Proteomics Analysis of Andrographolide-Induced Apoptosis via the Regulation of Tumor Suppressor p53 Proteolysis in Cervical Cancer-Derived Human Papillomavirus 16-Positive Cell Lines
Abstract
1. Introduction
2. Results
2.1. Cytotoxicity of Androg on Cervical Cancer Cells
2.2. Androg Induced Apoptotic Cell Death in HPV16-Positive Cervical Cell Lines
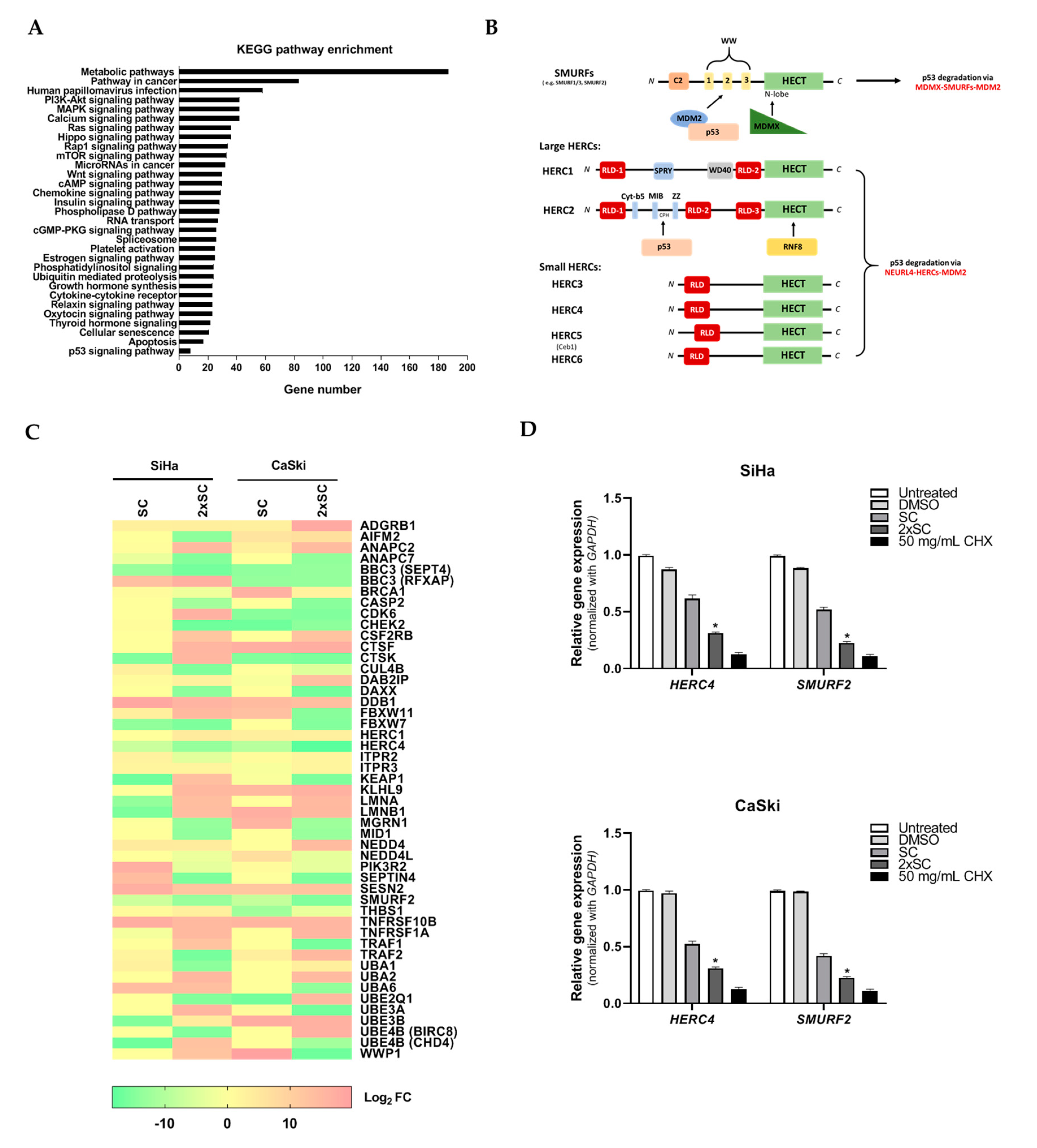
2.3. Proteomic Profiling of Androg-Treated HPV16-Positive Cervical Cells
2.4. Identification of Commonly Regulated Proteins in Androg-Treated HPV16-Positive Cervical Cells
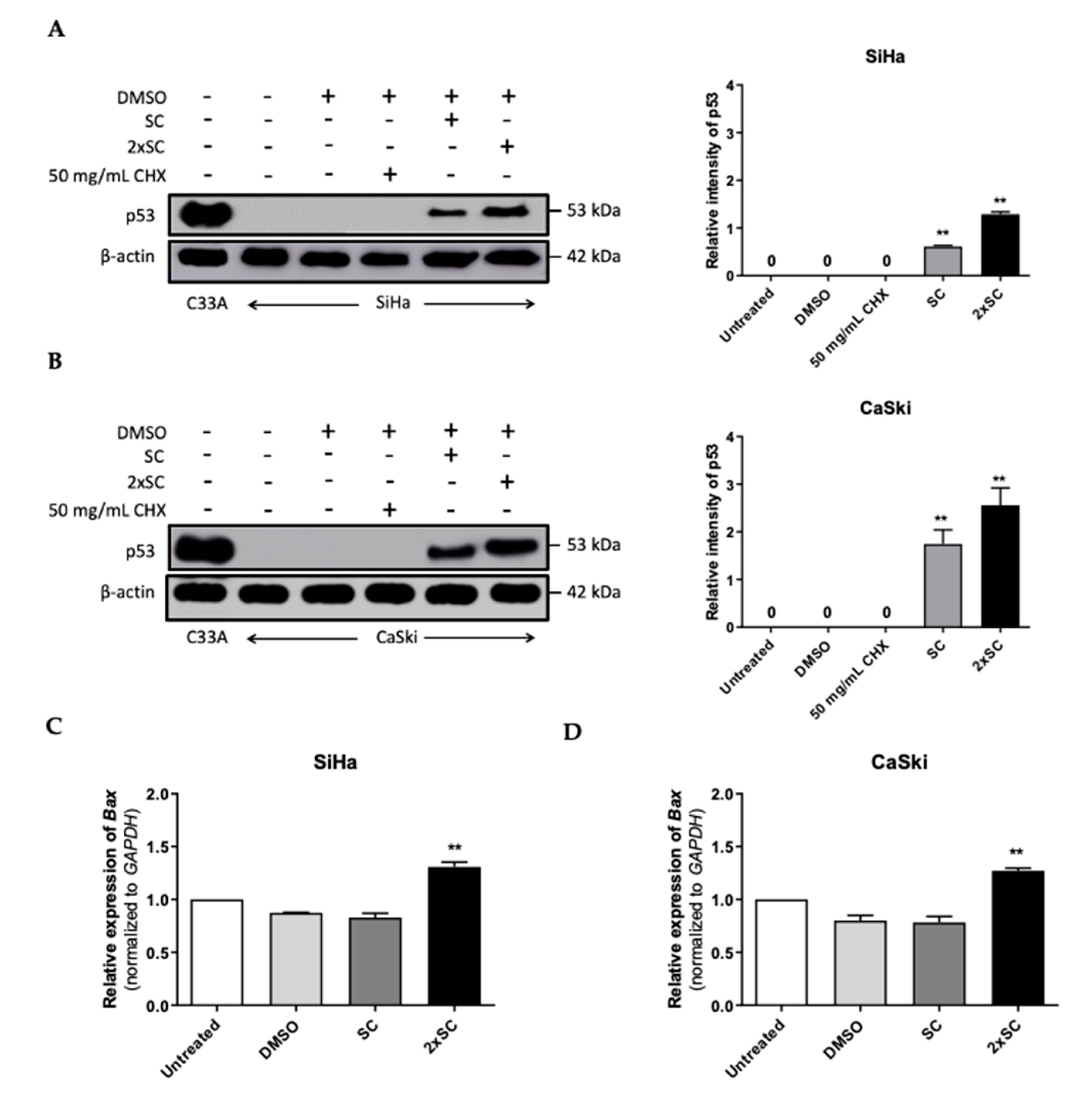
2.5. Androg Altered the Expression of HERC4 and SMURF2 from the Ubiquitin-Mediated Proteolysis Pathway and Restored p53 in HPV16-Positive Cervical Cells
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cultures
4.2. Andrographolide Compound and CC50 Determination
4.3. Detection of Apoptosis by Ethidium Bromide/Acridine Orange (EB/AO) Staining and Flow Cytometry
4.4. Determination of Genes Expression by Quantitative Reverse Transcriptase-PCR (qRT-PCR)
4.5. Measurement of p53 Protein Expression in Androg-Treated Cells by Immunoblotting
4.6. Preparation of Proteomic Samples
4.7. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
4.8. Data Processing of Mass Spectrometry (MS)
4.9. Analysis of LC-MS/MS Data
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Psyrri, A.; DiMaio, D. Human papillomavirus in cervical and head-and-neck cancer. Nat. Clin. Pr. Oncol. 2008, 5, 24–31. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.K.; Wise-Draper, T.M.; Wells, S.I. Human papillomavirus induced transformation in cervical and head and neck cancers. Cancers 2014, 6, 1793–1820. [Google Scholar] [CrossRef]
- Thierry, F. Transcriptional regulation of the papillomavirus oncogenes by cellular and viral transcription factors in cervical carcinoma. Virology 2009, 384, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Carson, A.; Khan, S.A. Characterization of transcription factor binding to human papillomavirus type 16 DNA during cellular differentiation. J. Virol. 2006, 80, 4356–4362. [Google Scholar] [CrossRef]
- Reinstein, E.; Scheffner, M.; Oren, M.; Ciechanover, A.; Schwartz, A. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: Targeting via ubiquitination of the N-terminal residue. Oncogene 2000, 19, 5944–5950. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Kundu, R. Human Papillomavirus E6 and E7: The Cervical Cancer Hallmarks and Targets for Therapy. Front Microbiol. 2019, 10, 3116. [Google Scholar] [CrossRef] [PubMed]
- Cutts, F.T.; Franceschi, S.; Goldie, S.; Castellsague, X.; de Sanjose, S.; Garnett, G.; Edmunds, W.J.; Claeys, P.; Goldenthal, K.L.; Harper, D.M.; et al. Human papillomavirus and HPV vaccines: A review. Bull. World Health Organ. 2007, 85, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Joura, E.A.; Giuliano, A.R.; Iversen, O.E.; Bouchard, C.; Mao, C.; Mehlsen, J.; Moreira, E.D., Jr.; Ngan, Y.; Petersen, L.K.; Lazcano-Ponce, E.; et al. A 9-valent HPV vaccine against infection and intraepithelial neoplasia in women. N. Engl. J. Med. 2015, 372, 711–723. [Google Scholar] [CrossRef]
- Farooq, Q.U.A.; Shaukat, Z.; Zhou, T.; Aiman, S.; Gong, W.; Li, C. Inferring Virus-Host relationship between HPV and its host Homo sapiens using protein interaction network. Sci. Rep. 2020, 10, 8719. [Google Scholar] [CrossRef]
- Huibregtse, J.M.; Scheffner, M.; Beaudenon, S.; Howley, P.M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 1995, 92, 2563–2567. [Google Scholar] [CrossRef]
- Jiang, P.; Yue, Y. Human papillomavirus oncoproteins and apoptosis (Review). ExpMed. 2014, 7, 3–7. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.K.; Li, Y.; Hafner, M.; Banerjee, N.S.; Tang, S.; Briskin, D.; Meyers, C.; Chow, L.T.; Xie, X.; et al. microRNAs are biomarkers of oncogenic human papillomavirus infections. Proc. Natl. Acad. Sci. USA 2014, 111, 4262–4267. [Google Scholar] [CrossRef]
- Ehrke-Schulz, E.; Heinemann, S.; Schulte, L.; Schiwon, M.; Ehrhardt, A. Adenoviral Vectors Armed with PAPILLOMAVIRUs Oncogene Specific CRISPR/Cas9 Kill Human-Papillomavirus-Induced Cervical Cancer Cells. Cancers 2020, 12, 1934. [Google Scholar] [CrossRef] [PubMed]
- Shankar, S.; Prasad, D.; Sanawar, R.; Das, A.V.; Pillai, M.R. TALEN based HPV-E7 editing triggers necrotic cell death in cervical cancer cells. Sci. Rep. 2017, 7, 5500. [Google Scholar] [CrossRef] [PubMed]
- Dymalla, S.; Scheffner, M.; Weber, E.; Sehr, P.; Lohrey, C.; Hoppe-Seyler, F.; Hoppe-Seyler, K. A novel peptide motif binding to and blocking the intracellular activity of the human papillomavirus E6 oncoprotein. J. Mol. Med. 2009, 87, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Jena, L.; Mohod, K.; Daf, S.; Varma, A.K. Virtual Screening for Potential Inhibitors of High-Risk Human Papillomavirus 16 E6 Protein. Interdiscip. Sci. 2015, 7, 136–142. [Google Scholar] [CrossRef]
- Khan, I.; Khan, F.; Farooqui, A.; Ansari, I.A. Andrographolide Exhibits Anticancer Potential Against Human Colon Cancer Cells by Inducing Cell Cycle Arrest and Programmed Cell Death via Augmentation of Intracellular Reactive Oxygen Species Level. Nutr. Cancer 2018, 70, 787–803. [Google Scholar] [CrossRef]
- Lee, M.J.; Rao, Y.K.; Chen, K.; Lee, Y.C.; Chung, Y.S.; Tzeng, Y.M. Andrographolide and 14-deoxy-11,12-didehydroandrographolide from Andrographis paniculata attenuate high glucose-induced fibrosis and apoptosis in murine renal mesangeal cell lines. J. Ethnopharmacol. 2010, 132, 497–505. [Google Scholar] [CrossRef]
- Kumar, R.A.; Sridevi, K.; Kumar, N.V.; Nanduri, S.; Rajagopal, S. Anticancer and immunostimulatory compounds from Andrographis paniculata. J. Ethnopharmacol. 2004, 92, 291–295. [Google Scholar] [CrossRef]
- Matsuda, T.; Kuroyanagi, M.; Sugiyama, S.; Umehara, K.; Ueno, A.; Nishi, K. Cell differentiation-inducing diterpenes from Andrographis paniculata Nees. Chem. Pharm. Bull. 1994, 42, 1216–1225. [Google Scholar] [CrossRef]
- Shi, M.D.; Lin, H.H.; Lee, Y.C.; Chao, J.K.; Lin, R.A.; Chen, J.H. Inhibition of cell-cycle progression in human colorectal carcinoma Lovo cells by andrographolide. Chem. Biol. Interact. 2008, 174, 201–210. [Google Scholar] [CrossRef]
- Satyanarayana, C.; Deevi, D.S.; Rajagopalan, R.; Srinivas, N.; Rajagopal, S. DRF 3188 a novel semi-synthetic analog of andrographolide: Cellular response to MCF 7 breast cancer cells. BMC Cancer 2004, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.C.; Jeon, H.J.; Kee, K.H.; Lee, M.J.; Hong, R.; Han, S.I. Andrographolide induces apoptotic and non-apoptotic death and enhances tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in gastric cancer cells. Oncol. Lett. 2017, 13, 3837–3844. [Google Scholar] [CrossRef] [PubMed]
- Ekalaksananan, T.; Sookmai, W.; Fangkham, S.; Pientong, C.; Aromdee, C.; Seubsasana, S.; Kongyingyoes, B. Activity of Andrographolide and Its Derivatives on HPV16 Pseudovirus Infection and Viral Oncogene Expression in Cervical Carcinoma Cells. Nutr. Cancer 2015, 67, 687–696. [Google Scholar] [CrossRef]
- Liu, S.H.; Lin, C.H.; Liang, F.P.; Chen, P.F.; Kuo, C.D.; Alam, M.M.; Maiti, B.; Hung, S.K.; Chi, C.W.; Sun, C.M.; et al. Andrographolide downregulates the v-Src and Bcr-Abl oncoproteins and induces Hsp90 cleavage in the ROS-dependent suppression of cancer malignancy. Biochem. Pharm. 2014, 87, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Nateewattana, J.; Dutta, S.; Reabroi, S.; Saeeng, R.; Kasemsook, S.; Chairoungdua, A.; Weerachayaphorn, J.; Wongkham, S.; Piyachaturawat, P. Induction of apoptosis in cholangiocarcinoma by an andrographolide analogue is mediated through topoisomerase II alpha inhibition. Eur. J. Pharm. 2014, 723, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, C.; Berman, S.H.; Babish, J.G.; Ma, X.; Shinto, L.; Dorr, M.; Wells, K.; Wenner, C.A.; Standish, L.J. A phase I trial of andrographolide in HIV positive patients and normal volunteers. Phytother. Res. 2000, 14, 333–338. [Google Scholar] [CrossRef]
- Lee, J.C.; Tseng, C.K.; Young, K.C.; Sun, H.Y.; Wang, S.W.; Chen, W.C.; Lin, C.K.; Wu, Y.H. Andrographolide exerts anti-hepatitis C virus activity by up-regulating haeme oxygenase-1 via the p38 MAPK/Nrf2 pathway in human hepatoma cells. Br. J. Pharm. 2014, 171, 237–252. [Google Scholar] [CrossRef]
- Ding, Y.; Chen, L.; Wu, W.; Yang, J.; Yang, Z.; Liu, S. Andrographolide inhibits influenza A virus-induced inflammation in a murine model through NF-kappaB and JAK-STAT signaling pathway. Microbes. Infect. 2017, 19, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Paemanee, A.; Hitakarun, A.; Wintachai, P.; Roytrakul, S.; Smith, D.R. A proteomic analysis of the anti-dengue virus activity of andrographolide. Biomed. Pharm. 2019, 109, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Wintachai, P.; Kaur, P.; Lee, R.C.; Ramphan, S.; Kuadkitkan, A.; Wikan, N.; Ubol, S.; Roytrakul, S.; Chu, J.J.; Smith, D.R. Activity of andrographolide against chikungunya virus infection. Sci. Rep. 2015, 5, 14179. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ma, Y.B.; Huang, X.Y.; Geng, C.A.; Zhao, Y.; Wang, L.J.; Guo, R.H.; Liang, W.J.; Zhang, X.M.; Chen, J.J. Synthesis, structure-activity relationships and biological evaluation of dehydroandrographolide and andrographolide derivatives as novel anti-hepatitis B virus agents. Bioorg. Med. Chem. Lett. 2014, 24, 2353–2359. [Google Scholar] [CrossRef]
- Fangkham, S.; Ekalaksananan, T.; Aromdee, C.; Seubsasana, S.; Kongyingyoes, B.; Patarapadungkit, N.; Pientong, C. The effect of andrographolide on Human papillomavirus type 16 (HPV16) positive cervical cancer cells (SiHa). Int. J. Infect. Dis. 2012, 16, E80. [Google Scholar] [CrossRef][Green Version]
- Ebner, F.A.; Sailer, C.; Eichbichler, D.; Jansen, J.; Sladewska-Marquardt, A.; Stengel, F.; Scheffner, M. A ubiquitin variant-based affinity approach selectively identifies substrates of the ubiquitin ligase E6AP in complex with HPV-11 E6 or HPV-16 E6. J. Biol. Chem. 2020, 295, 15070–15082. [Google Scholar] [CrossRef]
- Wang, J.; Tan, X.F.; Nguyen, V.S.; Yang, P.; Zhou, J.; Gao, M.; Li, Z.; Lim, T.K.; He, Y.; Ong, C.S.; et al. A quantitative chemical proteomics approach to profile the specific cellular targets of andrographolide, a promising anticancer agent that suppresses tumor metastasis. Mol. Cell Proteom. 2014, 13, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Pappa, K.I.; Lygirou, V.; Kontostathi, G.; Zoidakis, J.; Makridakis, M.; Vougas, K.; Daskalakis, G.; Polyzos, A.; Anagnou, N.P. Proteomic Analysis of Normal and Cancer Cervical Cell Lines Reveals Deregulation of Cytoskeleton-associated Proteins. Cancer Genom. Proteom. 2017, 14, 253–266. [Google Scholar] [CrossRef]
- Ciniglia, C.; Pinto, G.; Sansone, C.; Pollio, A. Acridine orange/Ethidium bromide double staining test: A simple In-vitro assay to detect apoptosis induced by phenolic compounds in plant cells. Allelopath. J. 2010, 26, 301–308. [Google Scholar]
- Ribble, D.; Goldstein, N.B.; Norris, D.A.; Shellman, Y.G. A simple technique for quantifying apoptosis in 96-well plates. BMC Biotechnol. 2005, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.L.; Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef]
- Haupt, S.; Berger, M.; Goldberg, Z.; Haupt, Y. Apoptosis—The p53 network. J. Cell Sci. 2003, 116, 4077–4085. [Google Scholar] [CrossRef]
- Schuler, M.; Green, D.R. Mechanisms of p53-dependent apoptosis. Biochem. Soc. Trans. 2001, 29, 684–688. [Google Scholar] [CrossRef] [PubMed]
- Thornborrow, E.C.; Patel, S.; Mastropietro, A.E.; Schwartzfarb, E.M.; Manfredi, J.J. A conserved intronic response element mediates direct p53-dependent transcriptional activation of both the human and murine bax genes. Oncogene 2002, 21, 990–999. [Google Scholar] [CrossRef]
- Wongjampa, W.; Ekalaksananan, T.; Chopjitt, P.; Chuerduangphui, J.; Kleebkaow, P.; Patarapadungkit, N.; Pientong, C. Suppression of miR-22, a tumor suppressor in cervical cancer, by human papillomavirus 16 E6 via a p53/miR-22/HDAC6 pathway. PLoS ONE 2018, 13, e0206644. [Google Scholar] [CrossRef]
- Jaudan, A.; Sharma, S.; Malek, S.N.A.; Dixit, A. Induction of apoptosis by pinostrobin in human cervical cancer cells: Possible mechanism of action. PLoS ONE 2018, 13, e0191523. [Google Scholar] [CrossRef]
- Lomeli, N.; Di, K.; Czerniawski, J.; Guzowski, J.F.; Bota, D.A. Cisplatin-induced mitochondrial dysfunction is associated with impaired cognitive function in rats. Free Radic. Biol. Med. 2017, 102, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Luo, H.; Zhang, W.; Shen, Z.; Hu, X.; Zhu, X. Molecular mechanisms of cisplatin resistance in cervical cancer. Drug Des. Devel. 2016, 10, 1885–1895. [Google Scholar] [CrossRef]
- Pisani, P.; Bray, F.; Parkin, D.M. Estimates of the world-wide prevalence of cancer for 25 sites in the adult population. Int. J. Cancer 2002, 97, 72–81. [Google Scholar] [CrossRef]
- Liao, H.C.; Chou, Y.J.; Lin, C.C.; Liu, S.H.; Oswita, A.; Huang, Y.L.; Wang, Y.L.; Syu, J.L.; Sun, C.M.; Leu, C.M.; et al. Andrographolide and its potent derivative exhibit anticancer effects against imatinib-resistant chronic myeloid leukemia cells by downregulating the Bcr-Abl oncoprotein. Biochem. Pharm. 2019, 163, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.J.; Zhang, X.S.; He, D.L. Andrographolide sensitizes prostate cancer cells to TRAIL-induced apoptosis. Asian J. 2018, 20, 200–204. [Google Scholar] [CrossRef]
- Wang, Z.M.; Kang, Y.H.; Yang, X.; Wang, J.F.; Zhang, Q.; Yang, B.X.; Zhao, K.L.; Xu, L.P.; Yang, L.P.; Ma, J.X.; et al. Andrographolide radiosensitizes human esophageal cancer cell line ECA109 to radiation in vitro. Dis. Esophagus 2016, 29, 54–61. [Google Scholar] [CrossRef]
- Hsieh, M.J.; Chen, J.C.; Yang, W.E.; Chien, S.Y.; Chen, M.K.; Lo, Y.S.; Hsi, Y.T.; Chuang, Y.C.; Lin, C.C.; Yang, S.F. Dehydroandrographolide inhibits oral cancer cell migration and invasion through NF-kappa B-, AP-1-, and SP-1-modulated matrix metalloproteinase-2 inhibition. Biochem. Pharm. 2017, 130, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Alzaharna, M.; Alqouqa, I.; Cheung, H.Y. Taxifolin synergizes Andrographolide-induced cell death by attenuation of autophagy and augmentation of caspase dependent and independent cell death in HeLa cells. PLoS ONE 2017, 12, e0171325. [Google Scholar] [CrossRef] [PubMed]
- Bratton, D.L.; Fadok, V.A.; Richter, D.A.; Kailey, J.M.; Frasch, S.C.; Nakamura, T.; Henson, P.M. Polyamine regulation of plasma membrane phospholipid flip-flop during apoptosis. J. Biol. Chem. 1999, 274, 28113–28120. [Google Scholar] [CrossRef]
- Gupta, I.; Singh, K.; Varshney, N.K.; Khan, S. Delineating Crosstalk Mechanisms of the Ubiquitin Proteasome System That Regulate Apoptosis. Front Cell Dev. Biol. 2018, 6. [Google Scholar] [CrossRef]
- Sanchez-Tena, S.; Cubillos-Rojas, M.; Schneider, T.; Rosa, J.L. Functional and pathological relevance of HERC family proteins: A decade later. Cell Mol. Life Sci. 2016, 73, 1955–1968. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, S. Structural mechanisms of HECT-type ubiquitin ligases. Biol. Chem. 2018, 399, 127–145. [Google Scholar] [CrossRef] [PubMed]
- Beaudenon, S.; Huibregtse, J.M. HPV E6, E6AP and cervical cancer. BMC Biochem. 2008, 9, S4. [Google Scholar] [CrossRef] [PubMed]
- Emanuelli, A.; Borroni, A.P.; Apel-Sarid, L.; Shah, P.A.; Ayyathan, D.M.; Koganti, P.; Levy-Cohen, G.; Blank, M. Smurf2-Mediated Stabilization of DNA Topoisomerase IIalpha Controls Genomic Integrity. Cancer Res. 2017, 77, 4217–4227. [Google Scholar] [CrossRef]
- Koganti, P.; Levy-Cohen, G.; Blank, M. Smurfs in Protein Homeostasis, Signaling, and Cancer. Front. Oncol. 2018, 8, 295. [Google Scholar] [CrossRef] [PubMed]
- Emanuelli, A.; Manikoth Ayyathan, D.; Koganti, P.; Shah, P.A.; Apel-Sarid, L.; Paolini, B.; Detroja, R.; Frenkel-Morgenstern, M.; Blank, M. Altered Expression and Localization of Tumor Suppressive E3 Ubiquitin Ligase SMURF2 in Human Prostate and Breast Cancer. Cancers 2019, 11, 556. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jho, E.H. The protein stability of Axin, a negative regulator of Wnt signaling, is regulated by Smad ubiquitination regulatory factor 2 (Smurf2). J. Biol. Chem. 2010, 285, 36420–36426. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Huang, J.H.; Sampson, E.R.; Kim, K.O.; Zuscik, M.J.; O’Keefe, R.J.; Chen, D.; Rosier, R.N. Smurf2 induces degradation of GSK-3beta and upregulates beta-catenin in chondrocytes: A potential mechanism for Smurf2-induced degeneration of articular cartilage. Exp. Cell Res. 2009, 315, 2386–2398. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar] [PubMed]
- Kaewseekhao, B.; Roytrakul, S.; Yingchutrakul, Y.; Salao, K.; Reechaipichitkul, W.; Faksri, K. Proteomic analysis of infected primary human leucocytes revealed PSTK as potential treatment-monitoring marker for active and latent tuberculosis. PLoS ONE 2020, 15, e0231834. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Ruprecht, B.; Di Bernardo, J.; Wang, Z.; Mo, X.; Ursu, O.; Christopher, M.; Fernandez, R.B.; Zheng, L.; Dill, B.D.; Wang, H.; et al. A mass spectrometry-based proteome map of drug action in lung cancer cell lines. Nat. Chem. Biol. 2020, 16, 1111–1119. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
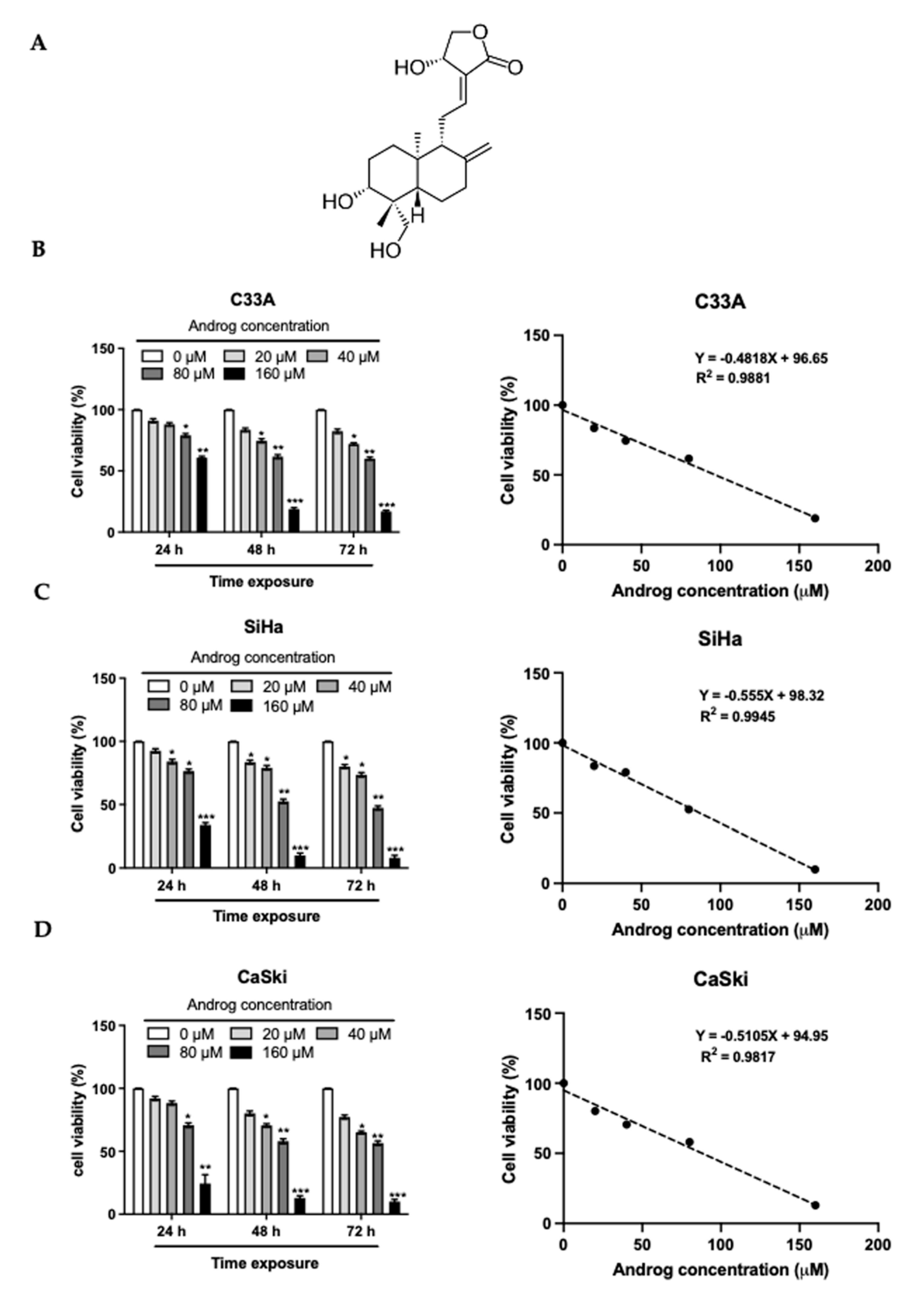



| C33A | SiHa | CaSki | ||||||
|---|---|---|---|---|---|---|---|---|
| CC50 (μM) | SC (μM) | 2xSC (μM) | CC50 (μM) | SC (μM) | 2xSC (μM) | CC50 (μM) | SC (μM) | 2xSC (μM) |
| 96.05 | 21.28 | 42.56 | 85.59 | 21.44 | 42.88 | 87.52 | 18.05 | 36.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Udomwan, P.; Pientong, C.; Tongchai, P.; Burassakarn, A.; Sunthamala, N.; Roytrakul, S.; Suebsasana, S.; Ekalaksananan, T. Proteomics Analysis of Andrographolide-Induced Apoptosis via the Regulation of Tumor Suppressor p53 Proteolysis in Cervical Cancer-Derived Human Papillomavirus 16-Positive Cell Lines. Int. J. Mol. Sci. 2021, 22, 6806. https://doi.org/10.3390/ijms22136806
Udomwan P, Pientong C, Tongchai P, Burassakarn A, Sunthamala N, Roytrakul S, Suebsasana S, Ekalaksananan T. Proteomics Analysis of Andrographolide-Induced Apoptosis via the Regulation of Tumor Suppressor p53 Proteolysis in Cervical Cancer-Derived Human Papillomavirus 16-Positive Cell Lines. International Journal of Molecular Sciences. 2021; 22(13):6806. https://doi.org/10.3390/ijms22136806
Chicago/Turabian StyleUdomwan, Pariyakorn, Chamsai Pientong, Panwad Tongchai, Ati Burassakarn, Nuchsupha Sunthamala, Sittiruk Roytrakul, Supawadee Suebsasana, and Tipaya Ekalaksananan. 2021. "Proteomics Analysis of Andrographolide-Induced Apoptosis via the Regulation of Tumor Suppressor p53 Proteolysis in Cervical Cancer-Derived Human Papillomavirus 16-Positive Cell Lines" International Journal of Molecular Sciences 22, no. 13: 6806. https://doi.org/10.3390/ijms22136806
APA StyleUdomwan, P., Pientong, C., Tongchai, P., Burassakarn, A., Sunthamala, N., Roytrakul, S., Suebsasana, S., & Ekalaksananan, T. (2021). Proteomics Analysis of Andrographolide-Induced Apoptosis via the Regulation of Tumor Suppressor p53 Proteolysis in Cervical Cancer-Derived Human Papillomavirus 16-Positive Cell Lines. International Journal of Molecular Sciences, 22(13), 6806. https://doi.org/10.3390/ijms22136806







