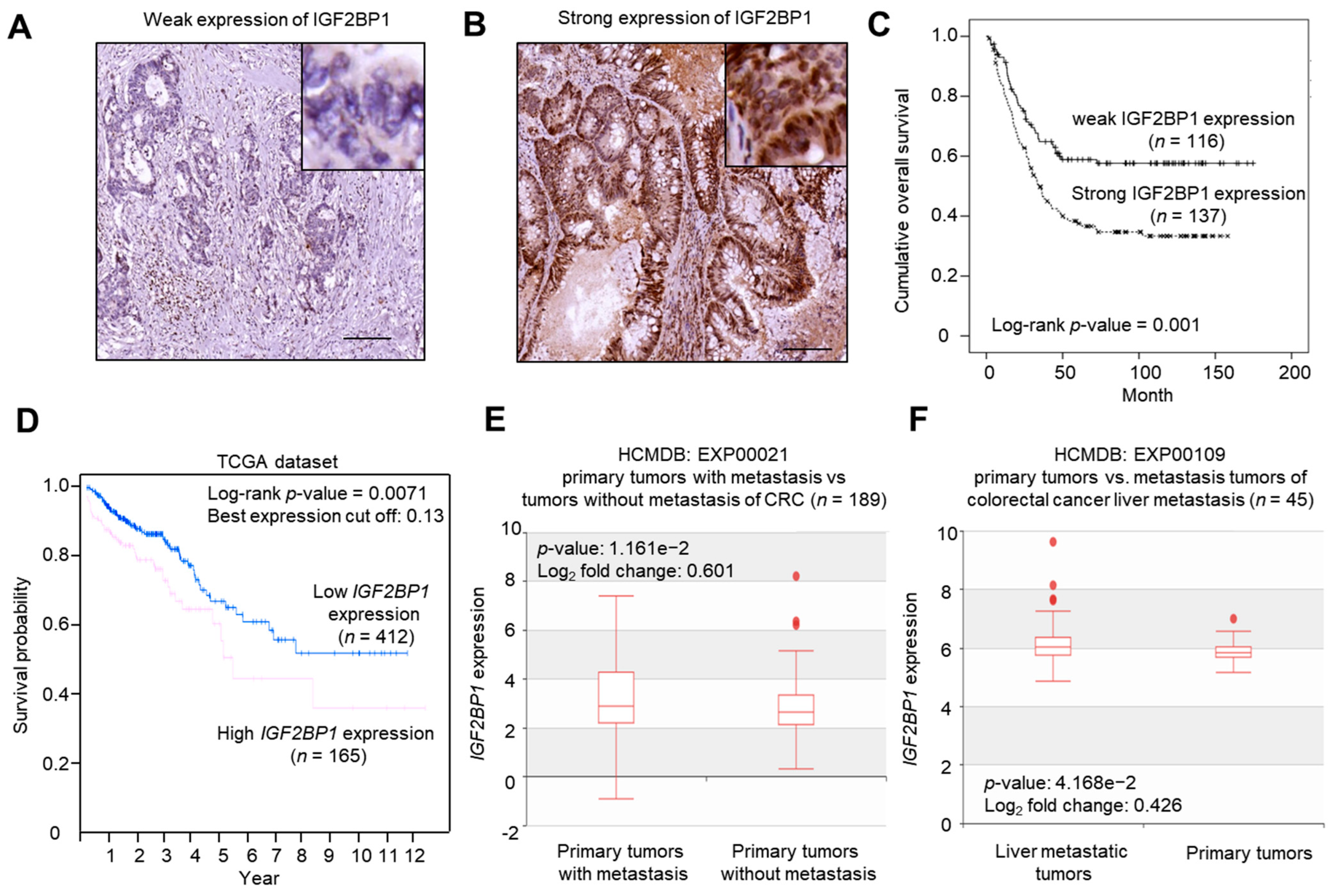
2.2. IGF2BP1 Expression Was Associated with Late Stage (III/IV) Status and Poor Overall Survival in CRC Patients
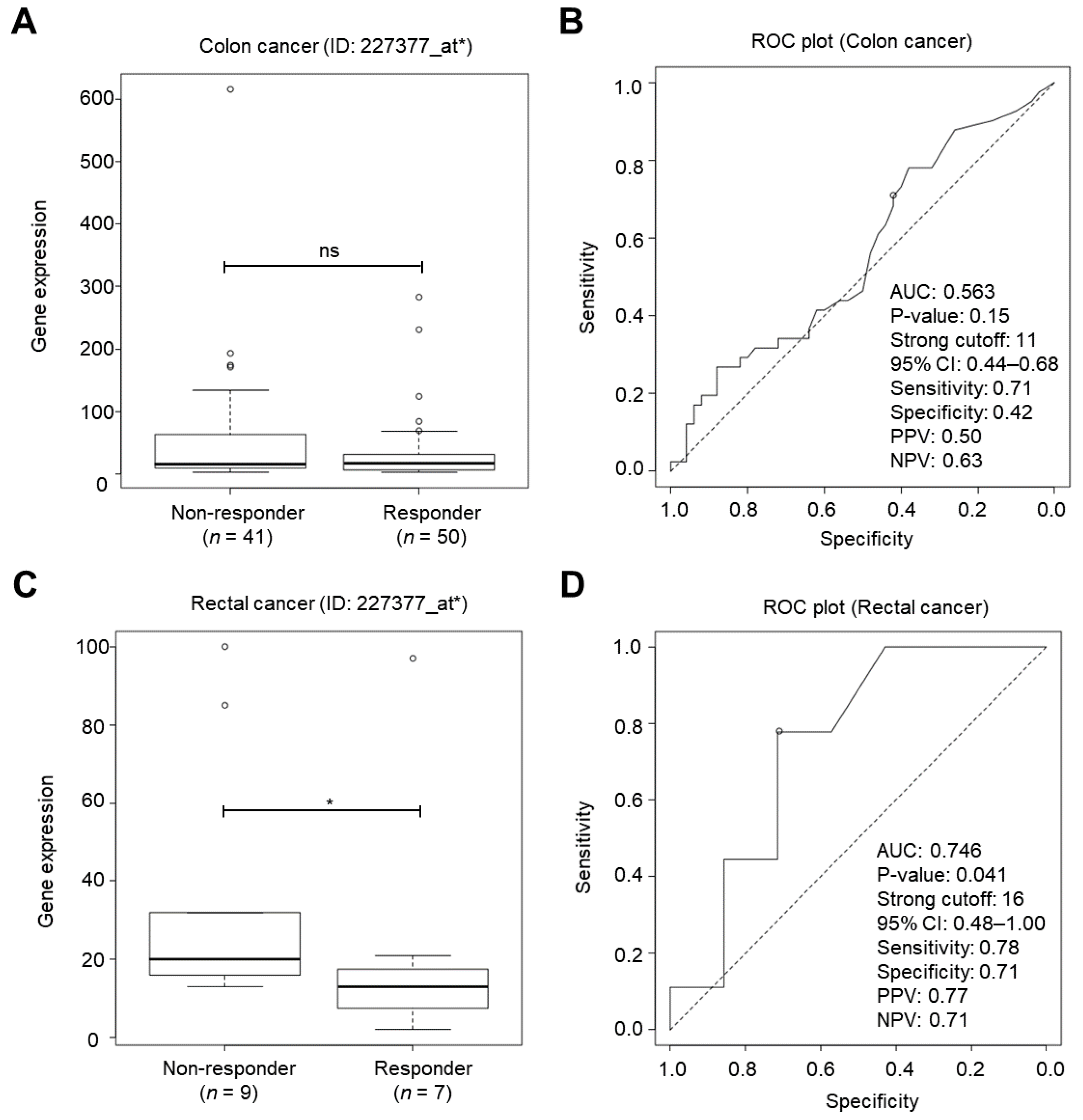
2.3. IGF2BP1 Was a Cancer Biomarker for Classifying Chemotherapy Responsiveness in Rectal Cancer Patients
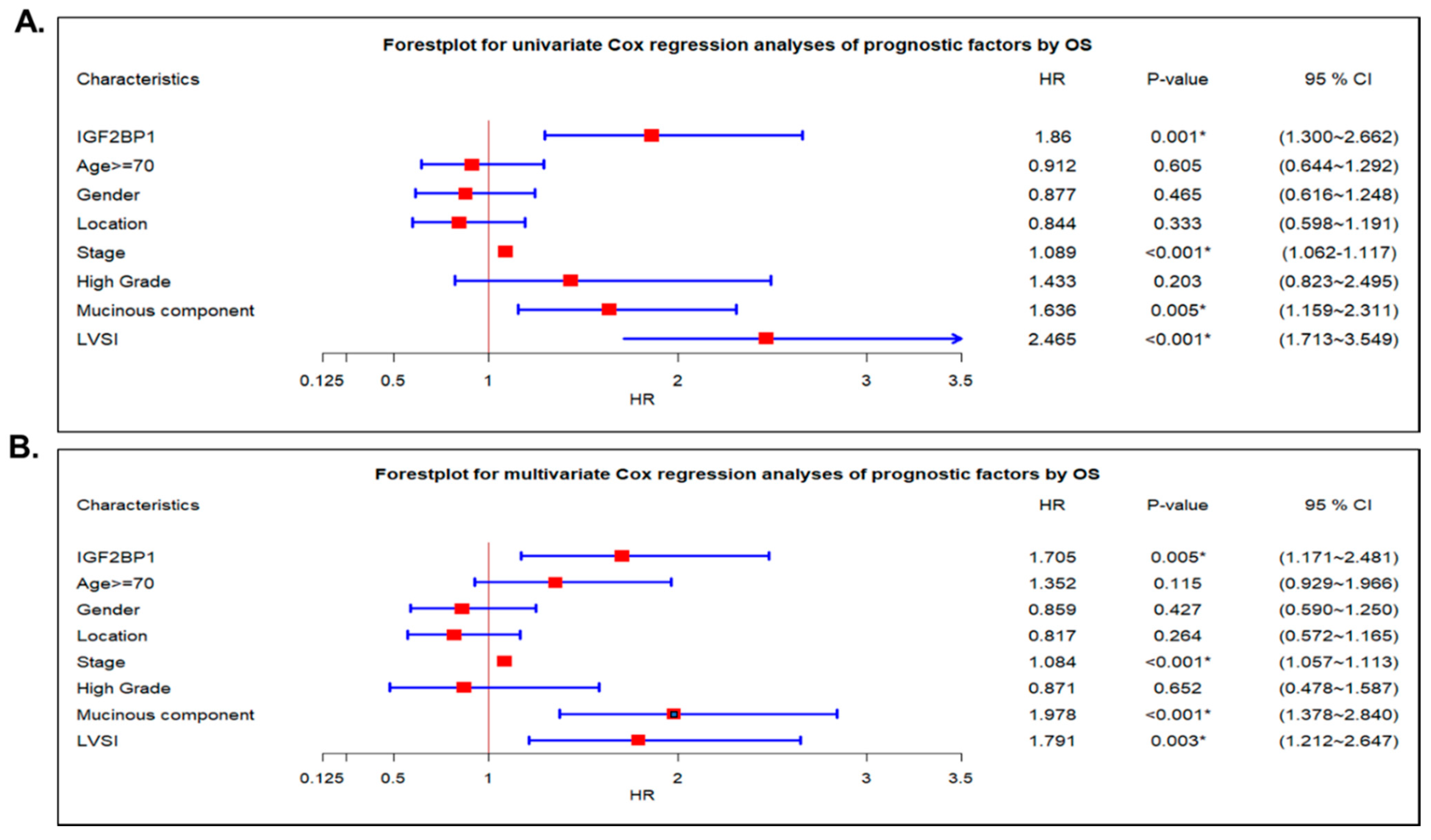
2.4. IGF2BP1 Was an Independent Prognostic Cancer Biomarker in CRC Patients
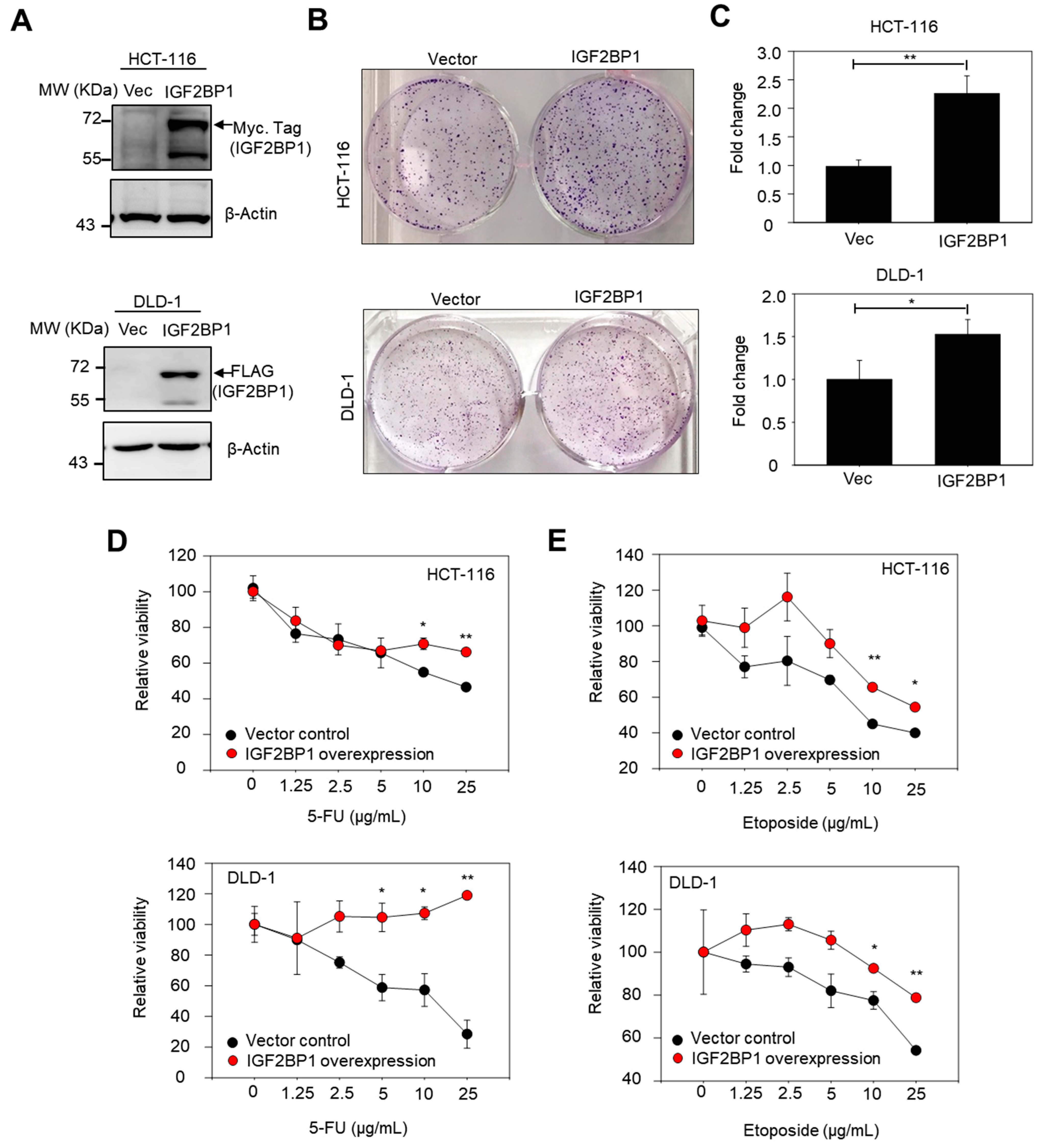
2.5. Overexpression of IGF2BP1 Increased Colony-Forming Capacity and Conferred Drug Resistance in CRC Cell Lines
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Immunohistochemistry
4.3. Bioinformatic Analysis
4.4. Cell Lines and Transfection
4.5. Colony Formation Assay
4.6. Cell Viability Assay
4.7. Western Blotting
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Yoshino, T.; Arnold, D.; Taniguchi, H.; Pentheroudakis, G.; Yamazaki, K.; Xu, R.H.; Kim, T.W.; Ismail, F.; Tan, I.B.; Yeh, K.H.; et al. Pan-Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: A JSMO-ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann. Oncol. 2018, 29, 44–70. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef]
- Schmoll, H.J.; Van Cutsem, E.; Stein, A.; Valentini, V.; Glimelius, B.; Haustermans, K.; Nordlinger, B.; van de Velde, C.J.; Balmana, J.; Regula, J.; et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann. Oncol. 2012, 23, 2479–2516. [Google Scholar] [CrossRef]
- Muller, M.F.; Ibrahim, A.E.; Arends, M.J. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016, 469, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Purnak, T.; Ozaslan, E.; Efe, C. Molecular basis of colorectal cancer. N. Engl. J. Med. author reply 1246–1247. 2010, 362, 1246. [Google Scholar]
- Denduluri, S.K.; Idowu, O.; Wang, Z.; Liao, Z.; Yan, Z.; Mohammed, M.K.; Ye, J.; Wei, Q.; Wang, J.; Zhao, L.; et al. Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes. Dis. 2015, 2, 13–25. [Google Scholar] [CrossRef] [Green Version]
- LeRoith, D.; Roberts, C.T., Jr. The insulin-like growth factor system and cancer. Cancer Lett. 2003, 195, 127–137. [Google Scholar] [CrossRef] [Green Version]
- Mahaira, L.G.; Katsara, O.; Pappou, E.; Iliopoulou, E.G.; Fortis, S.; Antsaklis, A.; Fotinopoulos, P.; Baxevanis, C.N.; Papamichail, M.; Perez, S.A. IGF2BP1 expression in human mesenchymal stem cells significantly affects their proliferation and is under the epigenetic control of TET1/2 demethylases. Stem. Cells Dev. 2014, 23, 2501–2512. [Google Scholar] [CrossRef]
- Lapidus, K.; Wyckoff, J.; Mouneimne, G.; Lorenz, M.; Soon, L.; Condeelis, J.S.; Singer, R.H. ZBP1 enhances cell polarity and reduces chemotaxis. J. Cell Sci. 2007, 120 Pt 18, 3173–3178. [Google Scholar] [CrossRef] [Green Version]
- Pollack, M.N. Insulin, insulin-like growth factors, insulin resistance, and neoplasia. Am. J. Clin. Nutr. 2007, 86, s820–s822. [Google Scholar] [CrossRef] [Green Version]
- Pollak, M. Insulin, insulin-like growth factors and neoplasia. Best Pract. Res. Clin. Endocrinol. Metab. 2008, 22, 625–638. [Google Scholar] [CrossRef]
- Gutschner, T.; Hammerle, M.; Pazaitis, N.; Bley, N.; Fiskin, E.; Uckelmann, H.; Heim, A.; Grobeta, M.; Hofmann, N.; Geffers, R.; et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology 2014, 59, 1900–1911. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, H.; Guo, X.; Zhu, Z.; Cai, H.; Kong, X. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J. Hematol. Oncol. 2018, 11, 88. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, K.E.; Noubissi, F.K.; Katti, P.S.; Hahn, C.M.; Davey, S.R.; Lundsmith, E.T.; Klein-Szanto, A.J.; Rhim, A.D.; Spiegelman, V.S.; Rustgi, A.K. IMP1 promotes tumor growth, dissemination and a tumor-initiating cell phenotype in colorectal cancer cell xenografts. Carcinogenesis 2013, 34, 2647–2654. [Google Scholar] [CrossRef] [Green Version]
- Mongroo, P.S.; Noubissi, F.K.; Cuatrecasas, M.; Kalabis, J.; King, C.E.; Johnstone, C.N.; Bowser, M.J.; Castells, A.; Spiegelman, V.S.; Rustgi, A.K. IMP-1 displays cross-talk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2. Cancer Res. 2011, 71, 2172–2182. [Google Scholar] [CrossRef] [Green Version]
- Stohr, N.; Huttelmaier, S. IGF2BP1: A post-transcriptional "driver" of tumor cell migration. Cell Adh. Migr. 2012, 6, 312–318. [Google Scholar] [CrossRef] [Green Version]
- Stohr, N.; Kohn, M.; Lederer, M.; Glass, M.; Reinke, C.; Singer, R.H.; Huttelmaier, S. IGF2BP1 promotes cell migration by regulating MK5 and PTEN signaling. Genes. Dev. 2012, 26, 176–189. [Google Scholar] [CrossRef] [Green Version]
- Fakhraldeen, S.A.; Clark, R.J.; Roopra, A.; Chin, E.N.; Huang, W.; Castorino, J.; Wisinski, K.B.; Kim, T.; Spiegelman, V.S.; Alexander, C.M. Two Isoforms of the RNA Binding Protein, Coding Region Determinant-binding Protein (CRD-BP/IGF2BP1), Are Expressed in Breast Epithelium and Support Clonogenic Growth of Breast Tumor Cells. J. Biol. Chem. 2015, 290, 13386–13400. [Google Scholar] [CrossRef] [Green Version]
- Jeng, Y.M.; Wang, T.H.; Lu, S.H.; Yuan, R.H.; Hsu, H.C. Prognostic significance of insulin-like growth factor II mRNA-binding protein 3 expression in gastric adenocarcinoma. Br. J. Surg. 2009, 96, 66–73. [Google Scholar] [CrossRef]
- Bellezza, G.; Cavaliere, A.; Sidoni, A. IMP3 expression in non-small cell lung cancer. Hum. Pathol. 2009, 40, 1205–1206. [Google Scholar] [CrossRef]
- Pryor, J.G.; Bourne, P.A.; Yang, Q.; Spaulding, B.O.; Scott, G.A.; Xu, H. IMP-3 is a novel progression marker in malignant melanoma. Mod. Pathol. 2008, 21, 431–437. [Google Scholar] [CrossRef] [Green Version]
- Vikesaa, J.; Hansen, T.V.; Jonson, L.; Borup, R.; Wewer, U.M.; Christiansen, J.; Nielsen, F.C. RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J. 2006, 25, 1456–1468. [Google Scholar] [CrossRef] [Green Version]
- Dai, N. The Diverse Functions of IMP2/IGF2BP2 in Metabolism. Trends Endocrinol. Metab. 2020, 31, 670–679. [Google Scholar] [CrossRef]
- Wang, J.; Chen, L.; Qiang, P. The role of IGF2BP2, an m6A reader gene, in human metabolic diseases and cancers. Cancer Cell Int. 2021, 21, 99. [Google Scholar] [CrossRef]
- Sachdev, D.; Yee, D. Disrupting insulin-like growth factor signaling as a potential cancer therapy. Mol. Cancer Ther. 2007, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Mu, Q.; Huang, H. The Roles of Insulin-Like Growth Factor 2 mRNA-Binding Protein 2 in Cancer and Cancer Stem Cells. Stem. Cells Int. 2018, 2018, 4217259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Loree, J.M.; Pereira, A.A.L.; Lam, M.; Willauer, A.N.; Raghav, K.; Dasari, A.; Morris, V.K.; Advani, S.; Menter, D.G.; Eng, C.; et al. Classifying Colorectal Cancer by Tumor Location Rather than Sidedness Highlights a Continuum in Mutation Profiles and Consensus Molecular Subtypes. Clin. Cancer Res. 2018, 24, 1062–1072. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zheng, Y.; Liu, H.; Li, T. Modulation of IGF2BP1 by long non-coding RNA HCG11 suppresses apoptosis of hepatocellular carcinoma cells via MAPK signaling transduction. Int. J. Oncol. 2017, 51, 791–800. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Wang, J.Z.; Chen, D.; He, Y.T.; Meng, N.; Chen, M.; Lu, R.X.; Chen, X.H.; Zhang, X.L.; Yan, G.R. An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat. Commun. 2020, 11, 1685. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, K.E.; Chatterji, P.; Lundsmith, E.T.; Andres, S.F.; Giroux, V.; Hicks, P.D.; Noubissi, F.K.; Spiegelman, V.S.; Rustgi, A.K. Loss of Stromal IMP1 Promotes a Tumorigenic Microenvironment in the Colon. Mol. Cancer Res. 2015, 13, 1478–1486. [Google Scholar] [CrossRef] [Green Version]
- Rasola, A.; Fassetta, M.; De Bacco, F.; D’Alessandro, L.; Gramaglia, D.; Di Renzo, M.F.; Comoglio, P.M. A positive feedback loop between hepatocyte growth factor receptor and beta-catenin sustains colorectal cancer cell invasive growth. Oncogene 2007, 26, 1078–1087. [Google Scholar] [CrossRef] [Green Version]
- Luraghi, P.; Reato, G.; Cipriano, E.; Sassi, F.; Orzan, F.; Bigatto, V.; De Bacco, F.; Menietti, E.; Han, M.; Rideout, W.M.; et al. MET signaling in colon cancer stem-like cells blunts the therapeutic response to EGFR inhibitors. Cancer Res 2014, 74, 1857–1869. [Google Scholar] [CrossRef] [Green Version]
- Goke, M.; Kanai, M.; Podolsky, D.K. Intestinal fibroblasts regulate intestinal epithelial cell proliferation via hepatocyte growth factor. Am. J. Physiol. 1998, 274, G809–G818. [Google Scholar] [CrossRef]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef]
- Qin, X.; Sun, L.; Wang, J. Restoration of microRNA-708 sensitizes ovarian cancer cells to cisplatin via IGF2BP1/Akt pathway. Cell Biol. Int. 2017, 41, 1110–1118. [Google Scholar] [CrossRef]
- Faye, M.D.; Beug, S.T.; Graber, T.E.; Earl, N.; Xiang, X.; Wild, B.; Langlois, S.; Michaud, J.; Cowan, K.N.; Korneluk, R.G.; et al. IGF2BP1 controls cell death and drug resistance in rhabdomyosarcomas by regulating translation of cIAP1. Oncogene 2015, 34, 1532–1541. [Google Scholar] [CrossRef]
- Kim, T.; Havighurst, T.; Kim, K.; Albertini, M.; Xu, Y.G.; Spiegelman, V.S. Targeting insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in metastatic melanoma to increase efficacy of BRAF(V600E) inhibitors. Mol. Carcinog. 2018, 57, 678–683. [Google Scholar] [CrossRef]
- Teng, H.W.; Yang, S.H.; Lin, J.K.; Chen, W.S.; Lin, T.C.; Jiang, J.K.; Yen, C.C.; Li, A.F.; Chen, P.C.; Lan, Y.T.; et al. CIP2A is a predictor of poor prognosis in colon cancer. J. Gastrointest. Surg. 2012, 16, 1037–1047. [Google Scholar] [CrossRef]
- Ho, H.L.; Chou, T.Y.; Yang, S.H.; Jiang, J.K.; Chen, W.S.; Chao, Y.; Teng, H.W. PD-L1 is a double-edged sword in colorectal cancer: The prognostic value of PD-L1 depends on the cell type expressing PD-L1. J. Cancer Res. Clin. Oncol. 2019, 145, 1785–1794. [Google Scholar] [CrossRef]
- Zheng, G.; Ma, Y.; Zou, Y.; Yin, A.; Li, W.; Dong, D. HCMDB: The human cancer metastasis database. Nucleic Acids Res. 2018, 46, D950–D955. [Google Scholar] [CrossRef] [Green Version]
- Fekete, J.T.; Gyorffy, B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3,104 breast cancer patients. Int. J. Cancer 2019, 145, 3140–3151. [Google Scholar] [CrossRef]
| Characteristics | Weak | Strong | p Value | |||
|---|---|---|---|---|---|---|
| n | (%) | n | (%) | |||
| Age (y/o) | <70 | 59 | 40.4% | 87 | 59.6% | 0.043 * |
| ≥70 | 57 | 53.3% | 50 | 46.7% | ||
| Gender | Female | 37 | 41.6% | 52 | 58.4% | 0.315 |
| Male | 79 | 48.2% | 85 | 51.8% | ||
| Pathology | Adenocarcinoma | 102 | 45.5% | 122 | 54.5% | 0.553 |
| Carcinoma | 0 | 0.0% | 1 | 100.0% | ||
| Mucinous adenocarcinoma | 14 | 51.9% | 13 | 48.1% | ||
| Signet ring cell | 0 | 0.0% | 1 | 100.0% | ||
| Location | Left | 49 | 42.6% | 66 | 57.4% | 0.345 |
| Right | 67 | 48.6% | 71 | 51.4% | ||
| Stage (AJCC VII) | I/II | 51 | 60.0% | 34 | 40.0% | 0.001 * |
| III/IV | 65 | 38.7% | 103 | 61.3% | ||
| Grade | Low | 106 | 45.9% | 125 | 54.1% | 0.969 |
| High | 10 | 45.5% | 12 | 54.5% | ||
| Mucinous component | No | 70 | 44.0% | 89 | 56.0% | 0.449 |
| Yes | 46 | 48.9% | 48 | 51.1% | ||
| LVSI | No | 93 | 47.4% | 103 | 52.6% | 0.344 |
| Yes | 23 | 40.4% | 34 | 59.6% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.-M.; Lin, C.-C.; Chen, W.-S.; Jiang, J.-K.; Yang, S.-H.; Chang, S.-C.; Ho, C.-L.; Yang, C.-C.; Huang, S.-C.; Chao, Y.; et al. Insulin-Like Growth Factor 2 mRNA-Binding Protein 1 (IGF2BP1) Is a Prognostic Biomarker and Associated with Chemotherapy Responsiveness in Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 6940. https://doi.org/10.3390/ijms22136940
Chen H-M, Lin C-C, Chen W-S, Jiang J-K, Yang S-H, Chang S-C, Ho C-L, Yang C-C, Huang S-C, Chao Y, et al. Insulin-Like Growth Factor 2 mRNA-Binding Protein 1 (IGF2BP1) Is a Prognostic Biomarker and Associated with Chemotherapy Responsiveness in Colorectal Cancer. International Journal of Molecular Sciences. 2021; 22(13):6940. https://doi.org/10.3390/ijms22136940
Chicago/Turabian StyleChen, Hung-Ming, Chun-Chi Lin, Wei-Shone Chen, Jeng-Kai Jiang, Shung-Haur Yang, Shih-Ching Chang, Ching-Liang Ho, Chung-Chi Yang, Shih-Ching Huang, Yee Chao, and et al. 2021. "Insulin-Like Growth Factor 2 mRNA-Binding Protein 1 (IGF2BP1) Is a Prognostic Biomarker and Associated with Chemotherapy Responsiveness in Colorectal Cancer" International Journal of Molecular Sciences 22, no. 13: 6940. https://doi.org/10.3390/ijms22136940
APA StyleChen, H.-M., Lin, C.-C., Chen, W.-S., Jiang, J.-K., Yang, S.-H., Chang, S.-C., Ho, C.-L., Yang, C.-C., Huang, S.-C., Chao, Y., Liao, T.-T., Hwang, W.-L., & Teng, H.-W. (2021). Insulin-Like Growth Factor 2 mRNA-Binding Protein 1 (IGF2BP1) Is a Prognostic Biomarker and Associated with Chemotherapy Responsiveness in Colorectal Cancer. International Journal of Molecular Sciences, 22(13), 6940. https://doi.org/10.3390/ijms22136940








