Metabolomic Reprogramming of C57BL/6-Macrophages during Early Infection with L. amazonensis
Abstract
:1. Introduction
2. Results
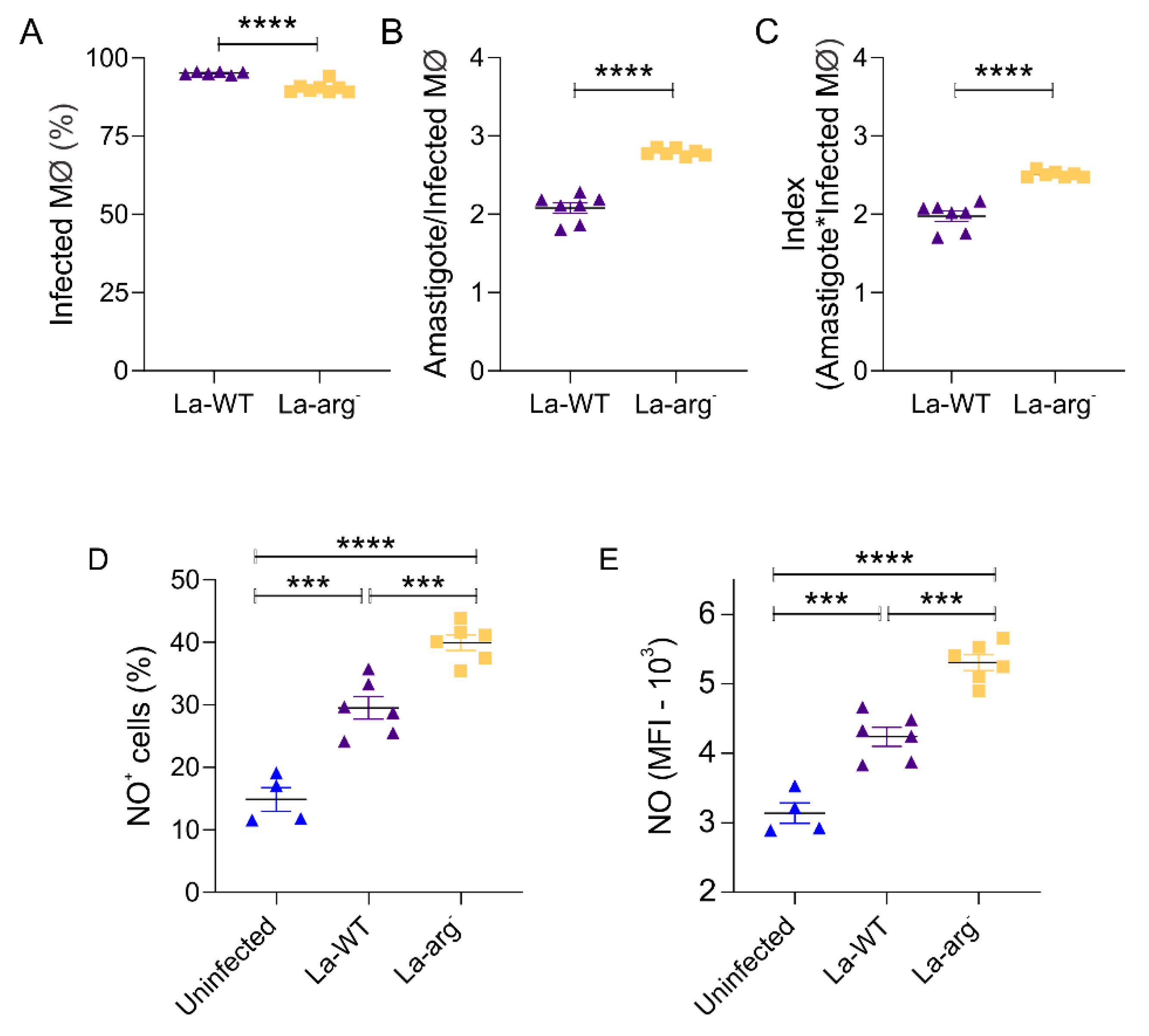
2.1. The Parasite Arginase Modulates NO Production and Infectivity in Leishmania-Infected Macrophages
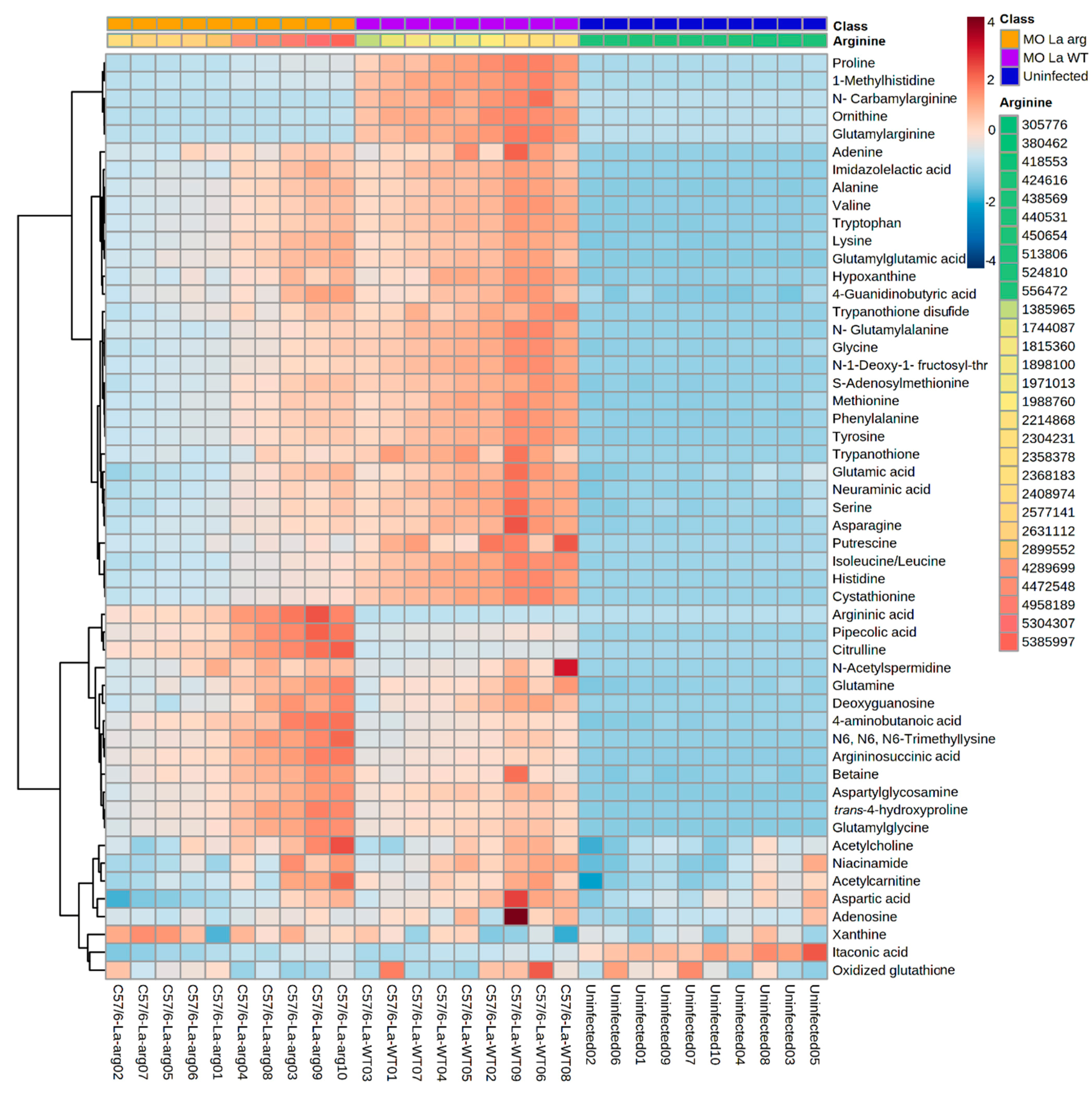
2.2. Statistical Analyses of the Metabolic Profile of Leishmania Infected C57BL/6-Macrophages
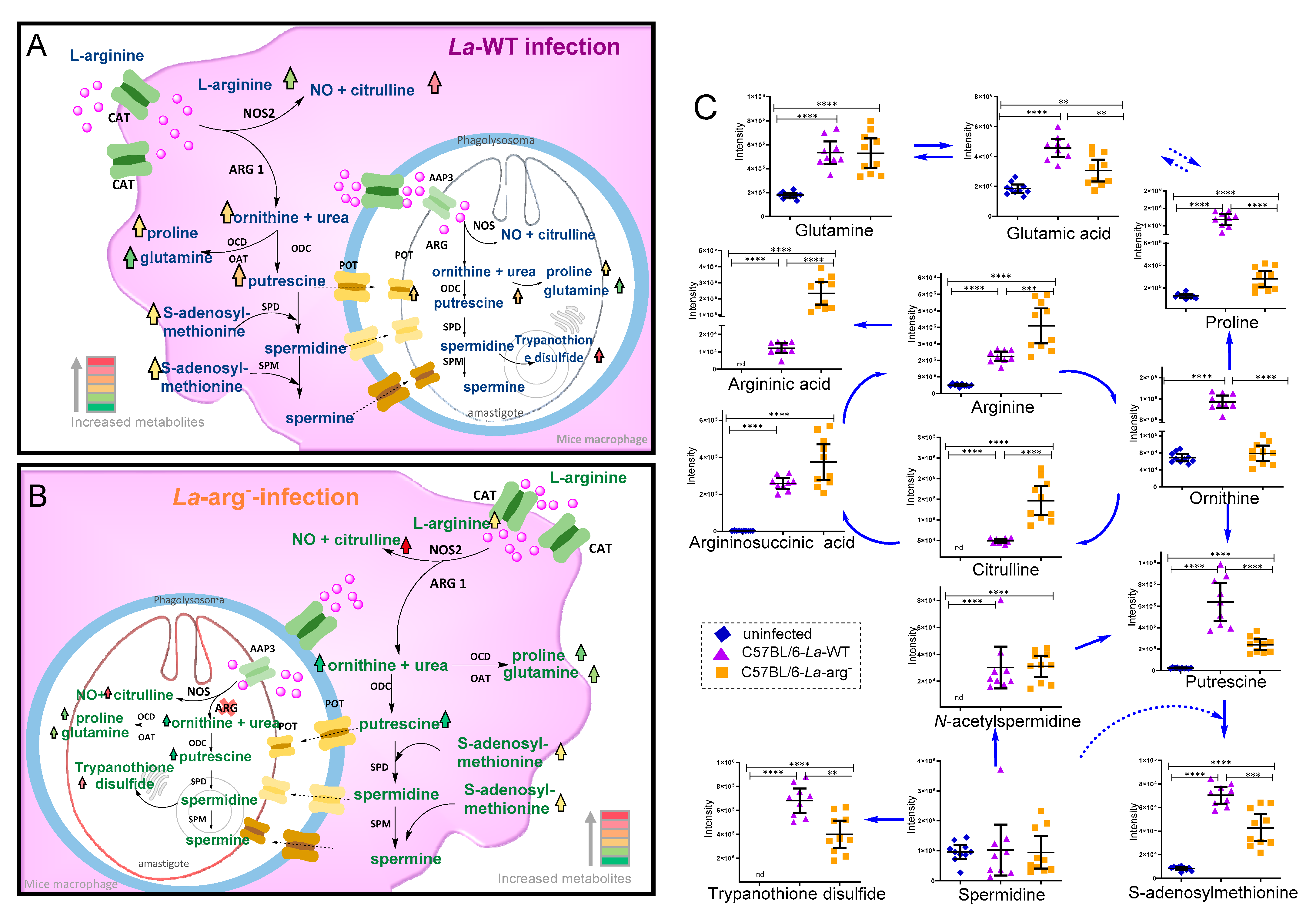
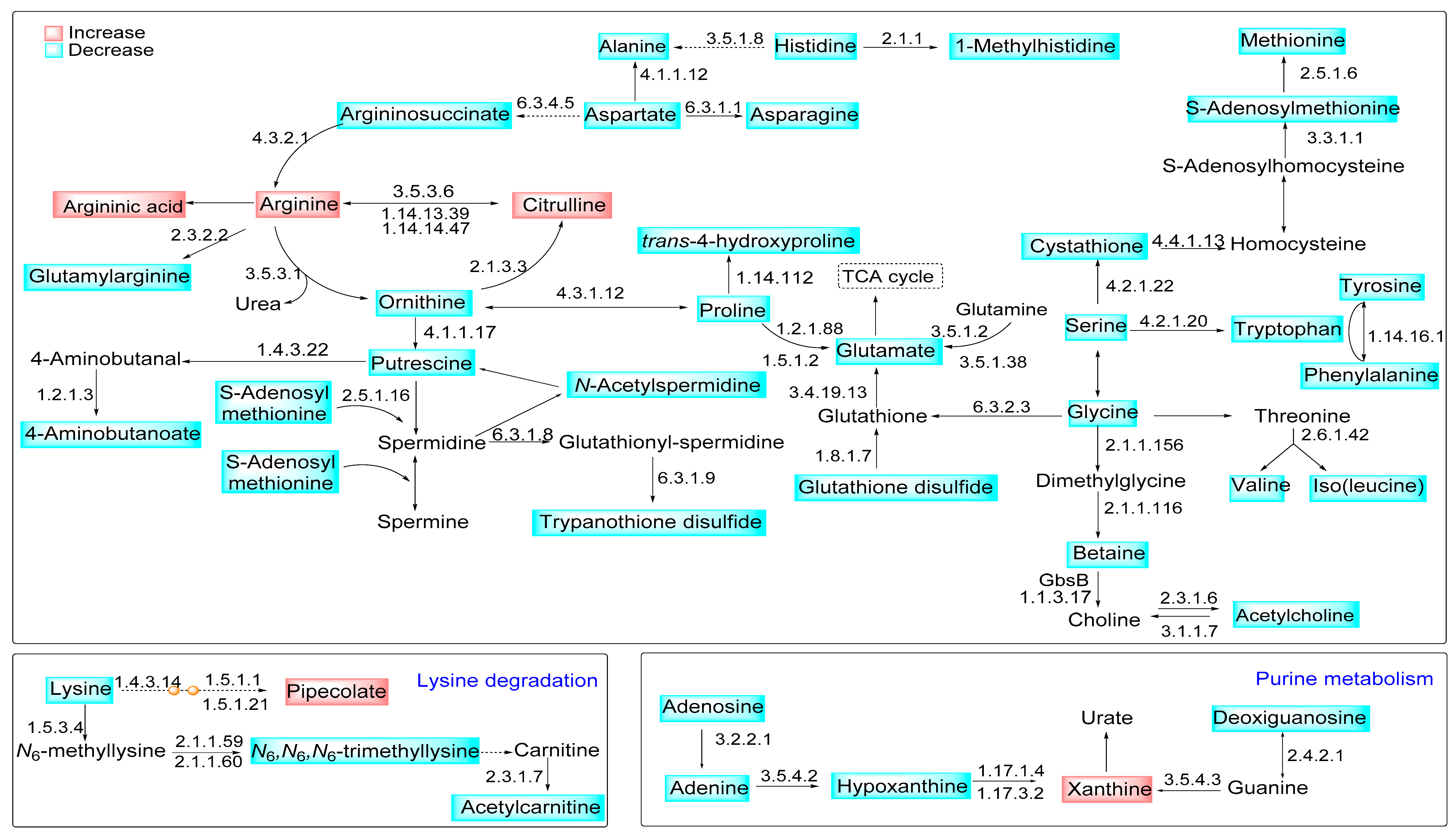
2.3. Parasite Arginase Activity Leads to Differential Metabolites Profiles of C57BL/6-Macrophages Infected with L. amazonensis
3. Discussion
4. Materials and Methods
4.1. In Vitro Macrophage Infections
4.2. Infectivity Quantification
4.3. NO Quantification
4.4. CE-MS Metabolic Fingerprinting
4.5. CE-MS Data Treatment
4.6. Statistical Analysis
4.7. Pathway Enrichment Analysis
4.8. Ethics Statement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Martinez, F.O.; Sica, A.; Mantovani, A.; Locati, M. Macrophage activation and polarization. Front. Biosci. 2008, 13, 453–461. [Google Scholar] [CrossRef] [Green Version]
- Vogel, D.Y.S.; Glim, J.E.; Stavenuiter, A.W.D.; Breur, M.; Heijnen, P.; Amor, S.; Dijkstra, C.D.; Beelen, R.H.J. Human macrophage polarization in vitro: Maturation and activation methods compared. Immunobiology 2014, 219, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Filardy, A.A.; Pires, D.R.; Nunes, M.P.; Takiya, C.M.; Freire-de-Lima, C.G.; Ribeiro-Gomes, F.L.; DosReis, G.A. Proinflammatory clearance of apoptotic neutrophils induces an IL-12 low IL-10 high regulatory phenotype in macrophages. J. Immunol. 2010, 185, 2044–2050. [Google Scholar] [CrossRef] [Green Version]
- Boucher, J.L.; Moali, C.; Tenu, J.P. Nitric oxide biosynthesis, nitric oxide synthase inhibitors and arginase competition for L-arginine utilization. Cell. Mol. Life Sci. 1999, 55, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Hrabak, A.; Bajor, T.; Temesi, A.; Meszaros, G. The inhibitory effect of nitrite, a stable product of nitric oxide (NO) formation, on arginase. FEBS Lett. 1996, 390, 203–206. [Google Scholar] [CrossRef] [Green Version]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization Trends Immunol. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Van den Bossche, J.; Baardman, J.; de Winther, M.P.J. Metabolic characterization of polarized M1 and M2 bone marrow-derived macrophages using real-time extracellular flux analysis. J. Vis. Exp. 2015, 28, e53424. [Google Scholar] [CrossRef] [Green Version]
- Covarrubias, A.J.; Aksoylar, H.I.; Horng, T. Control of macrophage metabolism and activation by mTOR and Akt signaling. Semin. Immunol. 2015, 27, 286–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vergadi, E.; Ieronymaki, E.; Lyroni, K.; Vaporidi, K.; Tsatsanis, C. Akt signaling pathway in macrophage activation and M1/M2 polarization. J. Immunol. 2017, 198, 1006–1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rafferty, S.; Malech, H.L. High reductase activity of recombinant NOS2 flavoprotein domain lacking the calmodulin binding regulatory sequence. Biochem. Biophys. Res. Commun. 1996, 220, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Huang, S.C.C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B.; et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015, 42, 419–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blagih, J.; Jones, R.G. Polarizing macrophages through reprogramming of glucose metabolism Cell. Metab. 2012, 15, 793–795. [Google Scholar] [CrossRef] [Green Version]
- Koo, S.j.; Garg, N.J. Metabolic programming of macrophage functions and pathogens control. Redox Biol. 2019, 24, 101198. [Google Scholar] [CrossRef]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Kishton, R.J.; Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016, 16, 553–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Liang, H.; Zen, K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol. 2014, 15, 614. [Google Scholar] [CrossRef] [Green Version]
- Whyte, C.S.; Ruckerl, D.; Bishop, E.; Barker, R.N.; Rees, A.; Allen, J.; Wilson, H.M. Regulation of alternative (M2) macrophage activation by Suppressor of Cytokine Signalling (SOCS) 1. Immunology 2010, 131, 86. [Google Scholar]
- Moore, K.J.; Sheedy, F.J.; Fisher, E.A. Macrophages in atherosclerosis: A dynamic balance. Nat. Rev. Immunol. 2013, 13, 709–721. [Google Scholar] [CrossRef]
- Mills, C.D. M1 and M2 macrophages: Oracles of health and disease. Crit. Rev. Immunol. 2012, 32, 463–488. [Google Scholar] [CrossRef] [Green Version]
- Burza, S.; Croft, S.L.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- Murray, H.W.W.; Berman, J.D.D.; Davies, C.R.R.; Saravia, N.G.G. Advances in leishmaniasis. Lancet 2005, 366, 1561–1577. [Google Scholar] [CrossRef]
- Aoki, J.I.; Muxel, S.M.; Zampieri, R.A.; Müller, K.E.; Nerland, A.H.; Floeter-Winter, L.M. Differential immune response modulation in early Leishmania amazonensis infection of BALB/c and C57BL/6 macrophages based on transcriptome profiles. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Muxel, S.M.; Laranjeira-Silva, M.F.; Zampieri, R.A.; Floeter-Winter, L.M. Leishmania (Leishmania) amazonensis induces macrophage miR-294 and miR-721 expression and modulates infection by targeting NOS2 and L-arginine metabolism. Sci. Rep. 2017, 7, 44141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muxel, S.M.; Mamani-Huanca, M.; Aoki, J.I.; Zampieri, R.A.; Floeter-Winter, L.M.; López-Gonzálvez, Á.; Barbas, C. Metabolomic profile of BALB/c macrophages infected with Leishmania amazonensis: Deciphering l-arginine metabolism. Int. J. Mol. Sci. 2019, 20, 6248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velasquez, L.G.; Galuppo, M.K.E.; De Rezende, E.; Brandao, W.N.; Peron, J.P.; Uliana, S.R.; Duarte, M.I.; Stolf, B.S. Distinct courses of infection with Leishmania (L.) amazonensis are observed in BALB/c, BALB/c nude and C57BL/6 mice. Parasitology 2016, 143, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Sun, J.; Soong, L. Impaired expression of inflammatory cytokines and chemokines at early stages of infection with Leishmania amazonensis. Infect. Immun. 2003, 71, 4278–4288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boitz, J.M.; Gilroy, C.A.; Olenyik, T.D.; Paradis, D.; Perdeh, J.; Dearman, K.; Davis, M.J.; Yates, P.A.; Li, Y.; Riscoe, M.K.; et al. Arginase is essential for survival of Leishmania donovani promastigotes but not intracellular amastigotes. Infect. Immun. 2017, 85, e00554-16. [Google Scholar] [CrossRef] [Green Version]
- Muxel, S.M.; Acuña, S.M.; Aoki, J.I.; Zampieri, R.A.; Floeter-Winter, L.M. Toll-like receptor and miRNA-let-7e expression alter the inflammatory response in leishmania amazonensis-infected macrophages Front. Immunol. 2018, 9, 2792. [Google Scholar] [CrossRef]
- Rath, M.; Müller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Front. Immunol. 2014, 5, 664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, S.C.; Tancer, M.J.; Polinsky, M.R.; Gibson, K.M.; Heby, O.; Ullman, B. Arginase plays a pivotal role in polyamine precursor metabolism in Leishmania. Characterization of gene deletion mutants. J. Biol. Chem. 2004, 279, 23668–23678. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, M.F.L.; Zampieri, R.A.; Muxel, S.M.; Beverley, S.M.; Floeter-Winter, L.M. Leishmania amazonensis arginase compartmentalization in the glycosome is important for parasite infectivity. PLoS ONE 2012, 7, e34022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoki, J.I.; Muxel, S.M.; Zampieri, R.A.; Laranjeira-Silva, M.F.; Müller, K.E.; Nerland, A.H.; Floeter-Winter, L.M. RNA-seq transcriptional profiling of Leishmania amazonensis reveals an arginase-dependent gene expression regulation. PLoS Negl. Trop. Dis. 2017, 11, e0006026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castilho-Martins, E.A.; Canuto, G.A.B.; Muxel, S.M.; daSilva, M.F.L.; Floeter-Winter, L.M.; del Aguila, C.; López-Gonzálvez, Á.; Barbas, C. Capillary electrophoresis reveals polyamine metabolism modulation in Leishmania (Leishmania) amazonensis wild-type and arginase-knockout mutants under arginine starvation. Electrophoresis 2015, 13, 2314–2323. [Google Scholar] [CrossRef] [PubMed]
- McConville, M.J. Metabolic crosstalk between Leishmania and the macrophage host. Trends Parasitol. 2016, 32, 666–668. [Google Scholar] [CrossRef] [PubMed]
- McConville, M.J.; de Souza, D.; Saunders, E.; Likic, V.A.; Naderer, T. Living in a phagolysosome; metabolism of Leishmania amastigotes. Trends Parasitol. 2007, 23, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Aoki, J.I.; Muxel, S.M.; Zampieri, R.A.; Acuña, S.M.; Fernandes, J.C.R.; Vanderlinde, R.H.; Sales, M.C.O.D.P.; Floeter-Winter, L.M. L-arginine availability and arginase activity: Characterization of amino acid permease 3 in Leishmania amazonensis. PLoS Negl. Trop. Dis. 2017, 11, e0006025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, R.W.; Pesce, J.T.; Ramalingam, T.; Wilson, M.S.; White, S.; Cheever, A.W.; Ricklefs, S.M.; Porcella, S.F.; Li, L.; Ellies, L.G.; et al. Cationic amino acid transporter-2 regulates immunity by modulating arginase activity. PLoS Pathog. 2008, 4, e1000023. [Google Scholar] [CrossRef] [Green Version]
- Wanasen, N.; Soong, L. L-arginine metabolism and its impact on host immunity against Leishmania infection. Immunol. Res. 2008, 41, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Laranjeira-Silva, M.F.; Zampieri, R.A.; Muxel, S.M.; Floeter-Winter, L.M.; Markus, R.P. Melatonin attenuates Leishmania (L.) amazonensis infection by modulating arginine metabolism. J. Pineal Res. 2015, 59, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Iniesta, V.; Gomez-Nieto, L.C.; Corraliza, I. The inhibition of arginase by N(omega)-hydroxy-l-arginine controls the growth of Leishmania inside macrophages. J. Exp. Med. 2001, 193, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Mamani-Huanca, M.; de la Fuente, A.G.; Otero, A.; Gradillas, A.; Godzien, J.; Barbas, C.; López-Gonzálvez, Á. Enhancing confidence of metabolite annotation in capillary electrophoresis-mass spectrometry untargeted metabolomics with relative migration time and in-source fragmentation. J. Chromatogr. A 2021, 1635, 461758. [Google Scholar] [CrossRef]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, mechanisms, and significance of macrophage plasticity. Annu. Rev. Pathol. Mech. Dis. 2020, 10, e0115855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, D.G.; O’Neill, L.A.J. Krebs cycle reborn in macrophage immunometabolism. Annu. Rev. Immunol. 2020, 38, 289–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muxel, S.M.; Aoki, J.I.; Fernandes, J.C.R.; Laranjeira-Silva, M.F.; Zampieri, R.A.; Acuña, S.M.; Müller, K.E.; Vanderlinde, R.H.; Floeter-Winter, L.M. Arginine and polyamines fate in leishmania infection. Front. Microbiol. 2018, 8, 2214. [Google Scholar] [CrossRef] [Green Version]
- Rosenzweig, D.; Smith, D.; Opperdoes, F.; Stern, S.; Olafson, R.W.; Zilberstein, D. Retooling Leishmania metabolism: From sand fly gut to human macrophage. FASEB J. 2008, 22, 590–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McConville, M.J.; Saunders, E.C.; Kloehn, J.; Dagley, M.J. Leishmania carbon metabolism in the macrophage phagolysosome-feast or famine? F1000Research 2015, 4, 938. [Google Scholar] [CrossRef] [PubMed]
- Saunders, E.C.; Naderer, T.; Chambers, J.; Landfear, S.M.; McConville, M.J. Leishmania mexicana can utilize amino acids as major carbon sources in macrophages but not in animal models. Mol. Microbiol. 2018, 108, 143–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canuto, G.A.B.A.; Castilho-Martins, E.A.A.; Tavares, M.; Lopez-Gonzalvez, A.; Rivas, L.; Barbas, C.; López-Gonzálvez, Á.; Rivas, L.; Barbas, C. CE-ESI-MS metabolic fingerprinting of Leishmania resistance to antimony treatment. Electrophoresis 2012, 33, 1901–1910. [Google Scholar] [CrossRef] [PubMed]
- Malta-Santos, H.; França-Costa, J.; Macedo, A.; Queiroz, A.T.L.; Fukutani, K.F.; Muxel, S.M.; Khouri, R.; van Weyenbergh, J.; Boaventura, V.; Barral, A.; et al. Differential expression of polyamine biosynthetic pathways in skin lesions and in plasma reveals distinct profiles in diffuse cutaneous leishmaniasis. Sci. Rep. 2020, 10, 10543. [Google Scholar] [CrossRef] [PubMed]
- Aoki, J.I.; Muxel, S.M.; Laranjeira-Silva, M.F.; Zampieri, R.A.; Müller, K.E.; Nerland, A.H.; Floeter-Winter, L.M. Dual transcriptome analysis reveals differential gene expression modulation influenced by Leishmania arginase and host genetic background. Microb. Genomics. 2020, 6, mgen000427. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Fernando, V.; Sharma, V.; Walia, Y.; Letson, J.; Furuta, S. Correction of arginine metabolism with sepiapterin—the precursor of nitric oxide synthase cofactor BH4—Induces immunostimulatory-shift of breast cancer. Biochem. Pharmacol. 2020, 176, 113887. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, C. Macrophages as host, effector and immunoregulatory cells in leishmaniasis: Impact of tissue micro-environment and metabolism. Cytokine X 2020, 12, 100041. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E.; McCann, P.P. Polyamine metabolism and function. Am. J. Physiol. Cell. Physiol. 1982, 243, C212–C221. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Thomas, T.J. Polyamines in cell growth and cell death: Molecular mechanisms and therapeutic applications. Cell. Mol. Life Sci. 2001, 58, 244–258. [Google Scholar] [CrossRef]
- Puleston, D.J.; Buck, M.D.; Geltink, R.I.K.; Kyle, R.L.; Caputa, G.; O’Sullivan, D.; Cameron, A.M.; Castoldi, A.; Musa, Y.; Kabat, A.M.; et al. Polyamines and eIF5A hypusination modulate mitochondrial respiration and macrophage activation. Cell. Metab. 2019, 30, 352–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gogoi, M.; Datey, A.; Wilson, K.T.; Chakravortty, D. Dual role of arginine metabolism in establishing pathogenesis. Curr. Opin. Microbiol. 2016, 29, 43–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Leal, O.; Barrero, C.A.; Clarkson, A.B.; Casero, R.A.; Merali, S. Polyamine-regulated translation of spermidine/spermine-N1-scetyltransferase. Mol. Cell. Biol. 2012, 32, 1453–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hesterberg, R.; Cleveland, J.; Epling-Burnette, P. Role of polyamines in immune cell functions. Med. Sci. 2018, 6, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puleston, D.J.; Villa, M.; Pearce, E.L. Ancillary activity: Beyond core metabolism in immune cells. Cell. Metab. 2017, 26, 131–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Bazer, F.W.; Datta, S.; Johnson, G.A.; Li, P.; Satterfield, M.C.; Spencer, T.E. Proline metabolism in the conceptus: Implications for fetal growth and development. Amino Acids. 2008, 35, 691–702. [Google Scholar] [CrossRef]
- Goodman, J.L.; Wang, S.; Alam, S.; Ruzicka, F.J.; Frey, P.A.; Wedekind, J.E. Ornithine cyclodeaminase: Structure, mechanism of action, and implications for the μ-crystallin family. Biochemistry 2004, 43, 13883–13891. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, D.A.; Forman, H.J. Cellular glutathione and thiols metabolism. Biochem. Pharmacol. 2002, 64, 1019–1026. [Google Scholar] [CrossRef]
- Pegg, A.E. Spermidine/spermine-N1-acetyltransferase: A key metabolic regulator. Am. J. Physiol. Endocrinol. Metab. 2008, 294, E995–E1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polticelli, F.; Salvi, D.; Mariottini, P.; Amendola, R.; Cervelli, M. Molecular evolution of the polyamine oxidase gene family in Metazoa. BMC Evol. Biol. 2012, 12, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Jell, J.; Merali, S.; Hensen, M.L.; Mazurchuk, R.; Spernyak, J.A.; Diegelman, P.; Kisiel, N.D.; Barrero, C.; Deeb, K.K.; Alhonen, L.; et al. Genetically altered expression of spermidine/spermine N1- acetyltransferase affects fat metabolism in mice via acetyl-CoA J. Biol. Chem. 2007, 282, 8404–8413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niiranen, K.; Keinänen, T.A.; Pirinen, E.; Heikkinen, S.; Tusa, M.; Fatrai, S.; Suppola, S.; Pietilä, M.; Uimari, A.; Laakso, M.; et al. Mice with targeted disruption of spermidine/spermine N 1 -acetyltransferase gene maintain nearly normal tissue polyamine homeostasis but show signs of insulin resistance upon aging. J. Cell. Mol. Med. 2006, 10, 815–827. [Google Scholar] [CrossRef] [Green Version]
- Takao, K.; Shibata, S.; Ozawa, T.; Wada, M.; Sugitia, Y.; Samejima, K.; Shirahata, A. A conceptual model of the polyamine binding site of N 1- acetylpolyamine oxidase developed from a study of polyamine derivatives. Amino Acids. 2009, 47, 925–935. [Google Scholar] [CrossRef]
- Ikeguchi, Y.; Bewley, M.C.; Pegg, A.E. Aminopropyltransferases: Function, structure and genetics. J. Biochem. 2006, 139, 161–169. [Google Scholar] [CrossRef]
- Cervelli, M.; Amendola, R.; Polticelli, F.; Mariottini, P. Spermine oxidase: Ten years after. Amino Acids. 2012, 42, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Lüersen, K. Leishmania major thialysine Nε-acetyltransferase: Identification of amino acid residues crucial for substrate binding. FEBS Lett. 2005, 579, 5347–5352. [Google Scholar] [CrossRef] [Green Version]
- Li, C.H.; Ohn, T.; Ivanov, P.; Tisdale, S.; Anderson, P. eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS ONE 2010, 5, e9942. [Google Scholar] [CrossRef] [Green Version]
- Saini, P.; Eyler, D.E.; Green, R.; Dever, T.E. Hypusine-containing protein eIF5A promotes translation elongation. Nature 2009, 459, 118–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chawla, B.; Jhingran, A.; Singh, S.; Tyagi, N.; Park, M.H.H.; Srinivasan, N.; Roberts, S.C.C.; Madhubala, R. Identification and characterization of a novel deoxyhypusine synthase in Leishmania donovani. J. Biol. Chem. 2010, 285, 453–463. [Google Scholar] [CrossRef] [Green Version]
- Diotallevi, M.; Checconi, P.; Palamara, A.T.T.; Celestino, I.; Coppo, L.; Holmgren, A.; Abbas, K.; Peyrot, F.; Mengozzi, M.; Ghezzi, P. Glutathione fine-tunes the innate immune response toward antiviral pathways in a macrophage cell line independently of its antioxidant properties. Front. Immunol. 2017, 8, 1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribas, V.; García-Ruiz, C.; Fernández-Checa, J.C. Glutathione and mitochondria. Front. Pharmacol. 2014, 5, 151. [Google Scholar] [CrossRef] [Green Version]
- Carter, K.C.; Sundar, S.; Spickett, C.; Pereira, O.C.; Mullen, A.B. The in vivo susceptibility of Leishmania donovani to sodium stibogluconate is drug specific and can be reversed by inhibiting glutathione biosynthesis. Antimicrob. Agents Chemother. 2003, 47, 1529–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, O.; Cappel, D.; Nocker, M.; Jäger, T.; Flohé, L.; Sotriffer, C.A.; Selzer, P.M. Molecular dynamics reveal binding mode of glutathionylspermidine by trypanothione synthetase. PLoS ONE 2013, 8, e56788. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, M.S.; Comini, M.A.; Resende, B.V.; Santi, A.M.M.; Zoboli, A.P.; Moreira, D.S.; Murta, S.M.F. Ornithine decarboxylase or gamma-glutamylcysteine synthetase overexpression protects Leishmania (Vianna) guyanensis against antimony. Exp. Parasitol. 2017, 175, 36–43. [Google Scholar] [CrossRef]
- Saudagar, P.; Dubey, V.K. Cloning, expression, characterization, and inhibition studies on Trypanothione Synthetase, a drug target enzyme, from Leishmania donovani. Biol. Chem. 2011, 392, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.N.; Andersson, B.; Pettersson, U.; Åslund, L. Trypanothione synthetase locus in Trypanosoma cruzi CL Brener strain shows an extensive allelic divergence. Acta Trop. 2003, 87, 269–278. [Google Scholar] [CrossRef]
- Comini, M.; Menge, U.; Flohé, L. Biosynthesis of trypanothione in Trypanosoma brucei brucei. Biol. Chem. 2003, 384, 653–656. [Google Scholar] [CrossRef]
- Oza, S.L.; Wyllie, S.; Fairlamb, A.H. Mapping the functional synthetase domain of trypanothione synthetase from Leishmania major. Mol. Biochem. Parasitol. 2006, 149, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Colotti, G.; Ilari, A. Polyamine metabolism in Leishmania: From arginine to trypanothione. Amino Acids. 2011, 40, 269–285. [Google Scholar] [CrossRef]
- Karreth, F.A.A.; Tay, Y.; Perna, D.; Ala, U.; Tan, S.M.M.; Rust, A.G.G.; Denicola, G.; Webster, K.A.A.; Weiss, D.; Perez-Mancera, P.A.A.; et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cells 2011, 147, 382–395. [Google Scholar] [CrossRef] [Green Version]
- Mesías, A.C.; Sasoni, N.; Arias, D.G.; Brandán, C.P.; Orban, O.C.F.; Kunick, C.; Robello, C.; Comini, M.A.; Garg, N.J.; Zago, M.P. Trypanothione synthetase confers growth, survival advantage and resistance to anti-protozoal drugs in Trypanosoma cruzi. Free Radic. Biol. Med. 2019, 130, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Saudagar, P.; Saha, P.; Saikia, A.K.; Dubey, V.K. Molecular mechanism underlying antileishmanial effect of oxabicyclo[3.3.1]nonanones: Inhibition of key redox enzymes of the pathogen. Eur. J. Pharm. Biopharm. 2013, 85, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Irigoín, F.; Inada, N.M.; Fernandes, M.P.; Piacenza, L.; Gadelha, F.R.; Vercesi, A.E.; Radi, R. Mitochondrial calcium overload triggers complement-dependent superoxide-mediated programmed cell death in Trypanosoma cruzi. Biochem. J. 2009, 418, 595–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mesías, A.C.; Garg, N.J.; Zago, M.P. Redox balance keepers and possible cell functions managed by redox homeostasis in trypanosoma cruzi, front. Cell. Infect. Microbiol. 2019, 9, 435. [Google Scholar] [CrossRef] [PubMed]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The metabolic signature of macrophage responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [Green Version]
- Richardson, J.L.; Nett, I.R.E.; Jones, D.C.; Abdille, M.H.; Gilbert, I.H.; Fairlamb, A.H. Improved tricyclic inhibitors of trypanothione reductase by screening and chemical synthesis. Chem. Med. Chem. 2009, 4, 1333. [Google Scholar] [CrossRef] [Green Version]
- Battelli, M.G.; Polito, L.; Bortolotti, M.; Bolognesi, A. Xanthine oxidoreductase-derived reactive species: Physiological and pathological effects. Oxid. Med. Cell. Longev. 2016, 3227579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battelli, M.G.; Polito, L.; Bortolotti, M.; Bolognesi, A. Xanthine oxidoreductase in drug metabolism: Beyond a role as a detoxifying enzyme. Curr. Med. Chem. 2016, 35, 4027–4036. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Name | Mass (Da) | RMT | %RSD (QC) | p-Value (a) | p FDR | p-Value (b) | % Change | p (corr) | VIP |
|---|---|---|---|---|---|---|---|---|---|
| Glycine | 75.0330 | 0.72 | 8.24 | 4.47 × 10−6 | 2.23E-05 | 4.33 × 10−5 | −41.30 | −0.84 | <1 |
| Putrescine | 88.1000 | 0.42 | 5.81 | 3.92 × 10−6 | 2.07E-05 | 2.17 × 10−5 | −58.46 | −0.80 | <1 |
| Alanine | 89.0483 | 0.77 | 7.31 | 1.85 × 10−5 | 4.74E-05 | 1.01 × 10−2 | −24.82 | −0.62 | 2.81 |
| 4-aminobutanoic acid | 103.0647 | 0.67 | 7.41 | 1.85 × 10−5 | 4.74E-05 | 1.01 × 10−2 | 57.39 | 0.57 | <1 |
| Serine | 105.0429 | 0.85 | 6.10 | 7.46 × 10−6 | 2.76E-05 | 2.60 × 104 | −37.88 | −0.79 | <1 |
| Proline | 115.0634 | 0.92 | 7.42 | 5.14 × 10−6 | 2.34E-05 | 2.17 × 10−5 | −73.83 | −0.96 | 1.73 |
| Valine | 117.0792 | 0.85 | 6.73 | 1.26 × 10−5 | 3.70E-05 | 2.99 × 103 | −30.00 | −0.69 | 2.06 |
| Pipecolic acid | 129.0789 | 0.87 | 6.71 | 4.98 × 10−6 | 2.34E-05 | 6.50 × 104 | 117.06 | 0.67 | 4.05 |
| Isoleucine/leucine | 131.0945 | 0.87 | 6.51 | 3.92 × 10−6 | 2.07E-05 | 2.17 × 10−5 | −56.42 | −0.91 | 4.16 |
| Asparagine | 132.0528 | 0.89 | 6.82 | 5.74 × 10−6 | 2.50E-05 | 1.52 × 104 | −41.67 | −0.78 | <1 |
| Ornithine | 132.0896 | 0.60 | 6.67 | 1.04 × 10−2 | 1.80E-04 | 2.17 × 10−5 | −92.47 | −0.99 | 2.00 |
| Aspartic acid | 133.0374 | 0.97 | 6.46 | 2.10 × 10−2 | 2.88E-02 | 1.72 × 10−2 | −19.58 | −0.56 | <1 |
| Hypoxanthine | 136.0385 | 1.01 | 5.95 | 1.69 × 10−5 | 4.56E-05 | 7.62 × 103 | −26.98 | −0.61 | <1 |
| Glutamic acid | 147.0532 | 0.92 | 5.46 | 6.34 × 10−5 | 1.17E-04 | 4.14 × 103 | −26.26 | −0.66 | 3.45 |
| Methionine | 149.0512 | 0.91 | 3.94 | 1.13 × 10−5 | 3.54E-05 | 2.10 × 103 | −34.19 | −0.75 | <1 |
| Xanthine | 152.0271 | 1.72 | 12.19 | 1.53 × 10−2 | 2.15E-02 | 2.79 × 10−2 | 19.21 | 0.50 | <1 |
| Histidine | 155.0696 | 0.64 | 6.61 | 3.92 × 10−6 | 2.07E-05 | 2.17 × 10−5 | −59.90 | −0.93 | 1.96 |
| Imidazolelactic acid | 156.0554 | 0.76 | 8.51 | 1.06 × 10−5 | 3.41E-05 | 7.62 × 10−3 | −27.91 | −0.61 | <1 |
| Phenylalanine | 165.0808 | 0.93 | 6.50 | 8.17 × 10−6 | 2.92E-05 | 6.50 × 10−4 | −36.86 | −0.75 | 1.05 |
| 1-Methylhistidine | 169.0856 | 0.66 | 10.42 | 2.32 × 10−6 | 1.77E-05 | 2.17 × 10−5 | −81.52 | −0.98 | <1 |
| Arginine | 174.1117 | 0.63 | 6.37 | 9.14 × 10−6 | 3.05E-05 | 9.74 × 10−4 | 100.48 | 0.66 | 3.77 |
| Citrulline | 175.0972 | 0.94 | 6.14 | 2.32 × 10−6 | 1.77E-05 | 2.17 × 10−5 | 220.70 | 0.82 | <1 |
| Argininic acid | 175.0975 | 0.79 | 5.67 | 2.32 × 10−6 | 1.77E-05 | 2.17 × 10−5 | 1919.49 | 0.86 | <1 |
| Tyrosine | 181.0739 | 0.96 | 5.81 | 1.26 × 10−5 | 3.70E-05 | 2.99 × 10−3 | −37.73 | −0.78 | <1 |
| Tryptophan | 204.0894 | 0.93 | 5.11 | 1.54 × 10−6 | 1.77E-05 | 2.17 × 10−5 | −27.53 | −0.70 | <1 |
| N-carbamyl-arginine (c) | 217.1191 | 0.84 | 22.26 | 2.65 × 10−6 | 1.77E-05 | 4.33 × 10−5 | La-WT (d) | <0.5 | <1 |
| N-glutamyl-alanine (c) | 218.0906 | 1.03 | 7.04 | 2.32 × 10−6 | 1.77E-05 | 2.17 × 10−5 | −51.06 | −0.86 | <1 |
| Cystathionine | 222.0679 | 0.85 | 4.06 | 6.61 × 10−6 | 2.64E-05 | 1.52 × 10−4 | −57.72 | −0.93 | <1 |
| Neuraminic acid (c) | 267.0961 | 1.13 | 7.88 | 2.65 × 10−6 | 1.77E-05 | 4.33 × 10−5 | −40.89 | −0.79 | <1 |
| N-(1-Deoxy-1-fructosyl)threonine (c) | 281.1118 | 1.13 | 7.78 | 9.14 × 10−6 | 3.05E-05 | 9.74 × 10−4 | −44.93 | −0.86 | <1 |
| Glutamylarginine (c) | 303.1550 | 0.76 | 9.35 | 6.27 × 10−6 | 2.61E-05 | 1.45 × 10−3 | La-WT (d) | <0.5 | <1 |
| S-Adenosylmethionine (c) | 398.1385 | 0.63 | 4.84 | 2.65 × 10−6 | 1.71E-05 | 4.33 × 10−5 | −33.16 | −0.74 | <1 |
| Trypanothione disulfide | 721.2896 | 0.82 | 25.13 | 3.92 × 10−6 | 2.07E-05 | 2.17 × 10−5 | −35.56 | −0.71 | <1 |
| Trypanothione | 723.3057 | 0.84 | 25.12 | 1.85 × 10−5 | 4.74E-05 | 1.01 × 10−2 | −52.10 | −0.80 | <1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mamani-Huanca, M.; Muxel, S.M.; Acuña, S.M.; Floeter-Winter, L.M.; Barbas, C.; López-Gonzálvez, Á. Metabolomic Reprogramming of C57BL/6-Macrophages during Early Infection with L. amazonensis. Int. J. Mol. Sci. 2021, 22, 6883. https://doi.org/10.3390/ijms22136883
Mamani-Huanca M, Muxel SM, Acuña SM, Floeter-Winter LM, Barbas C, López-Gonzálvez Á. Metabolomic Reprogramming of C57BL/6-Macrophages during Early Infection with L. amazonensis. International Journal of Molecular Sciences. 2021; 22(13):6883. https://doi.org/10.3390/ijms22136883
Chicago/Turabian StyleMamani-Huanca, Maricruz, Sandra Marcia Muxel, Stephanie Maia Acuña, Lucile Maria Floeter-Winter, Coral Barbas, and Ángeles López-Gonzálvez. 2021. "Metabolomic Reprogramming of C57BL/6-Macrophages during Early Infection with L. amazonensis" International Journal of Molecular Sciences 22, no. 13: 6883. https://doi.org/10.3390/ijms22136883
APA StyleMamani-Huanca, M., Muxel, S. M., Acuña, S. M., Floeter-Winter, L. M., Barbas, C., & López-Gonzálvez, Á. (2021). Metabolomic Reprogramming of C57BL/6-Macrophages during Early Infection with L. amazonensis. International Journal of Molecular Sciences, 22(13), 6883. https://doi.org/10.3390/ijms22136883








