Microbial Conjugation Studies of Licochalcones and Xanthohumol
Abstract
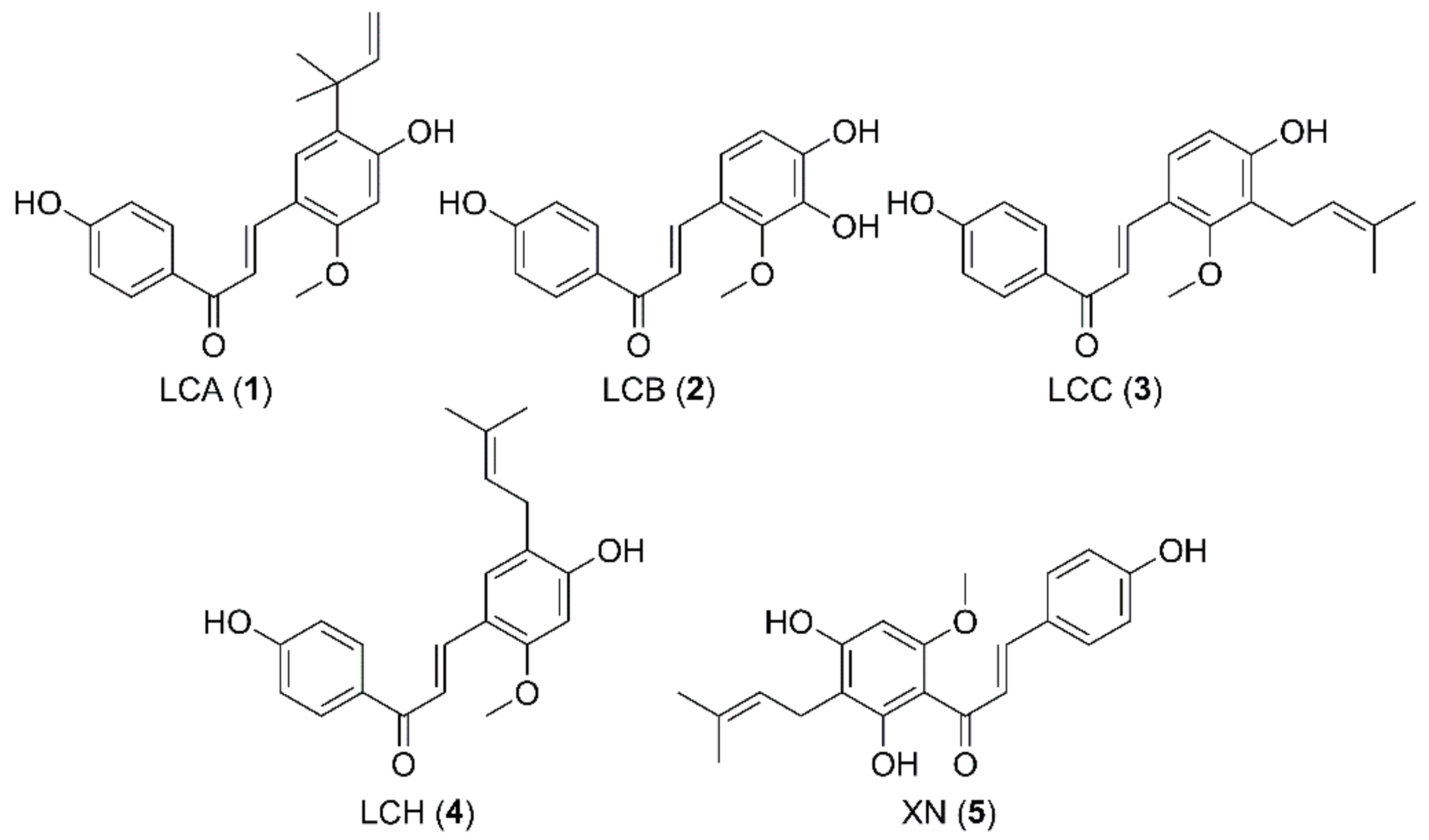
1. Introduction
2. Results and Discussion
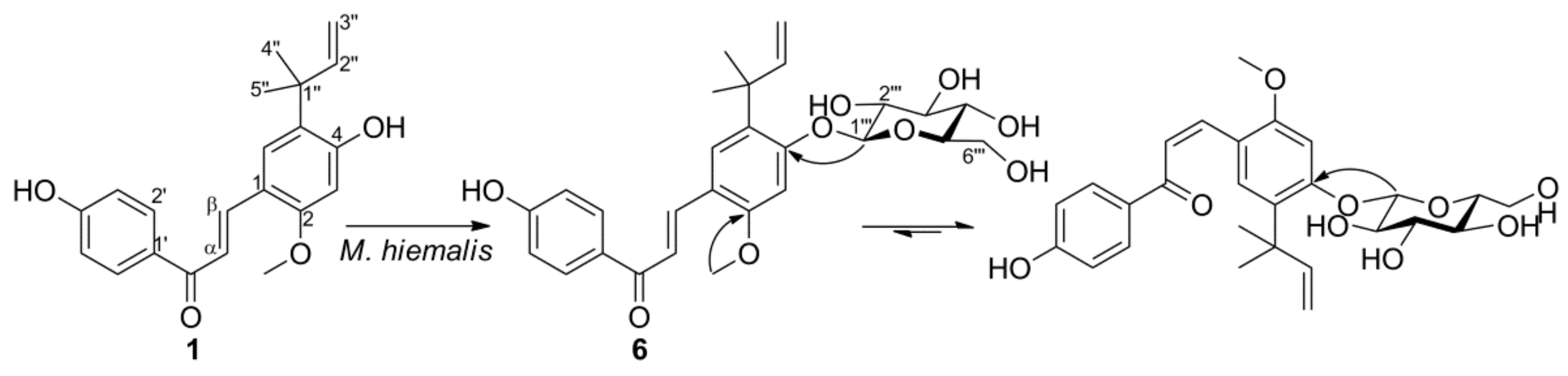
2.1. Microbial Transformation of Licochalcone A and Xanthohumol by M. hiemalis
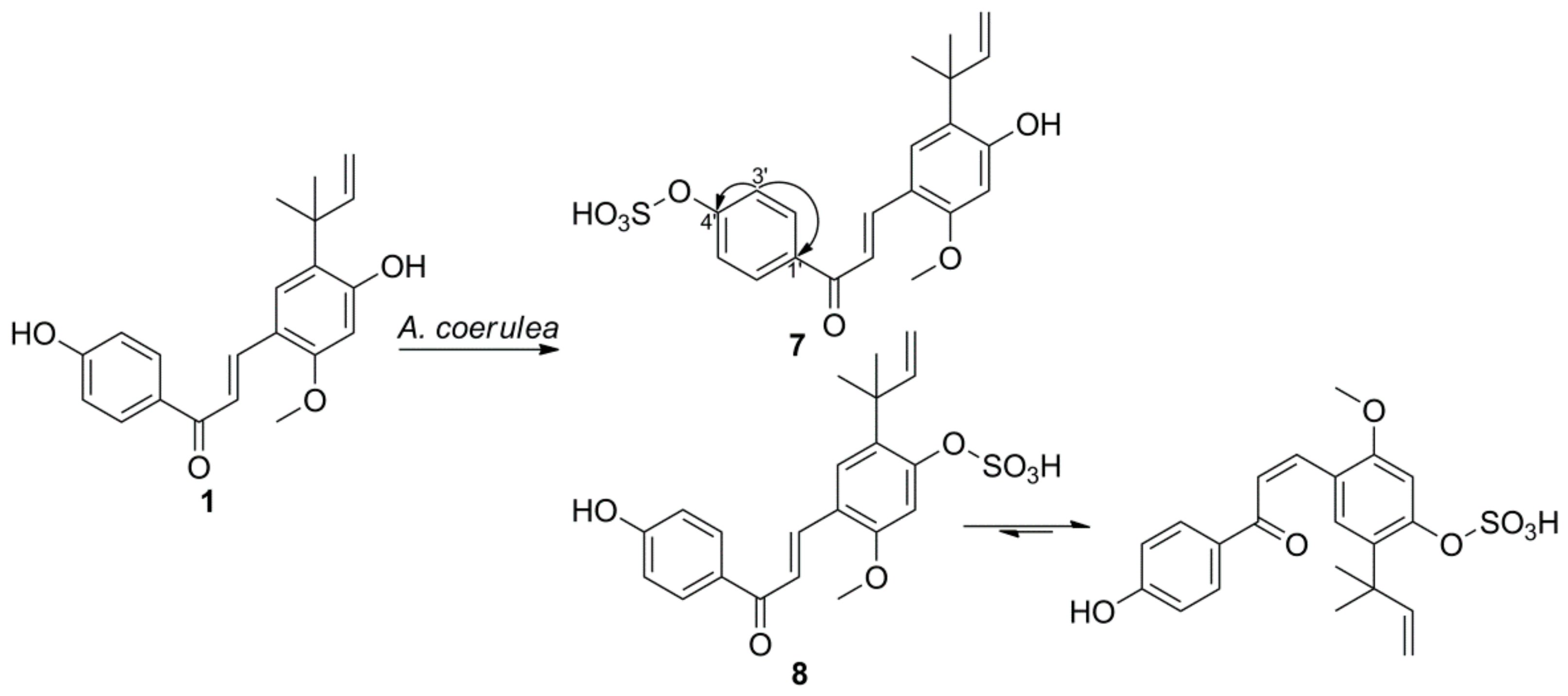
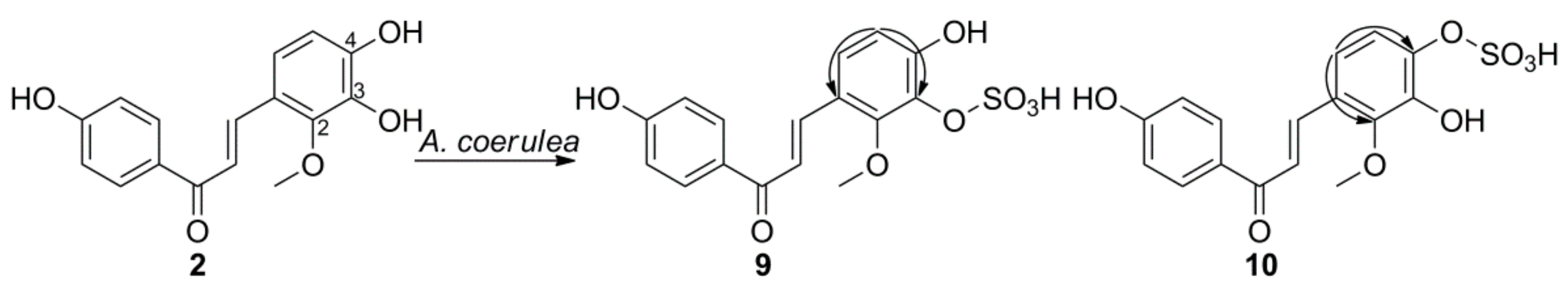
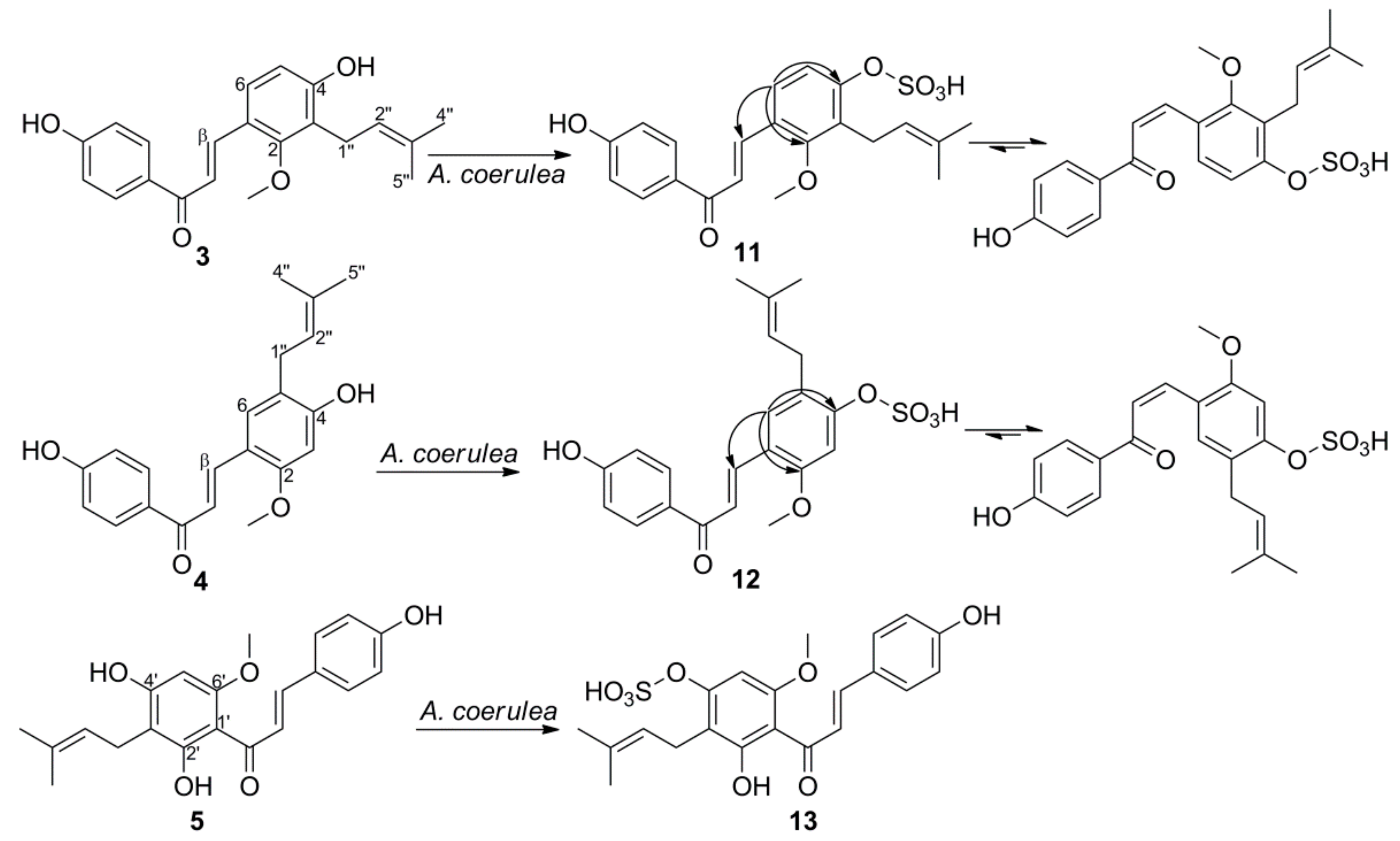
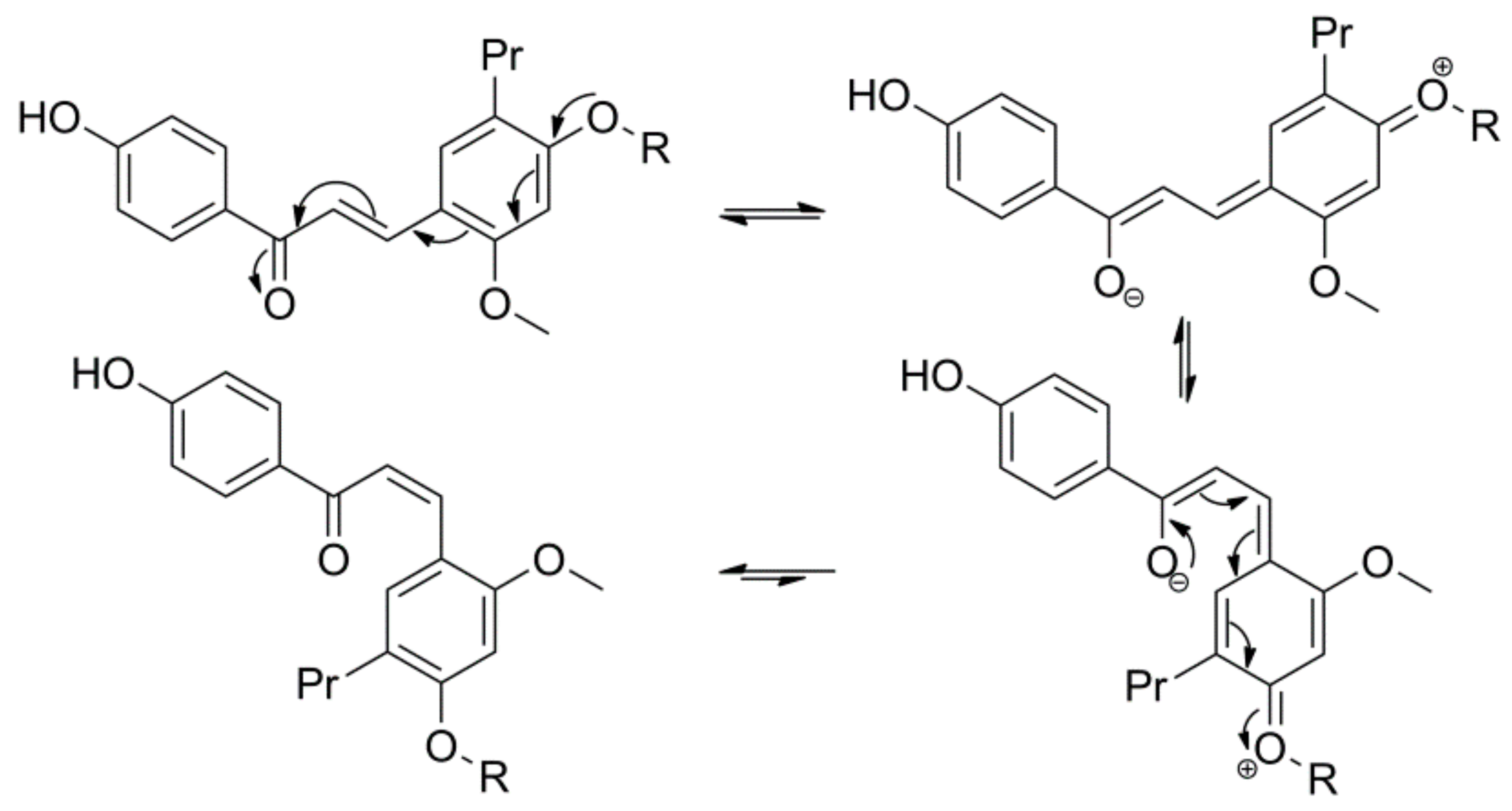
2.2. Microbial Transformation of Licochalcones A, B, C, H, and Xanthohumol by A. coerulea
3. Materials and Methods
3.1. Materials and Microorganisms
3.2. General Experimental Procedures
3.3. Screening Procedures
3.4. Biotransformation of 1 by M. hiemalis KCTC 26779
3.5. Biotransformation of 1–5 by A. coerulea KCTC 6936
3.5.1. Licochalcone A 4′-Sulfate (7)
3.5.2. Licochalcone A 4-Sulfate (8)
3.5.3. Licochalcone B 3-Sulfate (9)
3.5.4. Licochalcone B 4-Sulfate (10)
3.5.5. Licochalcone C 4-Sulfate (11)
3.5.6. Licochalcone H 4-Sulfate (12)
3.5.7. Xanthohumol 4′-Sulfate (13)
3.6. Cytotoxic Activity Evaluation
3.7. Hydrolysis of Compounds 6–13
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maria Pia, G.D.; Sara, F.; Mario, F.; Lorenza, S. Biological effects of licochalcones. Mini Rev. Med. Chem. 2019, 19, 647–656. [Google Scholar] [CrossRef]
- Wang, C.; Chen, L.; Xu, C.; Shi, J.; Chen, S.; Tan, M.; Chen, J.; Zou, L.; Chen, C.; Liu, Z.; et al. A comprehensive review for phytochemical, pharmacological, and biosynthesis studies on Glycyrrhiza spp. Am. J. Chin. Med. 2020, 48, 1–29. [Google Scholar] [CrossRef]
- Weng, Q.; Chen, L.; Ye, L.; Lu, X.; Yu, Z.; Wen, C.; Chen, Y.; Huang, G. Determination of licochalcone A in rat plasma by UPLC-MS/MS and its pharmacokinetics. Acta Chromatogr. 2019, 31, 262–265. [Google Scholar] [CrossRef]
- Nadelmann, L.; Tjørnelund, J.; Hansen, S.H.; Cornett, C.; Sidelmann, U.G.; Braumann, U.; Christensen, E.; Christensen, S.B. Synthesis, isolation and identification of glucuronides and mercapturic acids of a novel antiparasitic agent, licochalcone A. Xenobiotica 1997, 27, 667–680. [Google Scholar] [CrossRef]
- Huang, L.; Nikolic, D.; van Breemen, R.B. Hepatic metabolism of licochalcone A, a potential chemopreventive chalcone from licorice (Glycyrrhiza inflata), determined using liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2017, 409, 6937–6948. [Google Scholar] [CrossRef]
- Nadelmann, L.; Tjørnelund, J.; Christensen, E.; Hansen, S.H. High-performance liquid chromatographic determination of licochalcone A and its metabolites in biological fluids. J. Chromatogr. B Biomed. Sci. Appl. 1997, 695, 389–400. [Google Scholar] [CrossRef]
- Yilmazer, M.; Stevens, J.F.; Buhler, D.R. In vitro glucuronidation of xanthohumol, a flavonoid in hop and beer, by rat and human liver microsomes. FEBS Lett. 2001, 491, 252–256. [Google Scholar] [CrossRef]
- Yilmazer, M.; Stevens, J.F.; Deinzer, M.L.; Buhler, D.R. In vitro biotransformation of xanthohumol, a flavonoid from hops (Humulus lupulus), by rat liver microsomes. Drug Metab. Dispos. 2001, 29, 223–231. [Google Scholar] [PubMed]
- Nikolic, D.; Li, Y.; Chadwick, L.R.; Pauli, G.F.; van Breemen, R.B. Metabolism of xanthohumol and isoxanthohumol, prenylated flavonoids from hops (Humulus lupulus L.) by human liver microsomes. J. Mass Spectrom. 2005, 40, 289–299. [Google Scholar] [CrossRef]
- Ruefer, C.E.; Gerhäuser, C.; Frank, N.; Becker, H.; Kulling, S.E. In vitro phase II metabolism of xanthohumol by human UDP-glucuronosyltransferases and sulfotransferases. Mol. Nutr. Food Res. 2005, 49, 851–856. [Google Scholar] [CrossRef]
- Jirásko, R.; Holčapek, M.; Vrublová, E.; Ulrichová, J.; Šimánek, V. Identification of new phase II metabolites of xanthohumol in rat in vivo biotransformation of hop extracts using high-performance liquid chromatography electrospray ionization tandem mass spectrometry. J. Chromatogr. A 2010, 1217, 4100–4108. [Google Scholar] [CrossRef] [PubMed]
- Böhmdorfer, M.; Szakmary, A.; Schiestl, R.H.; Vaquero, J.; Riha, J.; Brenner, S.; Thalhammer, T.; Szekeres, T.; Jäger, W. Involvement of UDP-glucuronosyltransferases and sulfotransferases in the excretion and tissue distribution of resveratrol in mice. Nutrients 2017, 9, 1347. [Google Scholar] [CrossRef] [PubMed]
- Marhol, P.; Hartog, A.F.; van der Horst, M.A.; Wever, R.; Purchartová, K.; Fuksová, K.; Kuzma, M.; Cvačka, J.; Křen, V. Preparation of silybin and isosilybin sulfates by sulfotransferase from Desulfitobacterium hafniense. J. Mol. Catal. B. Enzym. 2013, 89, 24–27. [Google Scholar] [CrossRef]
- Sak, K. Chapter 6-Anticancer action of sulfated flavonoids as phase II metabolites. In Handbook of Food Bioengineering, Food Bioconversion; Grumezescu, A.M., Holban, A.M., Eds.; Academic Press: Cambridge, MA, USA; Elsevier Ltd.: Amsterdam, The Netherlands, 2017; Volume 2, pp. 207–236. [Google Scholar]
- Koizumi, M.; Shimizu, M.; Kobashi, K. Enzymatic sulfation of quercetin by arylsulfotransferase from a human intestinal bacterium. Chem. Pharm. Bull. 1990, 38, 794–796. [Google Scholar] [CrossRef] [PubMed]
- Correia-da-Silva, M.; Sousa, E.; Pinto, M.M.M. Emerging sulfated flavonoids and other polyphenols as drugs: Nature as an inspiration. Med. Res. Rev. 2014, 34, 223–279. [Google Scholar] [CrossRef] [PubMed]
- Roubalová, L.; Purchartová, K.; Papoušková, K.; Vacek, J.; Křen, V.; Ulrichová, J.; Vrba, J. Sulfation modulates the cell uptake, antiradical activity and biological effects of flavonoids in vitro: An examination of quercetin, isoquercitrin and taxifolin. Bioorg. Med. Chem. 2015, 23, 5402–5409. [Google Scholar] [CrossRef]
- Teles, Y.C.F.; Souza, M.S.R.; de Souza, M.F.V. Sulphated flavonoids: Biosynthesis, structures, and biological activities. Molecules 2018, 23, 480. [Google Scholar] [CrossRef]
- Arunrattiyakorn, P.; Suksamrarn, S.; Suwannasai, N.; Kanzaki, H. Microbial metabolism of α-mangostin isolated from Garcinia mangostana L. Phytochemistry 2011, 72, 730–734. [Google Scholar] [CrossRef]
- Kim, H.J.; Yim, S.-H.; Han, F.; Kang, B.Y.; Choi, H.J.; Jung, D.-W.; Williams, D.R.; Gustafson, K.R.; Kennelly, E.J.; Lee, I.-S. Biotransformed metabolites of the hop prenylflavanone isoxanthohumol. Molecules 2019, 24, 394. [Google Scholar] [CrossRef]
- Bartmańska, A.; Tronina, T.; Huszcza, E. Microbial sulfation of 8-prenylnaringenin. Z. Naturforsch 2013, 68c, 231–235. [Google Scholar] [CrossRef]
- Herath, W.; Mikell, J.R.; Hale, A.L.; Ferreira, D.; Khan, I.A. Microbial metabolism part 9. Structure and antioxidant significance of the metabolites of 5,7-dihydroxyflavone (chrysin), and 5- and 6-hydroxyflavones. Chem. Pharm. Bull. 2008, 56, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Mikell, J.R.; Khan, I.A. Bioconversion of 7-hydroxyflavanone: Isolation, characterization and bioactivity evaluation of twenty-one phase I and phase II microbial metabolites. Chem. Pharm. Bull. 2012, 60, 1139–1145. [Google Scholar] [CrossRef]
- Ibrahim, A.K.; Radwan, M.M.; Ahmed, S.A.; Slade, D.; Ross, S.A.; ElSohly, M.A.; Khan, I.A. Microbial metabolism of cannflavin A and B isolated from Cannabis sativa. Phytochemistry 2010, 71, 1014–1019. [Google Scholar] [CrossRef]
- Mikell, J.R.; Herath, W.; Khan, I.A. Microbial metabolism. Part 12. Isolation, characterization and bioactivity evaluation of eighteen microbial metabolites of 4′-hydroxyflavanone. Chem. Pharm. Bull. 2011, 59, 692–697. [Google Scholar] [CrossRef][Green Version]
- Herath, W.; Khan, I.A. Microbial metabolism. Part 13. Metabolites of hesperetin. Bioorg. Med. Chem. Lett. 2011, 21, 5784–5786. [Google Scholar] [CrossRef]
- Xiao, Y.; Han, F.; Lee, I.-S. Microbial transformation of licochalcones. Molecules 2020, 25, 60. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, I.-S. Microbial metabolism of the prenylated chalcone xanthohumol. J. Nat. Prod. 2006, 69, 1522–1524. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhang, P.; Cui, Y.; Li, K.; Qiao, X.; Zhang, Y.T.; Li, S.M.; Cox, R.J.; Wu, B.; Ye, M.; et al. Regio- and stereospecific O-glycosyation of phenolic compounds catalyzed by a fungal glycosyltransferases from Mucor hiemalis. Adv. Synth. Catal. 2017, 359, 995–1006. [Google Scholar] [CrossRef]
- Han, F.; Lee, I.-S. A new flavonol glycoside from the aerial parts of Epimedium koreanum Nakai. Nat. Prod. Res. 2017, 33, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.-R.S. Sulfation of naringenin by Cunninghamella elegans. Phytochemistry 2000, 53, 209–212. [Google Scholar] [CrossRef]
- Ibrahim, A.-R.S.; Galal, A.M.; Ahmed, M.S.; Mossa, G.S. O-Demethylation and sulfation of 7-methoxylated flavonones by Cunninghamella elegans. Chem. Pharm. Bull. 2003, 51, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Kajiyama, K.; Demizu, S.; Hiraga, Y.; Kinoshita, K.; Koyama, K.; Takahashi, K.; Tamura, Y.; Okada, K.; Kinoshita, T. Two prenylated retrochalcones from Glycyrrhiza inflata. Phytochemistry 1992, 31, 3229–3232. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, Y.; Paudel, S.; Yoon, G.; Cheon, S.H. Concise synthesis of licochalcone C and its regioisomer, licochalcone H. Arch. Pharm. Res. 2013, 36, 4132–4136. [Google Scholar] [CrossRef]
- Chen, Q.-H.; Fu, M.-L.; Chen, M.-M.; Liu, J.; Liu, X.-J.; He, G.-Q.; Pu, S.-C. Preparative isolation and purification of xanthohumol from hops (Humulus lupulus L.) by high-speed counter-current chromatography. Food Chem. 2012, 132, 619–623. [Google Scholar] [CrossRef]
- Basílio, N.; Bittar, S.A.; Mora, N.; Dangles, O.; Pina, F. Analogs of natural 3-deoxyanthocyanins: O-glucosides of the 4′,7-dihydroxyflavylium ion and the deep influence of glycosidation on color. Int. J. Mol. Sci. 2016, 17, 1751. [Google Scholar] [CrossRef] [PubMed]
- Simmler, C.; Lankin, D.C.; Nikolić, D.; van Breemen, R.B.; Pauli, G.F. Isolation and structural characterization of dihydrobenzofuran congers of licochalcone A. Fitoterapia 2017, 121, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Correia, M.S.P.; Lin, W.; Aria, A.J.; Jain, A.; Globisch, D. Rapid preparation of a large sulfated metabolite library for structure validation in human samples. Metabolites 2020, 10, 415. [Google Scholar] [CrossRef]
- Van der Horst, M.A.; Hartog, A.F.; El Morabet, R.; Marais, A.; Kircz, M.; Wever, R. Enzymatic sulfation of phenolic hydroxy groups of various plant metabolites by an arylsulfotransferase. Eur. J. Org. Chem. 2015, 2015, 534–541. [Google Scholar] [CrossRef]
- Bartmańska, A.; Tronina, T.; Huszcza, E. Transformation of 8-prenylnaringenin by Absidia coerulea and Beauveria bassiana. Bioorg. Med. Chem. Lett. 2012, 22, 6451–6453. [Google Scholar] [CrossRef]
- Fan, N.; Du, C.H.; Xu, J.Q.; Xu, Y.X.; Yu, B.Y.; Zhang, J. Glycosylation and sulfation of 4-methylumbelliferone by Gliocladium deliquescens NRRL 1086. Appl. Biochem. Microbiol. 2017, 53, 85–93. [Google Scholar] [CrossRef]
- Feng, J.; Liang, W.; Ji, S.; Qiao, X.; Zhang, Y.; Yu, S.; Ye, M. Microbial transformation of isoangustone A by Mucor hiemalis CGMCC 3.14114. J. Chin. Pharm. Sci. 2015, 24, 285–291. [Google Scholar] [CrossRef]
- Ruan, F.; Chen, R.; Li, J.; Zhang, M.; Xie, K.; Wang, Y.; Feng, R.; Dai, J. Sulfation of naringenin by Mucor sp. Zhongguo Zhongyao Zazhi 2014, 39, 2039–2042. [Google Scholar]
- Yuan, W.; Zhang, L.-P.; Cheng, K.-D.; Zhu, P.; Wang, Q.; He, H.-X.; Zhu, H.-X. Microbial O-demethylation, hydroxylation, sulfation, and ribosylation of a xanthone derivative from Halenia elliptica. J. Nat. Prod. 2006, 69, 811–814. [Google Scholar] [CrossRef]
- Xie, L.; Xiao, D.; Wang, X.; Wang, C.; Bai, J.; Yue, Q.; Yue, H.; Li, Y.; Molnár, I.; Xu, Y.; et al. Combinatorial biosynthesis of sulfated benzenediol lactones with a phenolic sulfotransferase from Fusarium graminearum PH-1. mSphere 2020, 5, e00949-20. [Google Scholar] [CrossRef]
- Wang, P.; Yuan, X.; Wang, Y.; Zhao, H.; Sun, X.; Zheng, Q. Licochalcone C induces apoptosis via B-cell lymphoma 2 family proteins in T24 cells. Mol. Med. Rep. 2015, 12, 7623–7628. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Yan, X.; Si, L.; Cao, C.; Yu, L.; Zheng, Q. Anti-proliferation effects of licochalcone B on human breast cancer MCF-7 cells. Zhongguo Shiyan Fangjixue Zazhi 2016, 22, 106–111. [Google Scholar]
- Kang, T.H.; Yoon, G.; Kang, I.-A.; Oh, H.-N.; Chae, J.-I.; Shim, J.-H. Natural compound licochalcone B induced extrinsic and intrinsic apoptosis in human skin melanoma (A375) and Squamous cell carcinoma (A431) cells. Phytother. Res. 2017, 31, 1858–1867. [Google Scholar] [CrossRef]
- Cho, J.J.; Chae, J.-I.; Yoon, G.; Ki, M.K.H.; Cho, J.H.; Cho, S.-S.; Cho, Y.S.; Shim, J.-H. Licochalcone A, a natural chalconoid isolated from Glycyrrhiza inflata root, induces apoptosis via Sp1 and Sp1 regulatory proteins in oral squamous cell carcinoma. Int. J. Oncol. 2014, 45, 667–674. [Google Scholar] [CrossRef]
- Oh, H.-N.; Yoon, G.; Shin, J.-C.; Park, S.-M.; Cho, S.-S.; Cho, J.H.; Lee, M.-H.; Liu, K.D.; Cho, Y.S.; Chae, J.-I.; et al. Licochalcone B induces apoptosis of human oral squamous cell carcinoma through the extrinsic- and intrinsic-signaling pathways. Int. J. Oncol. 2016, 48, 1749–1757. [Google Scholar] [CrossRef]
- Oh, H.-N.; Seo, J.-H.; Lee, M.-H.; Kim, C.; Kim, E.; Yoon, G.; Cho, S.-S.; Cho, Y.S.; Choi, H.W.; Shim, J.-H.; et al. Licochalcone C induces apoptosis in human oral squamous cell carcinoma cells by regulation of the JAK2/STAT3 signaling pathway. J. Cell. Biochem. 2018, 119, 10118–10130. [Google Scholar] [CrossRef]
- Oh, H.-N.; Oh, K.B.; Lee, M.-H.; Seo, J.-H.; Kim, E.; Yoon, G.; Cho, S.-S.; Cho, Y.S.; Choi, H.W.; Chae, J.-I.; et al. JAK2 regulation by licochalcone H inhibits the cell growth and induces apoptosis in human oral squamous cell carcinoma. Phytomedicine 2019, 52, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Szliszka, E.; Czuba, Z.P.; Mazur, B.; Sedek, L.; Paradysz, A.; Krol, W. Chalcones enhance TRAIL-induced apoptosis in prostate cancer cells. Int. J. Mol. Sci. 2010, 11, 1–13. [Google Scholar] [CrossRef]
| Compound | Cell Lines (IC50, μM) | ||
|---|---|---|---|
| A375P | MCF-7 | A549 | |
| 1 | 12.86 ± 3.42 | 19.16 ± 0.65 | 18.14 ± 1.26 |
| 5 | 11.02 ± 2.09 | 23.11 ± 1.36 | 26.93 ± 1.56 |
| 7 | 27.35 ± 1.44 | 30.85 ± 4.58 | 43.07 ± 1.63 |
| 13 | 41.04 ± 1.61 | 81.27 ± 3.05 | 99.40 ± 1.99 |
| DZ 1 | 9.86 ± 0.57 | 5.12 ± 0.44 | 4.01 ± 0.78 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, F.; Xiao, Y.; Lee, I.-S. Microbial Conjugation Studies of Licochalcones and Xanthohumol. Int. J. Mol. Sci. 2021, 22, 6893. https://doi.org/10.3390/ijms22136893
Han F, Xiao Y, Lee I-S. Microbial Conjugation Studies of Licochalcones and Xanthohumol. International Journal of Molecular Sciences. 2021; 22(13):6893. https://doi.org/10.3390/ijms22136893
Chicago/Turabian StyleHan, Fubo, Yina Xiao, and Ik-Soo Lee. 2021. "Microbial Conjugation Studies of Licochalcones and Xanthohumol" International Journal of Molecular Sciences 22, no. 13: 6893. https://doi.org/10.3390/ijms22136893
APA StyleHan, F., Xiao, Y., & Lee, I.-S. (2021). Microbial Conjugation Studies of Licochalcones and Xanthohumol. International Journal of Molecular Sciences, 22(13), 6893. https://doi.org/10.3390/ijms22136893






