The Roles Played by Long Non-Coding RNAs in Glioma Resistance
Abstract
:1. Introduction
2. Dysregulated lncRNA in Glioma
2.1. Upregulations of Oncogenic lncRNAs in Glioma
2.2. Downregulation of Tumor-Suppressive lncRNAs in Glioma
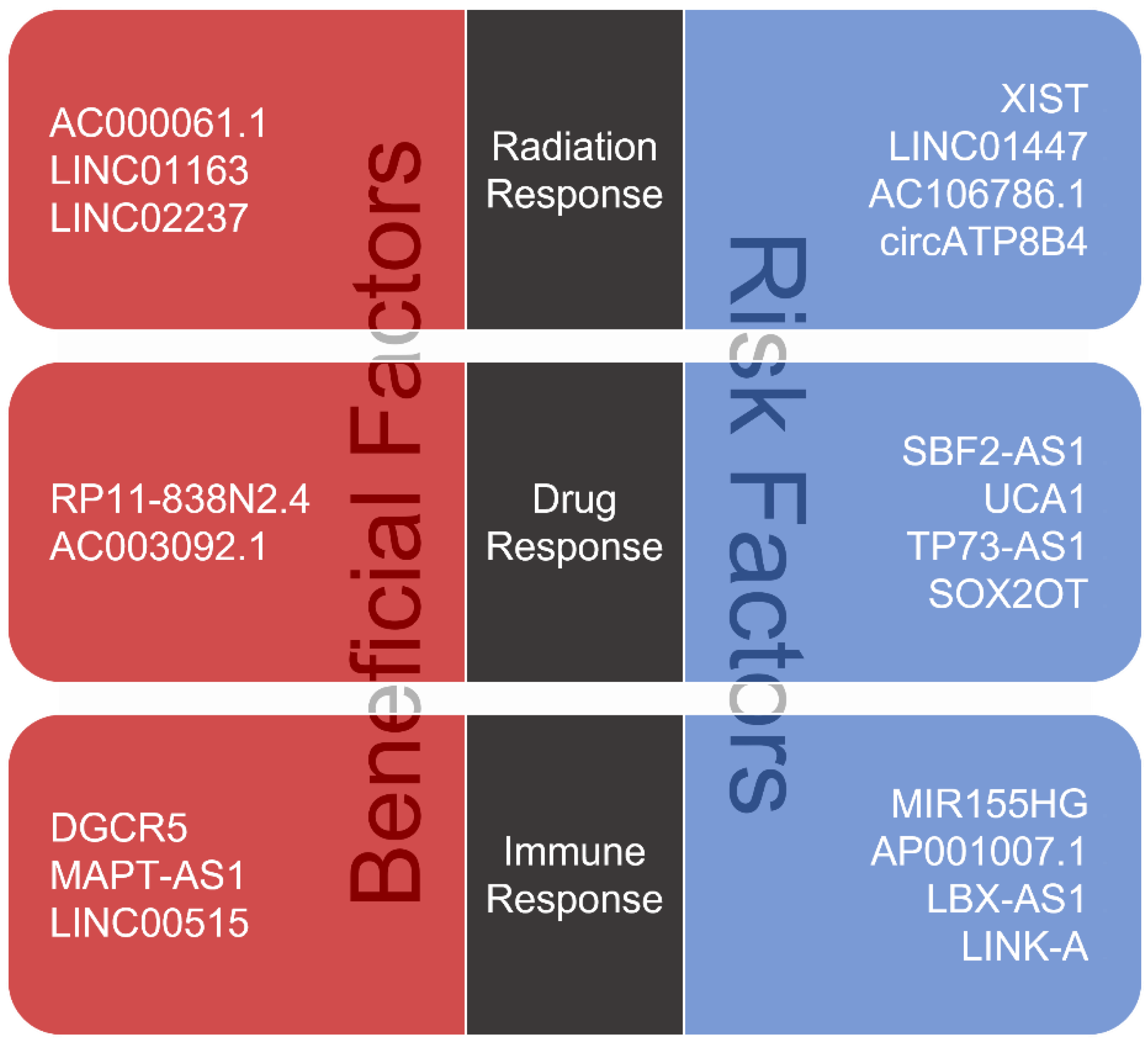
3. lncRNAs Associated with Therapeutic Resistance in Glioma
3.1. Roles of lncRNAs in Glioma Radiotherapy
3.2. Roles of lncRNAs in Glioma Chemotherapy
3.3. Roles of lncRNAs in Glioma Immunotherapy
4. Clinical Applications of lncRNAs in Glioma
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristensen, B.; Priesterbach-Ackley, L.; Petersen, J.; Wesseling, P. Molecular pathology of tumors of the central nervous system. Ann. Oncol. 2019, 30, 1265–1278. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [Green Version]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K. WHO Classification of Tumours of the Central Nervous System, 4th ed.; International Agency for Research on Cancer (IARC): Lyon, France, 2016. [Google Scholar]
- Schwartzbaum, J.A.; Fisher, J.L.; Aldape, K.D.; Wrensch, M. Epidemiology and molecular pathology of glioma. Nat. Clin. Pract. Neurol. 2006, 2, 494–503. [CrossRef]
- Alifieris, C.; Trafalis, D.T. Glioblastoma multiforme: Pathogenesis and treatment. Pharmacol. Ther. 2015, 152, 63–82. [Google Scholar] [CrossRef]
- Lim, M.; Xia, Y.; Bettegowda, C.; Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018, 15, 422–442. [Google Scholar] [CrossRef] [PubMed]
- Wiedmann, M.K.; Brunborg, C.; Di Ieva, A.; Lindemann, K.; Johannesen, T.B.; Vatten, L.; Helseth, E.; Zwart, J.A. The impact of body mass index and height on the risk for glioblastoma and other glioma subgroups: A large prospective cohort study. Neuro-Oncology 2016, 19, 976–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Meir, E.G.; Hadjipanayis, C.G.; Norden, A.D.; Shu, H.-K.; Wen, P.Y.; Olson, J.J. Exciting New Advances in Neuro-Oncology: The Avenue to a Cure for Malignant Glioma. CA A Cancer J. Clin. 2010, 60, 166–193. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [Green Version]
- Davis, M.E. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016, 20, S2–S8. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; McKay, R.M.; Parada, L.F. Malignant Glioma: Lessons from Genomics, Mouse Models, and Stem Cells. Cell 2012, 149, 36–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, W.X.; Koirala, P.; Mo, Y.Y. lncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Zhao, J.; Yeung, P.Y.; Zhang, Q.C.; Kwok, C.K. Revealing lncRNA Structures and Interactions by Sequencing-Based Approaches. Trends Biochem. Sci. 2019, 44, 33–52. [Google Scholar] [CrossRef]
- Banelli, B.; Forlani, A.; Allemanni, G.; Morabito, A.; Pistillo, M.P.; Romani, M. MicroRNA in Glioblastoma: An Overview. Int. J. Genom. 2017, 2017, 7639084. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.Z.; Tian, Y.F.; Wu, H.; Ouyang, S.Y.; Kuang, W.L. lncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1α/VEGF axis. Neoplasma 2020, 67, 111–118. [Google Scholar] [CrossRef]
- Gu, N.; Wang, X.; Di, Z.; Xiong, J.; Ma, Y.; Yan, Y.; Qian, Y.; Zhang, Q.; Yu, J. Silencing lncRNA FOXD2-AS1 inhibits prolifer-ation, migration, invasion and drug resistance of drug-resistant glioma cells and promotes their apoptosis via mi-croRNA-98-5p/CPEB4 axis. Aging 2019, 11, 10266–10283. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-P.; Li, H.-Q.; Chen, J.-X.; Kong, F.-G.; Mo, Z.-H.; Wang, J.-Z.; Huang, K.-M.; Li, X.-N.; Yan, Y. Overexpression of XIST facilitates cell proliferation, invasion and suppresses cell apoptosis by reducing radio-sensitivity of glioma cells via miR-329-3p/CREB1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3190–3203. [Google Scholar]
- Jing, S.-Y.; Lu, Y.-Y.; Yang, J.-K.; Deng, W.-Y.; Zhou, Q.; Jiao, B.-H. Expression of long non-coding RNA CRNDE in glioma and its correlation with tumor progression and patient survival. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3992–3996. [Google Scholar]
- Wu, F.; Zhang, C.; Cai, J.; Yang, F.; Liang, T.; Yan, X.; Wang, H.; Wang, W.; Chen, J.; Jiang, T. Upregulation of long noncoding RNA HOXA-AS3 promotes tumor progression and predicts poor prognosis in glioma. Oncotarget 2017, 8, 53110–53123. [Google Scholar] [CrossRef] [Green Version]
- Liang, Q.; Li, X.; Guan, G.; Xu, X.; Chen, C.; Cheng, P.; Cheng, W.; Wu, A. Long non-coding RNA, HOTAIRM1, promotes glioma malignancy by forming a ceRNA network. Aging 2019, 11, 6805–6838. [Google Scholar] [CrossRef]
- Birkó, Z.; Nagy, B.; Klekner, Á.; Virga, J. Novel Molecular Markers in Glioblastoma—Benefits of Liquid Biopsy. Int. J. Mol. Sci. 2020, 21, 7522. [Google Scholar] [CrossRef] [PubMed]
- Antunes-Ferreira, M.; Koppers-Lalic, D.; Würdinger, T. Circulating platelets as liquid biopsy sources for cancer detection. Mol. Oncol. 2021, 15, 1727–1743. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, J.T.; Tsirka, S.E. Advances in immunotherapeutic research for glioma therapy. J. Neurol. 2018, 265, 741–756. [Google Scholar] [CrossRef]
- Van den Bent, M.J. Chemotherapy for low-grade glioma: When, for whom, which regimen? Curr. Opin. Neurol. 2015, 28, 633–938. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.D. Recent Technical Advances and Indications for Radiation Therapy in Low-Grade Glioma. Semin. Radiat. Oncol. 2015, 25, 189–196. [Google Scholar] [CrossRef] [PubMed]
- De Andres-Pablo, A.; Morillon, A.; Wery, M. lncRNAs, lost in translation or licence to regulate? Curr. Genet. 2016, 63, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Do, H.; Kim, W. Roles of Oncogenic Long Non-coding RNAs in Cancer Development. Genom. Inform. 2018, 16, e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nat. Cell Biol. 2012, 493, 231–235. [Google Scholar] [CrossRef]
- Qian, K.; Liu, G.; Tang, Z.; Hu, Y.; Fang, Y.; Chen, Z.; Xu, X. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch. Biochem. Biophys. 2017, 615, 1–9. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, J.; Wen, L.; Lin, A. Membrane-lipid associated lncRNA: A new regulator in cancer signaling. Cancer Lett. 2018, 419, 27–29. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Dong, B.; Cao, J.; Mao, Y.; Guan, W.; Peng, Y.; Wang, S. Long non-coding RNA in glioma: Signaling pathways. Oncotarget 2017, 8, 27582–27592. [Google Scholar] [CrossRef] [Green Version]
- Bian, E.-B.; Li, J.; Xie, Y.-S.; Zong, G.; Li, J.; Zhao, B. lncRNAs: New Players in Gliomas, With Special Emphasis on the Interaction of lncRNAs With EZH2. J. Cell. Physiol. 2014, 230, 496–503. [Google Scholar] [CrossRef]
- Mu, M.; Niu, W.; Zhang, X.; Hu, S.; Niu, C. lncRNA BCYRN1 inhibits glioma tumorigenesis by competitively binding with miR-619-5p to regulate CUEDC2 expression and the PTEN/AKT/p21 pathway. Oncogene 2020, 39, 6879–6892. [Google Scholar] [CrossRef]
- Cheng, Z.; Luo, C.; Guo, Z. lncRNA-XIST/microRNA-126 sponge mediates cell proliferation and glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J. Cell Biochem. 2020, 121, 2170–2183. [Google Scholar] [CrossRef]
- Sun, L.; Hui, A.-M.; Su, Q.; Vortmeyer, A.; Kotliarov, Y.; Pastorino, S.; Passaniti, A.; Menon, J.; Walling, J.; Bailey, R.; et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006, 9, 287–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Sun, X.; Zhou, X.; Han, L.; Chen, L.; Shi, Z.; Zhang, A.; Ye, M.; Wang, Q.; Liu, C.; et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget 2015, 6, 537–546. [Google Scholar] [CrossRef] [Green Version]
- Xie, P.; Li, X.; Chen, R.; Liu, Y.; Liu, D.; Liu, W.; Cui, G.; Xu, J. Upregulation of HOTAIRM1 increases migration and invasion by glioblastoma cells. Aging 2020, 13, 2348–2364. [Google Scholar] [CrossRef] [PubMed]
- Lang, H.-L.; Hu, G.-W.; Zhang, B.; Kuang, W.; Chen, Y.; Wu, L.; Xu, G.-H. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol. Rep. 2017, 38, 785–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, J.; Zhang, L.; Chen, S.; Cao, H.; Xu, C.; Wang, X. lncRNA CCAT2 Enhanced Resistance of Glioma Cells Against Che-modrugs by Disturbing the Normal Function of miR-424. OncoTargets Ther. 2020, 13, 1431–1445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.-X.; Fei, X.-R.; Dong, Y.-F.; Cheng, C.-D.; Yang, Y.; Deng, X.-F.; Huang, H.-L.; Niu, W.-X.; Zhou, C.-X.; Xia, C.-Y.; et al. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget 2017, 8, 88163–88178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Wang, Y.; He, J.; Zhang, C.; Chen, J.; Shi, D. Long Noncoding RNA H19 Promotes Proliferation and Invasion in Human Glioma Cells by Downregulating miR-152. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2018, 26, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Yin, J.; Zeng, A.; Jin, X.; Zhang, Z.; Yan, W.; You, Y. H19 Functions as a Competing Endogenous RNA to Regulate EMT by Sponging miR-130a-3p in Glioma. Cell. Physiol. Biochem. 2018, 50, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Li, D.; Zhang, X.; Liu, N.; Chi, G.; Jin, X. lncRNA PVT1 Facilitates Tumorigenesis and Progression of Glioma via Regulation of MiR-128-3p/GREM1 Axis and BMP Signaling Pathway. Neurotherapeutics 2018, 15, 1139–1157. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhu, Q.; Guo, Y.; Xiao, Z.; Hu, L.; Xu, Q. lncRNA LINC00689 promotes the growth, metastasis and glycolysis of glioma cells by targeting miR-338-3p/PKM2 axis. Biomed. Pharmacother. 2019, 117, 109069. [Google Scholar] [CrossRef]
- Zhao, M.; Xu, J.; Zhong, S.; Liu, Y.; Xiao, H.; Geng, L.; Liu, H. Expression profiles and potential functions of circular RNAs in extracellular vesicles isolated from radioresistant glioma cells. Oncol. Rep. 2019, 41, 1893–1900. [Google Scholar] [CrossRef]
- Zhang, Z.; Yin, J.; Lu, C.; Wei, Y.; Zeng, A.; You, Y. Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemo-resistance to temozolomide in glioblastoma. J. Exp. Clin. Cancer Res. 2019, 38, 166. [Google Scholar] [CrossRef]
- Yu, H.; Zheng, J.; Liu, X.; Xue, Y.; Shen, S.; Zhao, L.; Li, Z.; Liu, Y. Transcription Factor NFAT5 Promotes Glioblastoma Cell-driven Angiogenesis via SBF2-AS1/miR-338-3p-Mediated EGFL7 Expression Change. Front. Mol. Neurosci. 2017, 10, 301. [Google Scholar] [CrossRef] [Green Version]
- Peng, L.; Chen, Z.; Chen, Y.; Wang, X.; Tang, N. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 2019, 8, 7161–7173. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xu, N.; Liu, B.; Huang, Y.; Zeng, H.; Yang, Z.; He, Z.; Guo, H. Long noncoding RNA RP11-838N2.4 enhances the cytotoxic effects of temozolomide by inhibiting the functions of miR-10a in glioblastoma cell lines. Oncotarget 2016, 7, 43835–43851. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Wu, Z.; Wu, T.; Huang, Y.; Cheng, Z.; Li, X.; Sun, T.; Xie, X.; Zhou, Y.; Du, Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016, 7, e2123. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Wang, Y.; Li, J.; Lv, M.; Niu, H.; Tian, Y. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am. J. Cancer Res. 2016, 6, 2561–2574. [Google Scholar]
- Young-Pearse, T.; Matsuda, T.; Cepko, C. The Noncoding RNA Taurine Upregulated Gene 1 Is Required for Differentiation of the Murine Retina. Curr. Biol. 2005, 15, 501–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhang, M.; An, G.; Ma, Q. lncRNA TUG1 acts as a tumor suppressor in human glioma by promoting cell apoptosis. Exp. Biol. Med. 2016, 241, 644–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.-Q.; Sun, S.; Lam, K.-F.; Kiang, K.M.-Y.; Pu, J.K.-S.; Ho, A.S.-W.; Lui, W.-M.; Fung, C.-F.; Wong, T.S.; Leung, G.K.K. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol. Dis. 2013, 58, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Piao, L.; Sun, G.; Lv, C.; Jing, Y.; Jin, R. Long Non-Coding RNA PART1 Exerts Tumor Suppressive Functions in Gli-oma via Sponging miR-190a-3p and Inactivation of PTEN/AKT Pathway. Oncotargets. Ther. 2020, 13, 1073–1086. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Xue, Y.; Wang, Q.; Zhou, X.; Liu, L.; Zhang, T.; Shang, C.; Ma, J.; Ma, T. Long non-coding RNA MIAT regulates blood tumor barrier permeability by functioning as a competing endogenous RNA. Cell Death Dis. 2020, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-P.; Shan, C.; Deng, X.-L.; Li, L.-Y.; Ma, W. Long non-coding RNA PAR5 inhibits the proliferation and progression of glioma through interaction with EZH2. Oncol. Rep. 2017, 38, 3177–3186. [Google Scholar] [CrossRef] [Green Version]
- Tang, T.; Wang, L.X.; Yang, M.L.; Zhang, R.M. lncRNA TPTEP1 inhibits stemness and radioresistance of glioma through miR-106a-5p-mediated P38 MAPK signaling. Mol. Med. Rep. 2020, 22, 4857–4867. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Huang, Y.-L. DGCR5 suppresses the EMT of pediatric primary glioblastoma multiforme cell and serves as a prognostic biomarker. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10024–10034. [Google Scholar]
- He, Z.; Long, J.; Yang, C.; Gong, B.; Cheng, M.; Wang, Q.; Tang, J. lncRNA DGCR5 plays a tumor-suppressive role in glioma via the miR-21/Smad7 and miR-23a/PTEN axes. Aging 2020, 12, 20285–20307. [Google Scholar] [CrossRef] [PubMed]
- Norden, A.D.; Wen, P.Y. Glioma Therapy in Adults. Neurologist 2006, 12, 279–292. [Google Scholar] [CrossRef]
- Xu, S.; Tang, L.; Li, X.; Fan, F.; Liu, Z. Immunotherapy for glioma: Current management and future application. Cancer Lett. 2020, 476, 1–12. [Google Scholar] [CrossRef]
- Lin, L.; Cai, J.; Jiang, C. Recent Advances in Targeted Therapy for Glioma. Curr. Med. Chem. 2017, 24, 1365–1381. [Google Scholar] [CrossRef] [PubMed]
- Tomiyama, A.; Ichimura, K. Signal transduction pathways and resistance to targeted therapies in glioma. Semin. Cancer Biol. 2019, 58, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Liu, J.; Quan, J.; Liu, W.; Tan, H.; Li, W. MicroRNAs as potential biomarkers for the diagnosis of glioma: A systematic review and meta-analysis. Cancer Sci. 2018, 109, 2651–2659. [Google Scholar] [CrossRef] [Green Version]
- Weidhaas, J.B.; Babar, I.; Nallur, S.M.; Trang, P.; Roush, S.; Boehm, M.; Gillespie, E.; Slack, F.J. MicroRNAs as Potential Agents to Alter Resistance to Cytotoxic Anticancer Therapy. Cancer Res. 2007, 67, 11111–11116. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Chen, G.; Zhu, W.; Shi, D.; Lv, L.; Zhang, C.; Liu, P.; Hu, W. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol. Rep. 2010, 23, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Jiapaer, S.; Furuta, T.; Tanaka, S.; Kitabayashi, T.; Nakada, M. Potential Strategies Overcoming the Temozolomide Resistance for Glioblastoma. Neurol. Med. Chir. 2018, 58, 405–421. [Google Scholar] [CrossRef] [Green Version]
- Gwak, H.S.; Kim, T.H.; Jo, G.H.; Kim, Y.J.; Kwak, H.J.; Kim, J.H.; Yin, J.; Yoo, H.; Lee, S.H.; Park, J.B. Silencing of microRNA-21 confers radio-sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS ONE 2012, 7, e47449. [Google Scholar] [CrossRef]
- Yue, X.; Lan, F.; Xia, T. Hypoxic Glioma Cell-Secreted Exosomal miR-301a Activates Wnt/β-catenin Signaling and Promotes Radiation Resistance by Targeting TCEAL7. Mol. Ther. 2019, 27, 1939–1949. [Google Scholar] [CrossRef]
- Yang, J.-K.; Yang, J.-P.; Tong, J.; Jing, S.-Y.; Fan, B.; Wang, F.; Sun, G.-Z.; Jiao, B.-H. Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J. Neuro-Oncol. 2017, 131, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Yang, L.; Zhang, X.; Peng, X.; Wei, S.; Su, D.; Zhai, Z.; Hua, X.; Li, H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018, 414, 107–115. [Google Scholar] [CrossRef]
- Wang, B.; Wang, K.; Jin, T.; Xu, Q.; He, Y.; Cui, B.; Wang, Y. NCK1-AS1 enhances glioma cell proliferation, radioresistance and chemoresistance via miR-22-3p/IGF1R ceRNA pathway. Biomed. Pharmacother. 2020, 129, 110395. [Google Scholar] [CrossRef]
- Wu, C.; Su, J.; Long, W.; Qin, C.; Wang, X.; Xiao, K.; Li, Y.; Xiao, Q.; Wang, J.; Pan, Y.; et al. LINC00470 promotes tumour proliferation and invasion, and attenuates chemosensitivity through the LINC00470/miR-134/Myc/ABCC1 axis in glioma. J. Cell. Mol. Med. 2020, 24, 12094–12106. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Baker, S.; Dahele, M.; Lagerwaard, F.J.; Senan, S. A critical review of recent developments in radiotherapy for non-small cell lung cancer. Radiat. Oncol. 2016, 11, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Huang, Z.; Xu, Y.; Chen, X.; Chen, T.; Ye, Y.; Ding, J.; Chen, Z.; Chen, L.; Qiu, X.; et al. A three-lncRNA signature predicts clinical outcomes in low-grade glioma patients after radiotherapy. Aging 2020, 12, 9188–9204. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Mehta, M.P. Low-Grade Glioma Radiotherapy Treatment and Trials. Neurosurg. Clin. N. Am. 2019, 30, 111–118. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, S.; Yang, B.; Mao, W.; Yang, X.; Cai, J. Molecular mechanisms of lncRNAs in regulating cancer cell radiosensitivity. Biosci. Rep. 2019, 39, BSR20190590. [Google Scholar] [CrossRef]
- Wang, C.; Yu, G.; Xu, Y.; Liu, C.; Sun, Q.; Li, W.; Sun, J.; Jiang, Y.; Ye, L. Knockdown of Long Non-Coding RNA HCP5 Increases Radiosensitivity Through Cellular Senescence by Regulating microRNA-128 in Gliomas. Cancer Manag. Res. 2021, 13, 3723–3737. [Google Scholar] [CrossRef] [PubMed]
- Naka-Kaneda, H.; Nakamura, S.; Igarashi, M.; Aoi, H.; Kanki, H.; Tsuyama, J.; Tsutsumi, S.; Aburatani, H.; Shimazaki, T.; Okano, H. The miR-17/106-p38 axis is a key regulator of the neurogenic-to-gliogenic transition in developing neural stem/progenitor cells. Proc. Natl. Acad. Sci. USA 2014, 111, 1604–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nat. Cell Biol. 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak-Wolf, A.; Maier, L.; Mackowiak, S.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nat. Cell Biol. 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Batchelor, T. Temozolomide for malignant brain tumours. Lancet 2000, 355, 1115–1116. [Google Scholar] [CrossRef]
- Yoshimoto, K.; Mizoguchi, M.; Hata, N.; Murata, H.; Hatae, R.; Amano, T.; Nakamizo, A.; Sasaki, T. Complex DNA repair pathways as possible therapeutic targets to overcome temozolomide resistance in glioblastoma. Front. Oncol. 2012, 2, 186. [Google Scholar] [CrossRef] [Green Version]
- Ochs, K.; Kaina, B. Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/3 regulated and Fas/caspase-8 independent. Cancer Res. 2000, 60, 5815–5824. [Google Scholar] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perazzoli, G.; Prados, J.; Ortiz, R.; Caba, O.; Cabeza, L.; Berdasco, M.; Gónzalez, B.; Melguizo, C. Temozolomide Resistance in Glioblastoma Cell Lines: Implication of MGMT, MMR, P-Glycoprotein and CD133 Expression. PLoS ONE 2015, 10, e0140131. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, K.; Scheithauer, B.W.; Lombardero, M.; McLendon, R.E.; Syro, L.V.; Uribe, H.; Ortiz, L.D.; Penagos, L.C. MGMT immunoexpression predicts responsiveness of pituitary tumors to temozolomide therapy. Acta Neuropathol. 2008, 115, 261–262. [Google Scholar] [CrossRef]
- Gerson, S.L. MGMT: Its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer 2004, 4, 296–307. [Google Scholar] [CrossRef]
- Shen, F.; Chang, H.; Gao, G.; Zhang, B.; Li, X.; Jin, B. Long noncoding RNA FOXD2-AS1 promotes glioma malignancy and tumorigenesis via targeting miR-185-5p/CCND2 axis. J. Cell Biochem. 2019, 120, 9324–9336. [Google Scholar] [CrossRef]
- Shangguan, W.; Lv, X.; Tian, N. FoxD2-AS1is a prognostic factor in glioma and promotes temozolomide resistance in a O6-methylguanine-DNA methyltransferase-dependent manner. Korean J. Physiol. Pharmacol. 2019, 23, 475–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Cai, J.; Chen, Q.; Han, B.; Meng, X.; Li, Y.; Li, Z.; Wang, R.; Lin, L.; Duan, C.; et al. Lnc-TALC promotes O6-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nat. Commun. 2019, 10, 2045. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Fang, S.; Cheng, Y.; Zhou, C.; Deng, F. The long non-coding RNA, urothelial carcinoma associated 1, promotes cell growth, invasion, migration, and chemo-resistance in glioma through Wnt/β-catenin signaling pathway. Aging 2019, 11, 8239–8253. [Google Scholar] [CrossRef] [PubMed]
- Ujifuku, K.; Mitsutake, N.; Takakura, S.; Matsuse, M.; Saenko, V.; Suzuki, K.; Hayashi, K.; Matsuo, T.; Kamada, K.; Nagata, I.; et al. miR-195, miR-455-3p and miR-10a∗ are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 2010, 296, 241–248. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.; Liu, B.; Lian, C.; Doycheva, D.M.; Fu, Z.; Liu, Y.; Zhou, J.; He, Z.; Yang, Z.; Huang, Q.; et al. Long noncoding RNA AC003092.1 promotes temozolomide chemosensitivity through miR-195/TFPI-2 signaling modulation in glioblastoma. Cell Death Dis. 2018, 9, 1139. [Google Scholar] [CrossRef]
- Mazor, G.; Levin, L.; Picard, D.; Ahmadov, U.; Carén, H.; Borkhardt, A.; Reifenberger, G.; Leprivier, G.; Remke, M.; Rotblat, B. The lncRNA TP73-AS1 is linked to aggressiveness in glioblastoma and promotes temozolomide resistance in glioblastoma cancer stem cells. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Liu, B.; Zhou, J.; Wang, C.; Chi, Y.; Wei, Q.; Fu, Z.; Lian, C.; Huang, Q.; Liao, C.; Yang, Z.; et al. lncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis. 2020, 11, 384. [Google Scholar] [CrossRef]
- Chen, Y.G.; Satpathy, A.; Chang, H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017, 18, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Hur, K.; Kim, S.-H.; Kim, J.-M. Potential Implications of Long Noncoding RNAs in Autoimmune Diseases. Immune Netw. 2019, 19, e4. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, T.; Zhou, W.; Li, J.; Li, X.; Wang, Q.; Jin, X.; Yin, J.; Chen, L.; Zhang, Y.; et al. Pan-cancer characterization of im-mune-related lncRNAs identifies potential oncogenic biomarkers. Nat. Commun. 2020, 11, 1000. [Google Scholar] [CrossRef] [Green Version]
- Poeta, V.M.; Massara, M.; Capucetti, A.; Bonecchi, R. Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Front. Immunol. 2019, 10, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, A.C.; Korkut, A.; Kanchi, R.S.; Hegde, A.M.; Lenoir, W.; Liu, W.; Liu, Y.; Fan, H.; Shen, H.; Ravikumar, V.; et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell 2018, 33, 690–705.e9. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Findlay, G.; Bandukwala, H.S.; Oberdoerffer, S.; Baust, B.; Li, Z.; Schmidt, V.; Hogan, P.G.; Sacks, D.B.; Rao, A. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc. Natl. Acad. Sci. USA 2011, 108, 11381–11386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Q.; Ye, Y.; Chan, L.-C.; Li, Y.; Liang, K.; Lin, A.; Egranov, S.D.; Zhang, Y.; Xia, W.; Gong, J.; et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat. Immunol. 2019, 20, 835–851. [Google Scholar] [CrossRef]
- Mirsafian, H.; Manda, S.S.; Mitchell, C.J.; Sreenivasamurthy, S.; Ripen, A.M.; Bin Mohamad, S.; Merican, A.F.; Pandey, A. Long non-coding RNA expression in primary human monocytes. Genomics 2016, 108, 37–45. [Google Scholar] [CrossRef]
- Wang, X.; Gao, M.; Ye, J.; Jiang, Q.; Yang, Q.; Zhang, C.; Wang, S.; Zhang, J.; Wang, L.; Wu, J.; et al. An Immune Gene-Related Five-lncRNA Signature for to Predict Glioma Prognosis. Front. Genet. 2020, 11, 612037. [Google Scholar] [CrossRef]
- Wen, J.; Wang, Y.; Luo, L.; Peng, L.; Chen, C.; Guo, J.; Ge, Y.; Li, W.; Jin, X. Identification and Verification on Prognostic Index of Lower-Grade Glioma Immune-Related lncRNAs. Front. Oncol. 2020, 10, 578809. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhu, J.; Chen, H.; Qian, J.; Zhang, L.; Wan, Z.; Chen, F.; Sun, S.; Li, W.; Luo, C. A novel lncRNA-LINC01116 regulates tumorigenesis of glioma by targeting VEGFA. Int. J. Cancer 2020, 146, 248–261. [Google Scholar] [CrossRef]
- Cheng, J.; Meng, J.; Zhu, L.; Peng, Y. Exosomal noncoding RNAs in Glioma: Biological functions and potential clinical ap-plications. Mol. Cancer 2020, 19, 66. [Google Scholar] [CrossRef] [PubMed]
| lncRNA | Interacting Molecules | Effects of lncRNA in Glioma Cells | Ref. |
|---|---|---|---|
| HOTAIRM1 | miR-495-3p and miR-129-5p | Promoting cell proliferation, EMT and TMZ resistance | [21] |
| CCAT2 | - | Suppressing endothelial cell apoptosis leading to angiogenesis | [40] |
| CRNDE | miR-136-5p | Promoting cell proliferation and invasion | [19,42] |
| H19 | miR-152 and miR-130a-3p | Promoting cell proliferation and EMT | [43,44] |
| PVT1 | miR-128-3p | Promoting cell proliferation, invasion and migration and inhibiting apoptosis | [45] |
| LINC00689 | miR-338-3p | Promoting cell growth, metastasis and glucose metabolism | [46] |
| XIST | miR-329-3p | Promoting cell proliferation, invasion and inhibiting cell apoptosis | [18] |
| circATP8B4 | miR-766-5p | Promoting cell proliferation and radioresistance | [47] |
| SBF2-AS1 | miR-151a-3p and miR-338-3p | Suppressing cell apoptosis and growth inhibition induced by TMZ, and stimulating cell viability, migration and tube formation of endothelial cells | [48,49] |
| MIR155HG | - | Promoting cell growth and expression of immune checkpoint inhibitors | [50] |
| lncRNA | Interacting Molecules | Effects of lncRNA in Glioma Cells | Ref. |
|---|---|---|---|
| RP11-838N2.4 | miR-10a | Enhancing sensitivity of TMZ | [51] |
| MALAT1 | miR-155 | Inhibiting cell viability and proliferation and invasion | [52] |
| TUG1 | caspase3, caspase 9, and BCL-2 | Inhibiting cell proliferation and promoting cell apoptosis | [55] |
| PART1 | miR-190a-3p | Inhibiting cell proliferation and promoting cell apoptosis | [57] |
| MIAT | miR-140-3p | Increasing blood–tumor barrier permeability | [58] |
| PAR5 | EZH2 | Inhibiting cell proliferation, invasion and migration | [59] |
| TPTEP1 | miR-106a-5p | Inhibiting cell stemness and radioresistance | [60] |
| DGCR5 | miR-21-3p and miR-23a-5p | Inhibiting Wnt/β-catenin signaing pathway and promoting cell apoptosis, migration and invasion | [61,62] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chae, Y.; Roh, J.; Kim, W. The Roles Played by Long Non-Coding RNAs in Glioma Resistance. Int. J. Mol. Sci. 2021, 22, 6834. https://doi.org/10.3390/ijms22136834
Chae Y, Roh J, Kim W. The Roles Played by Long Non-Coding RNAs in Glioma Resistance. International Journal of Molecular Sciences. 2021; 22(13):6834. https://doi.org/10.3390/ijms22136834
Chicago/Turabian StyleChae, Yeonsoo, Jungwook Roh, and Wanyeon Kim. 2021. "The Roles Played by Long Non-Coding RNAs in Glioma Resistance" International Journal of Molecular Sciences 22, no. 13: 6834. https://doi.org/10.3390/ijms22136834
APA StyleChae, Y., Roh, J., & Kim, W. (2021). The Roles Played by Long Non-Coding RNAs in Glioma Resistance. International Journal of Molecular Sciences, 22(13), 6834. https://doi.org/10.3390/ijms22136834







