Drosophila to Explore Nucleolar Stress
Abstract
:1. Introduction
2. Results
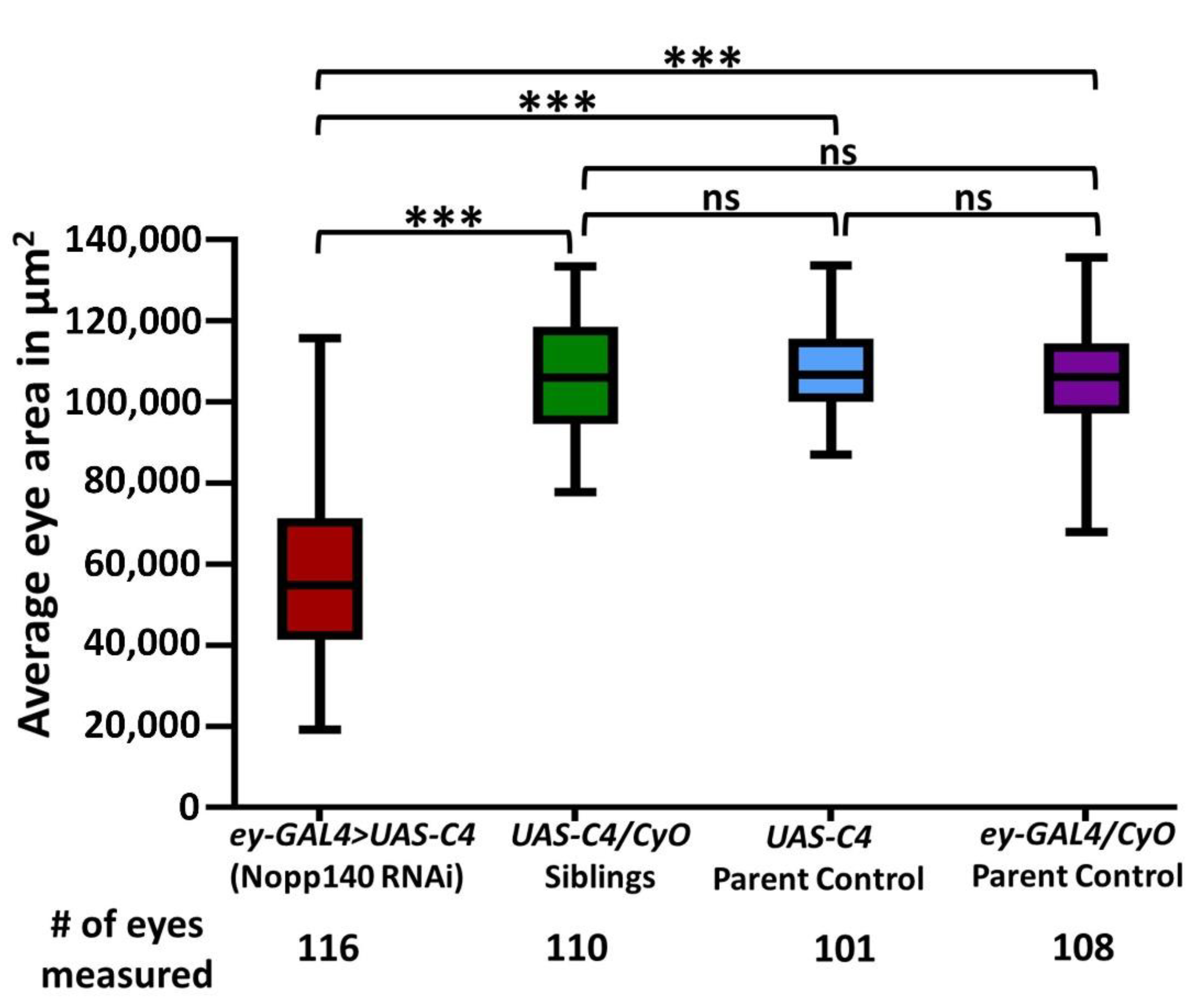
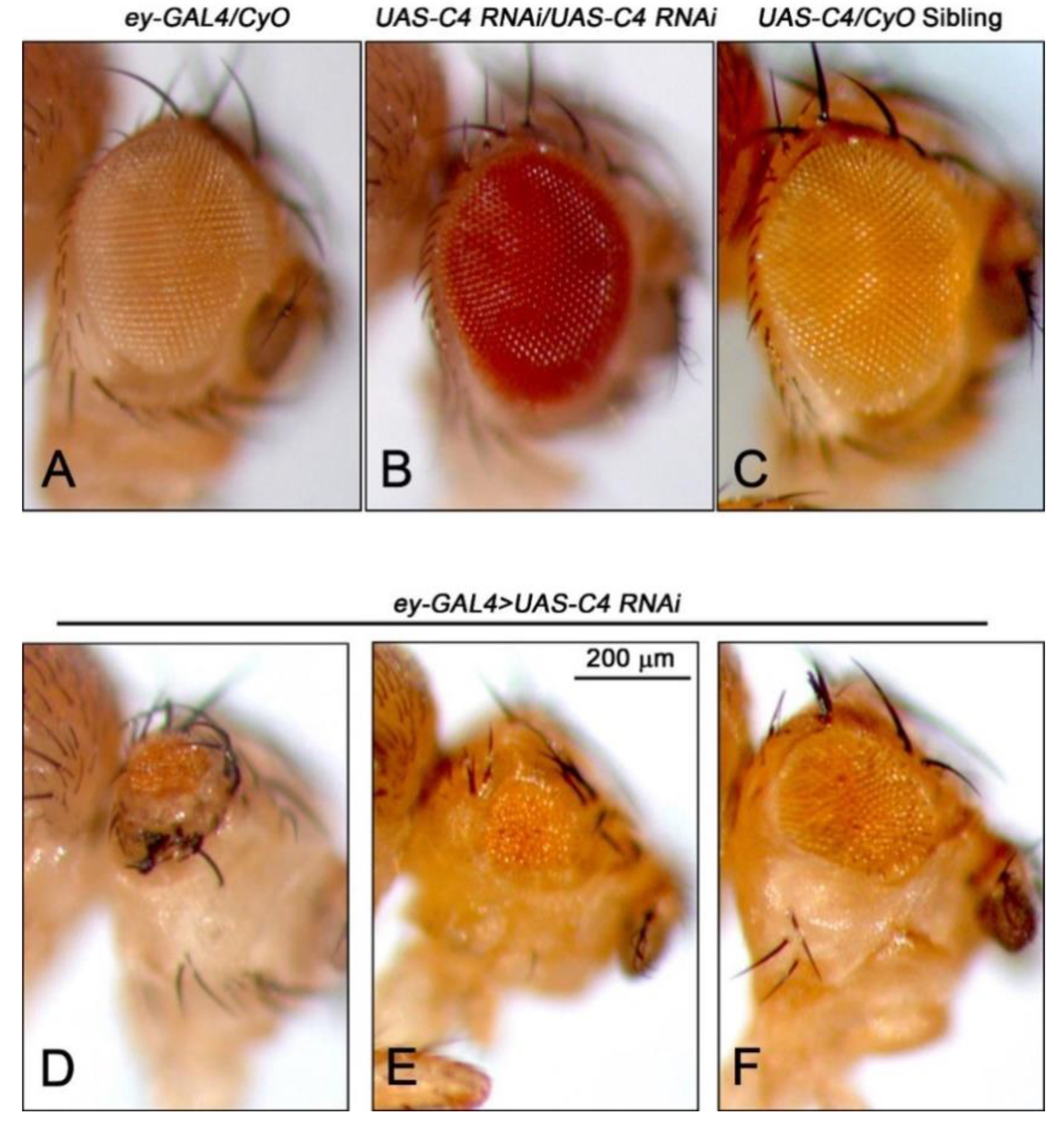
2.1. Depletion of Nopp140 in Eye Discs
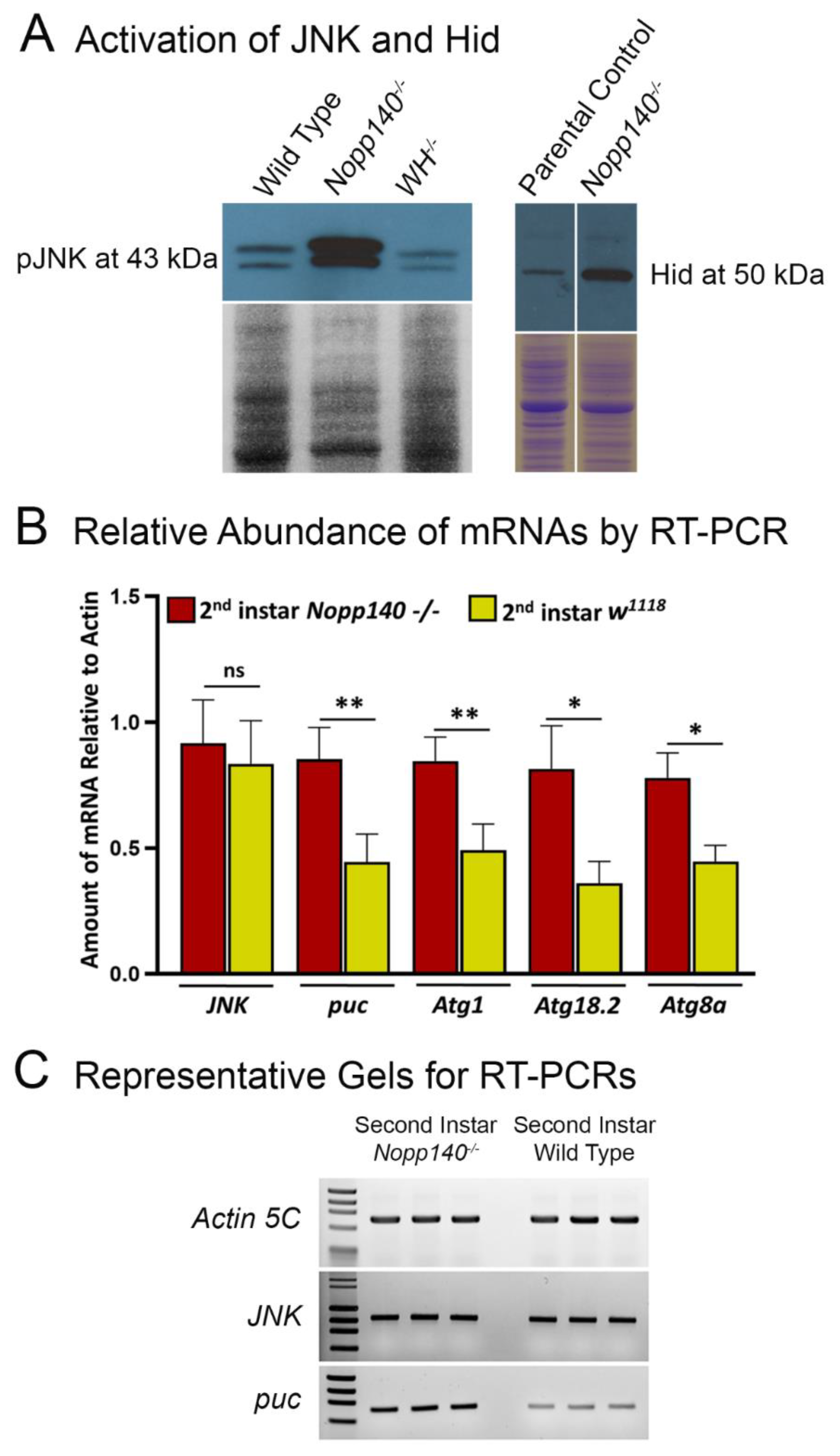
2.2. Stress-Related Proteins Are Upregulated in Homozygous Nopp140 Gene Knockout Larvae
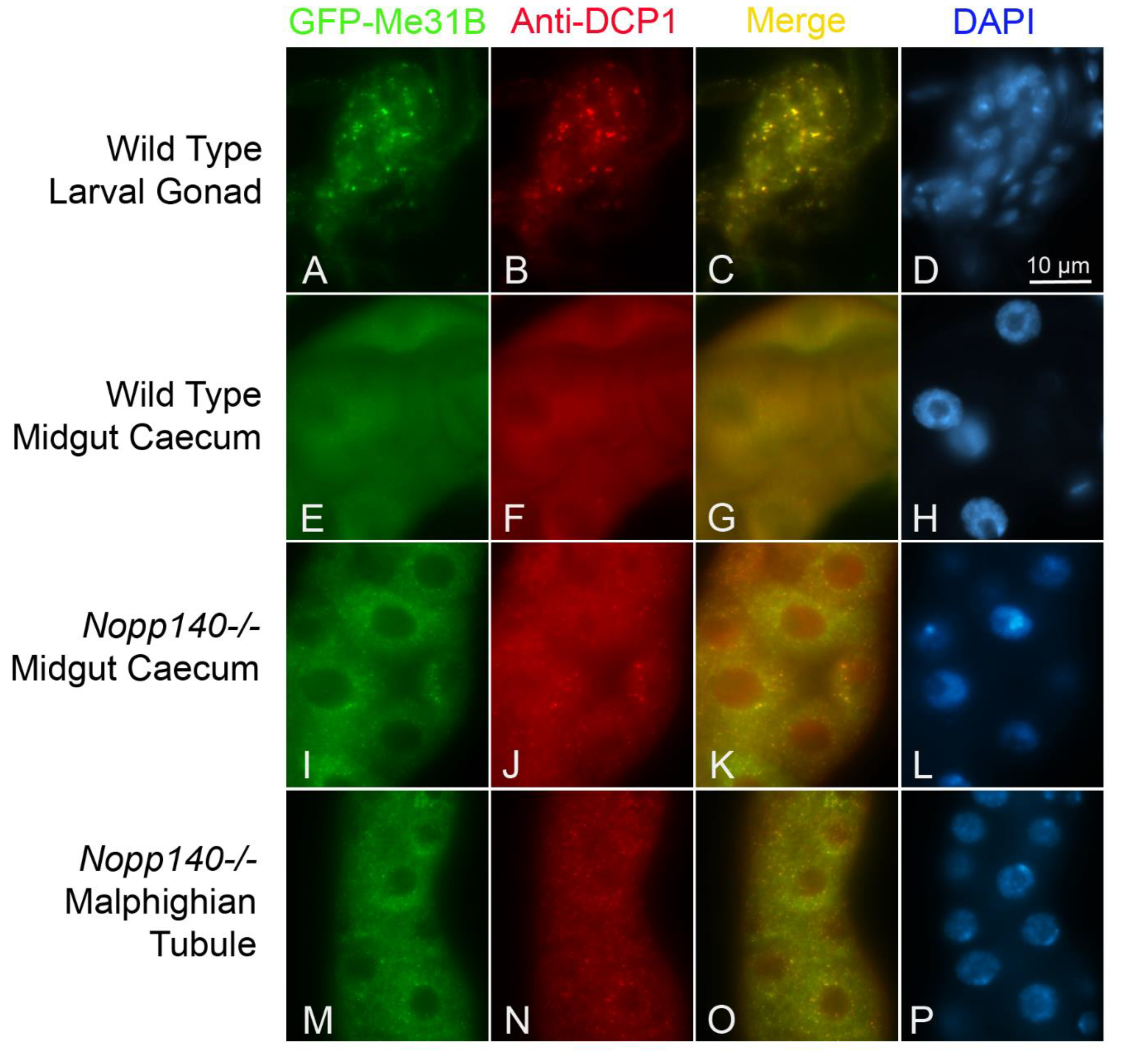
2.3. Processing(P)-Like Granules Appear in Polyploid Midgut of Nopp140-/- Larvae
2.4. Coilin Expression in Larval Neuroblasts
3. Discussion
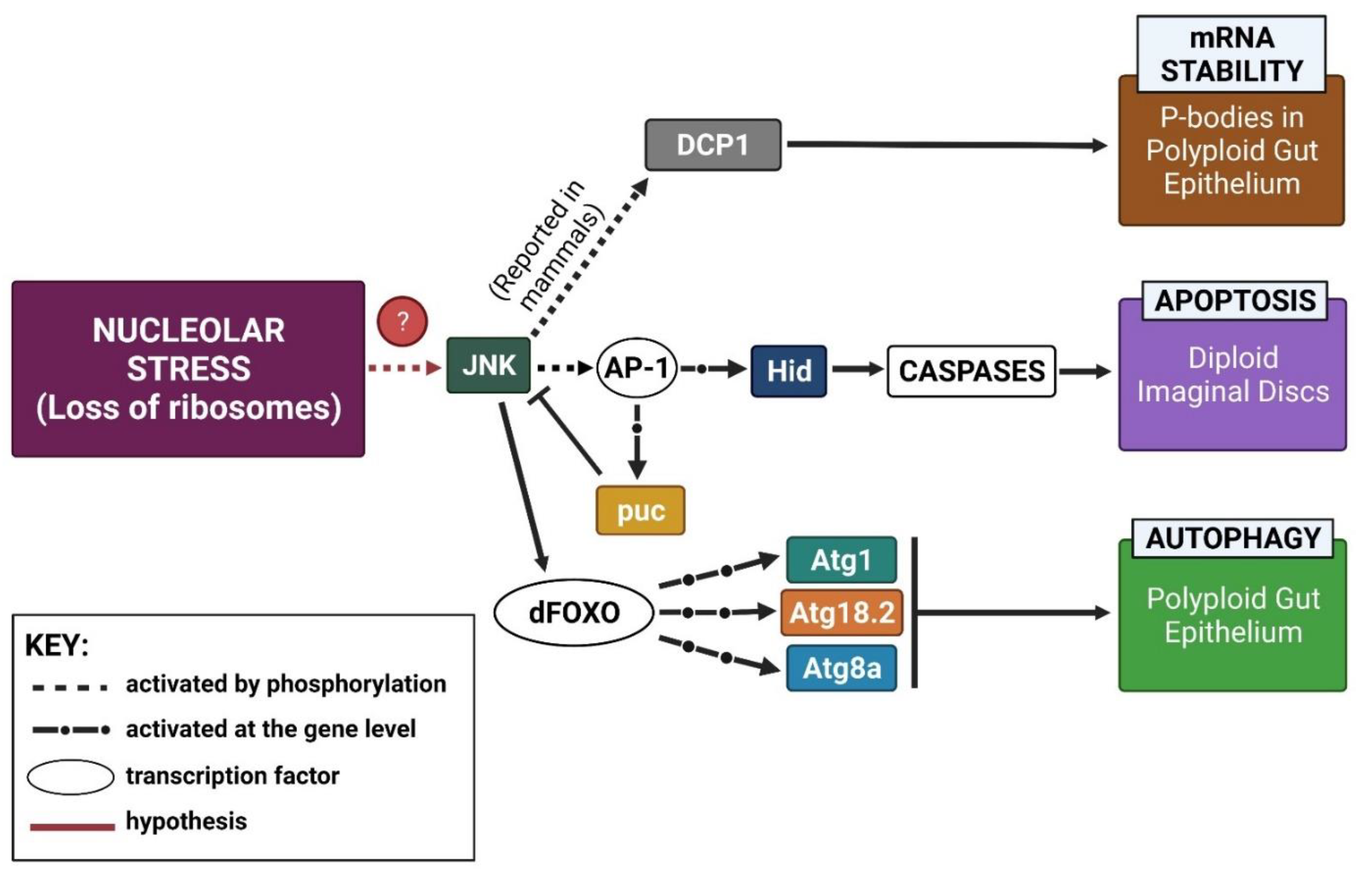
3.1. Working Model
3.2. Mushroom Body Lineages in Nopp140-/- Larvae Show Resilience to Nucleolar Stress
4. Materials and Methods
4.1. Fly Stocks
4.2. Assessing Eye Morphology
4.3. RT-PCRs
4.4. Antibodies and Immuno-Fluorescence
4.5. EdU Labeling
4.6. Transmission Electron Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.-M.; Lamond, A.I. The nucleolus under stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Grummt, I. The nucleolus—Guardian of cellular homeostasis and genome integrity. Chromosoma 2013, 122, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Pfister, A.S. Emerging role of the nucleolar stress response in autophagy. Front. Cell. Neurosci. 2019, 13, 156. [Google Scholar] [CrossRef] [Green Version]
- Brombin, A.; Joly, J.-S.; Jamen, F. New tricks for an old dog: Ribosome biogenesis contributes to stem cell homeostasis. Curr. Opin. Genet. Dev. 2015, 34, 61–70. [Google Scholar] [CrossRef]
- Honma, Y.; Kitamura, A.; Shioda, R.; Maruyama, H.; Ozaki, K.; Oda, Y.; Mini, T.; Jenö, P.; Maki, Y.; Yonezawa, K. TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients. EMBO J. 2006, 25, 3832–3842. [Google Scholar] [CrossRef] [Green Version]
- James, A.; Wang, Y.; Raje, H.; Rosby, R.; DiMario, P. Nucleolar stress with and without p53. Nucleus 2014, 5, 402–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, J.; Jones, N.C.; Sandell, L.L.; Jayasinghe, S.M.; Crane, J.; Rey, J.-P.; Dixon, M.J.; Trainor, P.A. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl. Acad. Sci. USA 2006, 103, 13403–13408. [Google Scholar] [CrossRef] [Green Version]
- Marygold, S.J.; Roote, J.; Reuter, G.; Lambertsson, A.; Ashburner, M.; Millburn, G.H.; Harrison, P.M.; Yu, Z.; Kenmochi, N.; Kaufman, T.C. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007, 8, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Narla, A.; Ebert, B.L. Ribosomopathies: Human disorders of ribosome dysfunction. Blood J. Am. Soc. Hematol. 2010, 115, 3196–3205. [Google Scholar] [CrossRef] [PubMed]
- Danilova, N.; Gazda, H.T. Ribosomopathies: How a common root can cause a tree of pathologies. Dis. Models Mech. 2015, 8, 1013–1026. [Google Scholar] [CrossRef] [Green Version]
- Vlachos, A.; Rosenberg, P.S.; Atsidaftos, E.; Alter, B.P.; Lipton, J.M. Incidence of neoplasia in Diamond Blackfan anemia: A report from the Diamond Blackfan Anemia Registry. Blood J. Am. Soc. Hematol. 2012, 119, 3815–3819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellis, S.R. Nucleolar stress in Diamond Blackfan anemia pathophysiology. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2014, 1842, 765–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landowski, M.; O’Donohue, M.-F.; Buros, C.; Ghazvinian, R.; Montel-Lehry, N.; Vlachos, A.; Sieff, C.A.; Newburger, P.E.; Niewiadomska, E.; Matysiak, M. Novel deletion of RPL15 identified by array-comparative genomic hybridization in Diamond–Blackfan anemia. Hum. Genet. 2013, 132, 1265–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipton, J.M.; Ellis, S.R. Diamond-Blackfan anemia: Diagnosis, treatment, and molecular pathogenesis. Hematol. Oncol. Clin. N. Am. 2009, 23, 261–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aggett, P.; Cavanagh, N.; Matthew, D.; Pincott, J.; Sutcliffe, J.; Harries, J. Shwachman’s syndrome. A review of 21 cases. Arch. Dis. Child. 1980, 55, 331–347. [Google Scholar] [CrossRef] [Green Version]
- Rujkijyanont, P.; Adams, S.L.; Beyene, J.; Dror, Y. Bone marrow cells from patients with Shwachman-Diamond syndrome abnormally express genes involved in ribosome biogenesis and RNA processing. Br. J. Haematol. 2009, 145, 806–815. [Google Scholar] [CrossRef]
- Jones, N.C.; Lynn, M.L.; Gaudenz, K.; Sakai, D.; Aoto, K.; Rey, J.-P.; Glynn, E.F.; Ellington, L.; Du, C.; Dixon, J. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 2008, 14, 125–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, E. The ribosomes of Drosophila. Mol. Gen. Genet. MGG 1974, 128, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Glover, D.M.; White, R.L.; Finnegan, D.J.; Hogness, D.S. Characterization of six cloned DNAs from Drosophila melanogaster, including one that contains the genes for rRNA. Cell 1975, 5, 149–157. [Google Scholar] [CrossRef]
- Long, E.O.; Dawid, I.B. Alternative pathways in the processing of ribosomal RNA precursor in Drosophila melanogaster. J. Mol. Biol. 1980, 138, 873–878. [Google Scholar] [CrossRef]
- DiMario, P.; James, A.; Raje, H. rDNA and Nucleologenesis in Drosophila. In Proteins of the Nucleolus; Springer: New York, NY, USA, 2013; pp. 39–78. [Google Scholar]
- Ritossa, F. The bobbed locus. In The Genetics and Biology of Drosophila; Ashburner, M., Novitski, E., Eds.; Academic Press: London, UK; New York, NY, USA, 1976; Volume 1b, pp. 801–846. [Google Scholar]
- Ashburner, M. Drosophila. A Laboratory Handbook; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Consortium, F. The FlyBase database of the Drosophila Genome Projects and community literature. Nucleic Acids Res. 1999, 27, 85–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambertsson, A. 3 The minute genes in Drosophila and their molecular functions. Adv. Genet. 1998, 38, 69–134. [Google Scholar] [PubMed]
- Sæbøe-Larssen, S.; Lyamouri, M.; Merriam, J.; Oksvold, M.P.; Lambertsson, A. Ribosomal protein insufficiency and the minute syndrome in Drosophila: A dose-response relationship. Genetics 1998, 148, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Rosby, R.; Cui, Z.; Rogers, E.; Delivron, M.A.; Robinson, V.L.; DiMario, P.J. Knockdown of the Drosophila GTPase nucleostemin 1 impairs large ribosomal subunit biogenesis, cell growth, and midgut precursor cell maintenance. Mol. Biol. Cell 2009, 20, 4424–4434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; DiMario, P. Loss of Drosophila nucleostemin 2 (NS2) blocks nucleolar release of the 60S subunit leading to ribosome stress. Chromosoma 2017, 126, 375–388. [Google Scholar] [CrossRef]
- Cui, Z.; DiMario, P.J. RNAi knockdown of Nopp140 induces Minute-like phenotypes in Drosophila. Mol. Biol. Cell 2007, 18, 2179–2191. [Google Scholar] [CrossRef] [Green Version]
- James, A.; Cindass, R., Jr.; Mayer, D.; Terhoeve, S.; Mumphrey, C.; DiMario, P. Nucleolar stress in Drosophila melanogaster: RNAi-mediated depletion of Nopp140. Nucleus 2013, 4, 123–133. [Google Scholar] [CrossRef] [Green Version]
- He, F.; James, A.; Raje, H.; Ghaffari, H.; DiMario, P. Deletion of Drosophila Nopp140 induces subcellular ribosomopathies. Chromosoma 2015, 124, 191–208. [Google Scholar] [CrossRef]
- Baral, S.S.; Lieux, M.E.; DiMario, P.J. Nucleolar stress in Drosophila neuroblasts, a model for human ribosomopathies. Biol. Open 2020, 9. [Google Scholar] [CrossRef] [Green Version]
- He, F.; DiMario, P. Structure and function of Nopp140 and Treacle. In The Nucleolus; Springer: New York, NY, USA, 2011; pp. 253–278. [Google Scholar]
- Quiring, R.; Walldorf, U.; Kloter, U.; Gehring, W.J. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science 1994, 265, 785–789. [Google Scholar] [CrossRef]
- Noveen, A.; Daniel, A.; Hartenstein, V. Early development of the Drosophila mushroom body: The roles of eyeless and dachshund. Development 2000, 127, 3475–3488. [Google Scholar] [CrossRef]
- Halder, G.; Callaerts, P.; Gehring, W.J. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 1995, 267, 1788–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, H. Regulation and function of the melanization reaction in Drosophila. Fly (Austin) 2009, 3, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Vlisidou, I.; Wood, W. Drosophila blood cells and their role in immune responses. FEBS J. 2015, 282, 1368–1382. [Google Scholar] [CrossRef] [PubMed]
- Bidla, G.; Dushay, M.S.; Theopold, U. Crystal cell rupture after injury in Drosophila requires the JNK pathway, small GTPases and the TNF homolog Eiger. J. Cell Sci. 2007, 120, 1209–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riesgo-Escovar, J.R.; Jenni, M.; Fritz, A.; Hafen, E. The Drosophila Jun-N-terminal kinase is required for cell morphogenesis but not for DJun-dependent cell fate specification in the eye. Genes Dev. 1996, 10, 2759–2768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNamee, L.M.; Brodsky, M.H. p53-independent apoptosis limits DNA damage-induced aneuploidy. Genetics 2009, 182, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Wang, M.C.; Bohmann, D. JNK protects Drosophila from oxidative stress by trancriptionally activating autophagy. Mech. Dev. 2009, 126, 624–637. [Google Scholar] [CrossRef]
- Nagy, P.; Varga, Á.; Kovács, A.L.; Takáts, S.; Juhász, G. How and why to study autophagy in Drosophila: It’s more than just a garbage chute. Methods 2015, 75, 151–161. [Google Scholar] [CrossRef]
- Barbee, S.A.; Estes, P.S.; Cziko, A.-M.; Hillebrand, J.; Luedeman, R.A.; Coller, J.M.; Johnson, N.; Howlett, I.C.; Geng, C.; Ueda, R. Staufen-and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 2006, 52, 997–1009. [Google Scholar] [CrossRef] [Green Version]
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22. [Google Scholar] [CrossRef]
- Kato, Y.; Nakamura, A. Roles of cytoplasmic RNP granules in intracellular RNA localization and translational control in the Drosophila oocyte. Dev. Growth Differ. 2012, 54, 19–31. [Google Scholar] [CrossRef]
- Hillebrand, J.; Pan, K.; Kokaram, A.; Barbee, S.; Parker, R.; Ramaswami, M. The Me31B DEAD-box helicase localizes to postsynaptic foci and regulates expression of a CaMKII reporter mRNA in dendrites of Drosophila olfactory projection neurons. Front. Neural Circuits 2010, 4, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallace, K.; Liu, T.H.; Vaessin, H. The pan-neural bHLH proteins DEADPAN and ASENSE regulate mitotic activity and cdk inhibitor dacapo expression in the Drosophila larval optic lobes. Genesis 2000, 26, 77–85. [Google Scholar] [CrossRef]
- Isaac, C.; Yang, Y.; Meier, U.T. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 1998, 142, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Cioce, M.; Lamond, A.I. Cajal bodies: A long history of discovery. Annu. Rev. Cell Dev. Biol. 2005, 21, 105–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade, L.; Chan, E.; Raska, I.; Peebles, C.L.; Roos, G.; Tan, E.M. Human autoantibody to a novel protein of the nuclear coiled body: Immunological characterization and cDNA cloning of p80-coilin. J. Exp. Med. 1991, 173, 1407–1419. [Google Scholar] [CrossRef] [Green Version]
- Raška, I.; Andrade, L.E.; Ochs, R.L.; Chan, E.K.; Chang, C.-M.; Roos, G.; Tan, E.M. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res. 1991, 195, 27–37. [Google Scholar] [CrossRef]
- Liu, J.-L.; Wu, Z.; Nizami, Z.; Deryusheva, S.; Rajendra, T.; Beumer, K.J.; Gao, H.; Matera, A.G.; Carroll, D.; Gall, J.G. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol. Biol. Cell 2009, 20, 1661–1670. [Google Scholar] [CrossRef] [Green Version]
- Martín-Blanco, E.; Gampel, A.; Ring, J.; Virdee, K.; Kirov, N.; Tolkovsky, A.M.; Martinez-Arias, A. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998, 12, 557–570. [Google Scholar] [CrossRef] [Green Version]
- Shlevkov, E.; Morata, G. A dp53/JNK-dependant feedback amplification loop is essential for the apoptotic response to stress in Drosophila. Cell Death Differ. 2012, 19, 451–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheth, U.; Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003, 300, 805–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCambridge, A.; Solanki, D.; Olchawa, N.; Govani, N.; Trinidad, J.C.; Gao, M. Comparative proteomics reveal Me31B’s interactome dynamics, expression regulation, and assembly mechanism into germ granules during Drosophila germline development. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.H.; Barbee, S.A.; Blankenship, J.T. GW-bodies and P-bodies constitute two separate pools of sequestered non-translating RNAs. PLoS ONE 2016, 11, e0150291. [Google Scholar] [CrossRef] [PubMed]
- Rzeczkowski, K.; Beuerlein, K.; Müller, H.; Dittrich-Breiholz, O.; Schneider, H.; Kettner-Buhrow, D.; Holtmann, H.; Kracht, M. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J. Cell Biol. 2011, 194, 581–596. [Google Scholar] [CrossRef] [Green Version]
- Shpargel, K.B.; Ospina, J.K.; Tucker, K.E.; Matera, A.G.; Hebert, M.D. Control of Cajal body number is mediated by the coilin C-terminus. J. Cell Sci. 2003, 116, 303–312. [Google Scholar] [CrossRef] [Green Version]
- Hebert, M.D.; Shpargel, K.B.; Ospina, J.K.; Tucker, K.E.; Matera, A.G. Coilin methylation regulates nuclear body formation. Dev Cell 2002, 3, 329–337. [Google Scholar] [CrossRef]
- De Cuevas, M.; Spradling, A.C. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development 1998, 125, 2781–2789. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
DeLeo, K.R.; Baral, S.S.; Houser, A.; James, A.; Sewell, P.; Pandey, S.; DiMario, P.J. Drosophila to Explore Nucleolar Stress. Int. J. Mol. Sci. 2021, 22, 6759. https://doi.org/10.3390/ijms22136759
DeLeo KR, Baral SS, Houser A, James A, Sewell P, Pandey S, DiMario PJ. Drosophila to Explore Nucleolar Stress. International Journal of Molecular Sciences. 2021; 22(13):6759. https://doi.org/10.3390/ijms22136759
Chicago/Turabian StyleDeLeo, Kathryn R., Sonu S. Baral, Alex Houser, Allison James, Phelan Sewell, Shova Pandey, and Patrick J. DiMario. 2021. "Drosophila to Explore Nucleolar Stress" International Journal of Molecular Sciences 22, no. 13: 6759. https://doi.org/10.3390/ijms22136759
APA StyleDeLeo, K. R., Baral, S. S., Houser, A., James, A., Sewell, P., Pandey, S., & DiMario, P. J. (2021). Drosophila to Explore Nucleolar Stress. International Journal of Molecular Sciences, 22(13), 6759. https://doi.org/10.3390/ijms22136759




