Soluble Cytoplasmic Expression and Purification of Immunotoxin HER2(scFv)-PE24B as a Maltose Binding Protein Fusion
Abstract
1. Introduction
2. Results
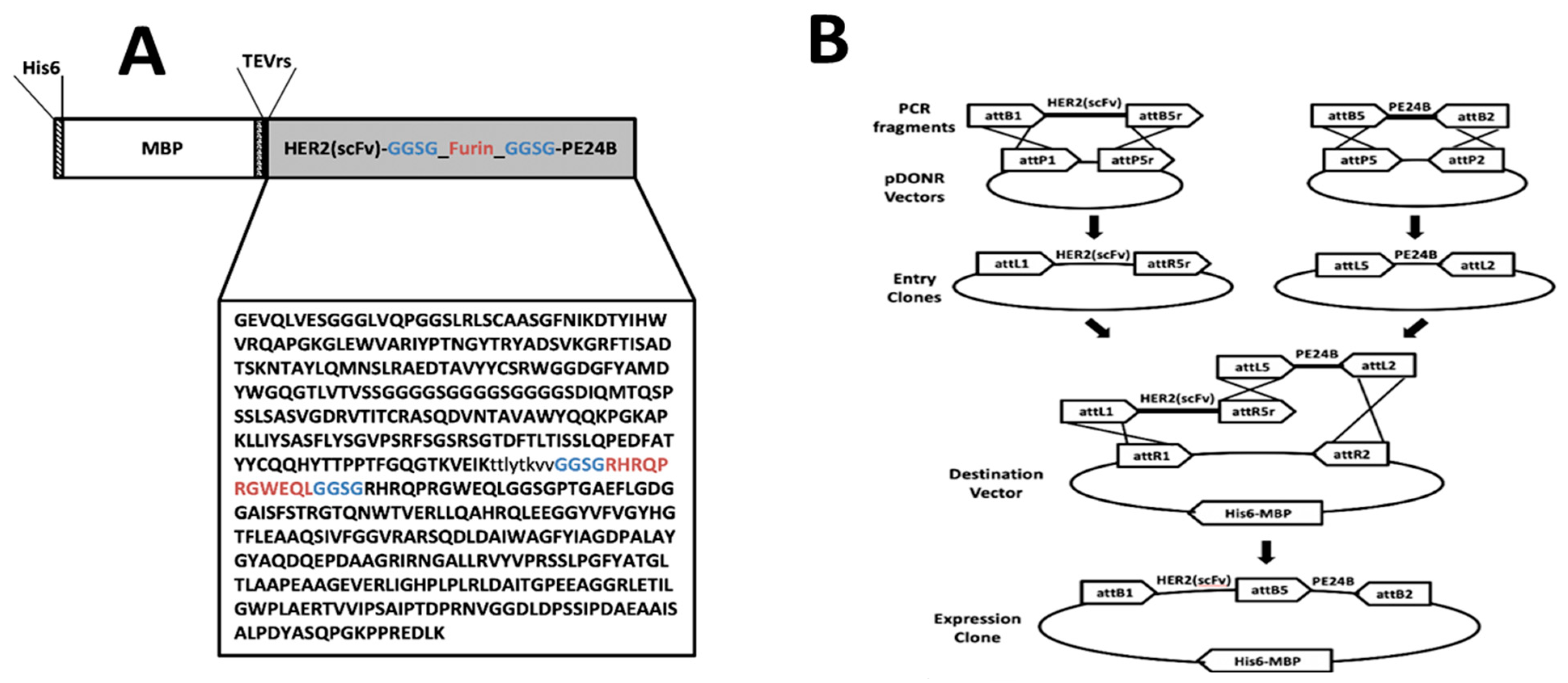
2.1. Design of Recombinant Immunotoxin MBP-HER2(scFv)-PE24B
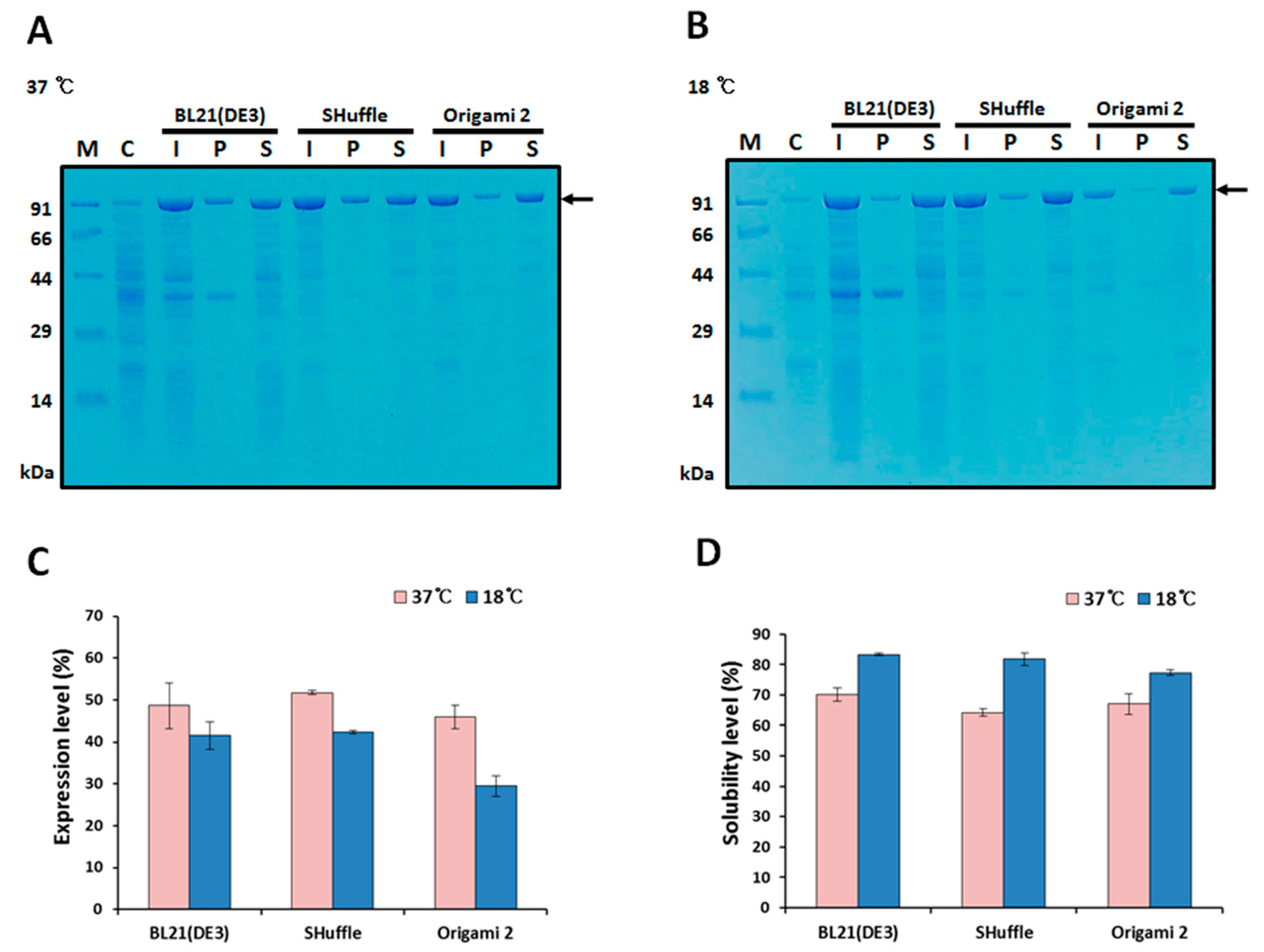
2.2. Expression and Solubility of MBP-HER2(scFv)-PE24B
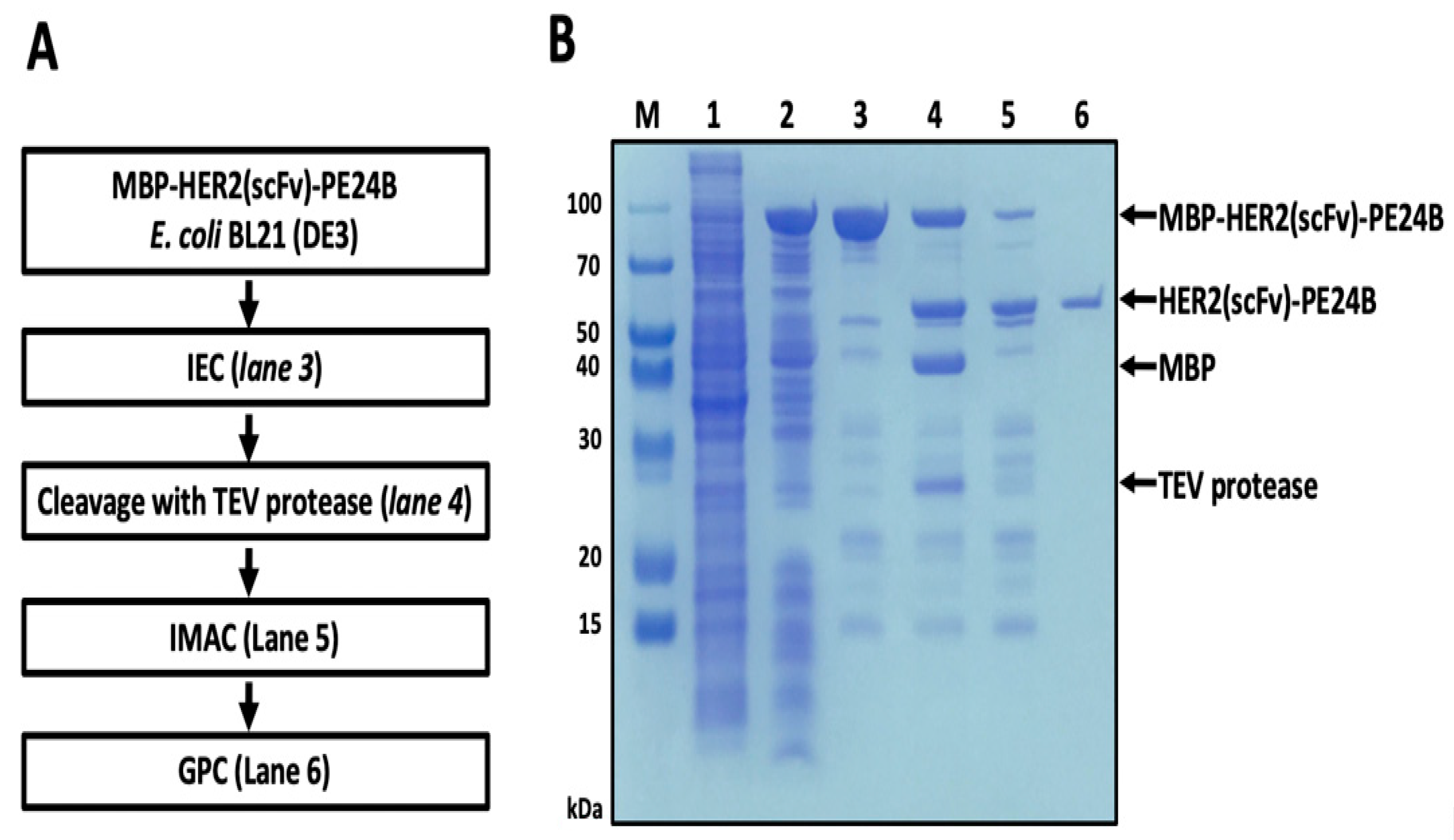
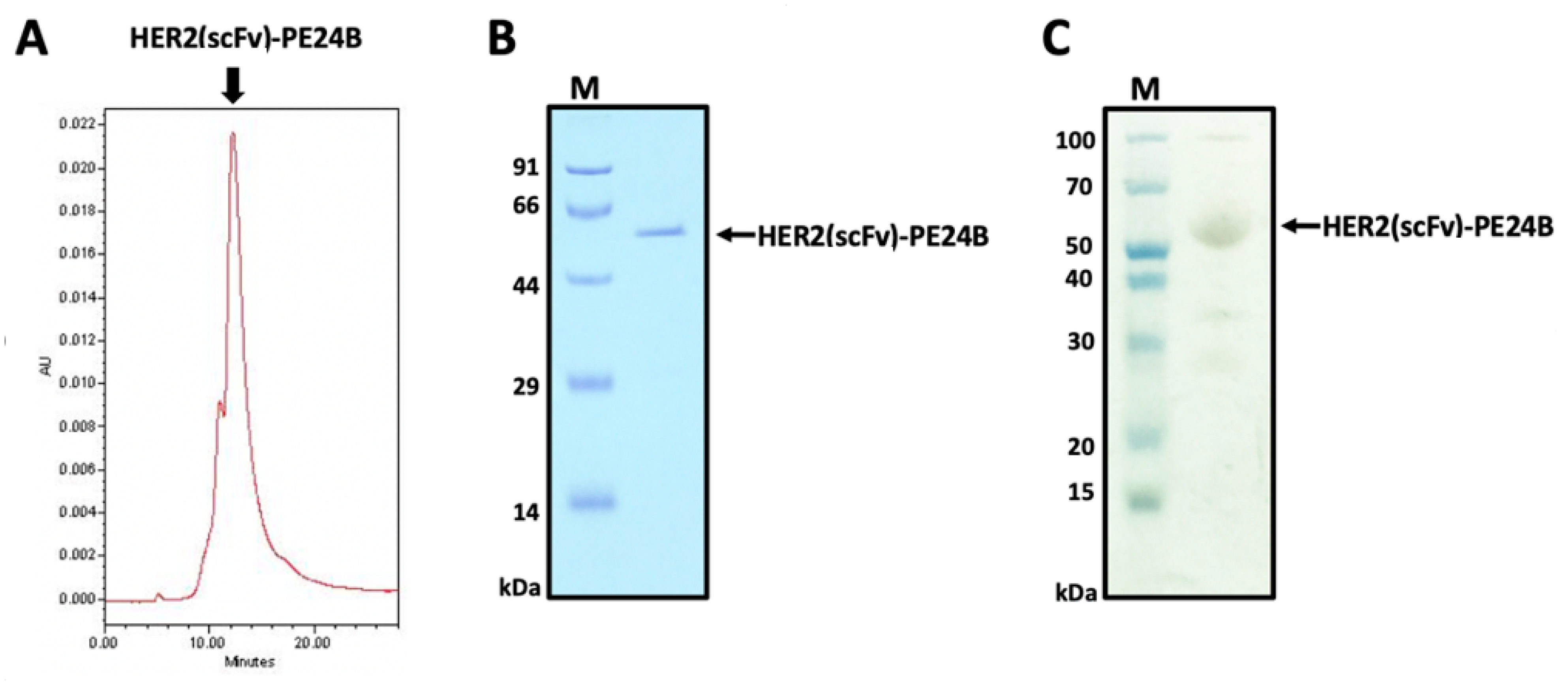
2.3. Purification of HER2(scFv)-PE24B
2.4. Verification of HER2(scFv)-PE24B by Mass Spectrometry
2.5. Purification of HER2(scFv)-GFP
2.6. HER2 Levels on the Surface of Breast Cancer Cell Lines
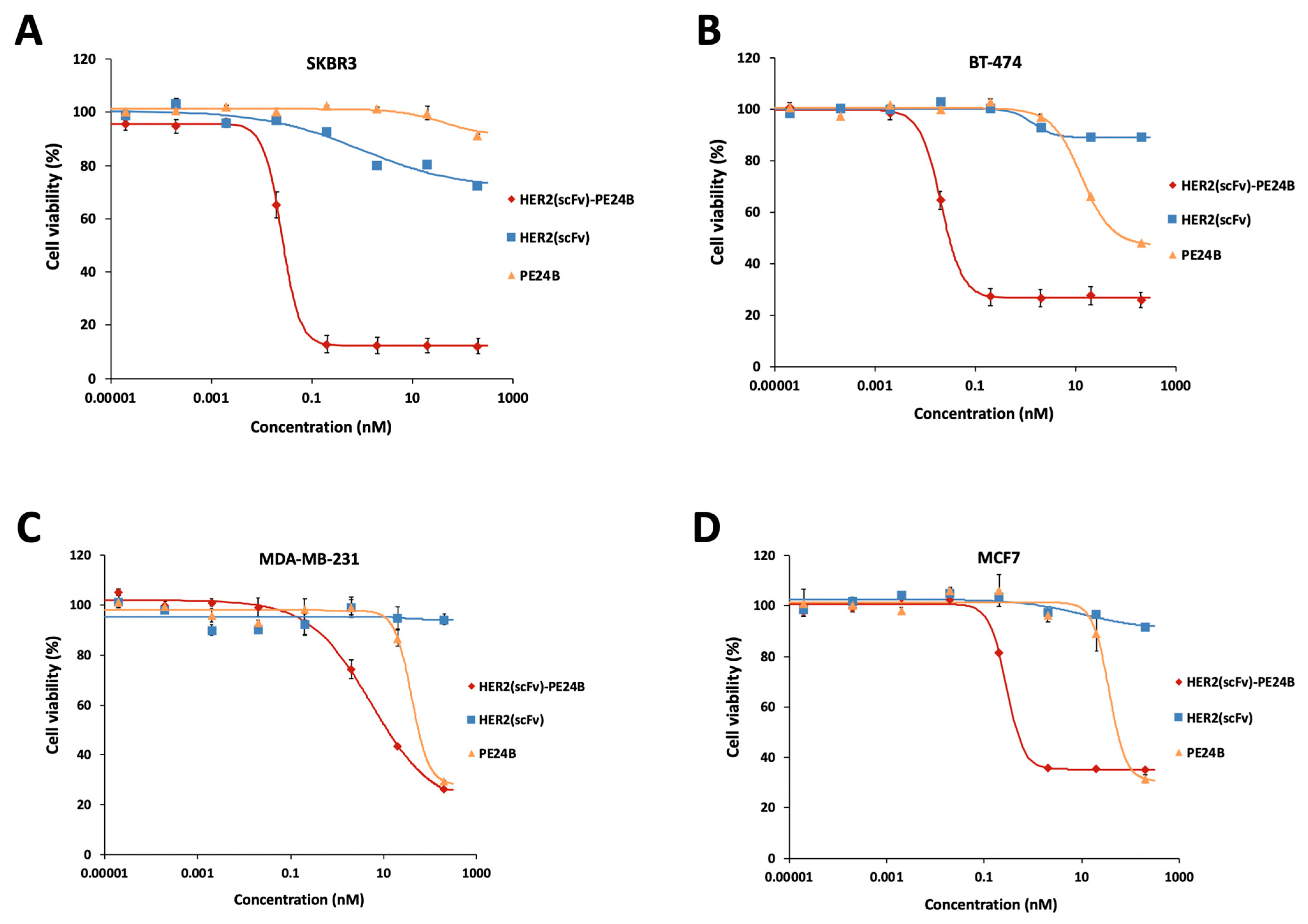
2.7. Cytotoxicity of HER2(scFv)-PE24B against High HER2-Expressing and Low HER2-Expressing Cell Lines
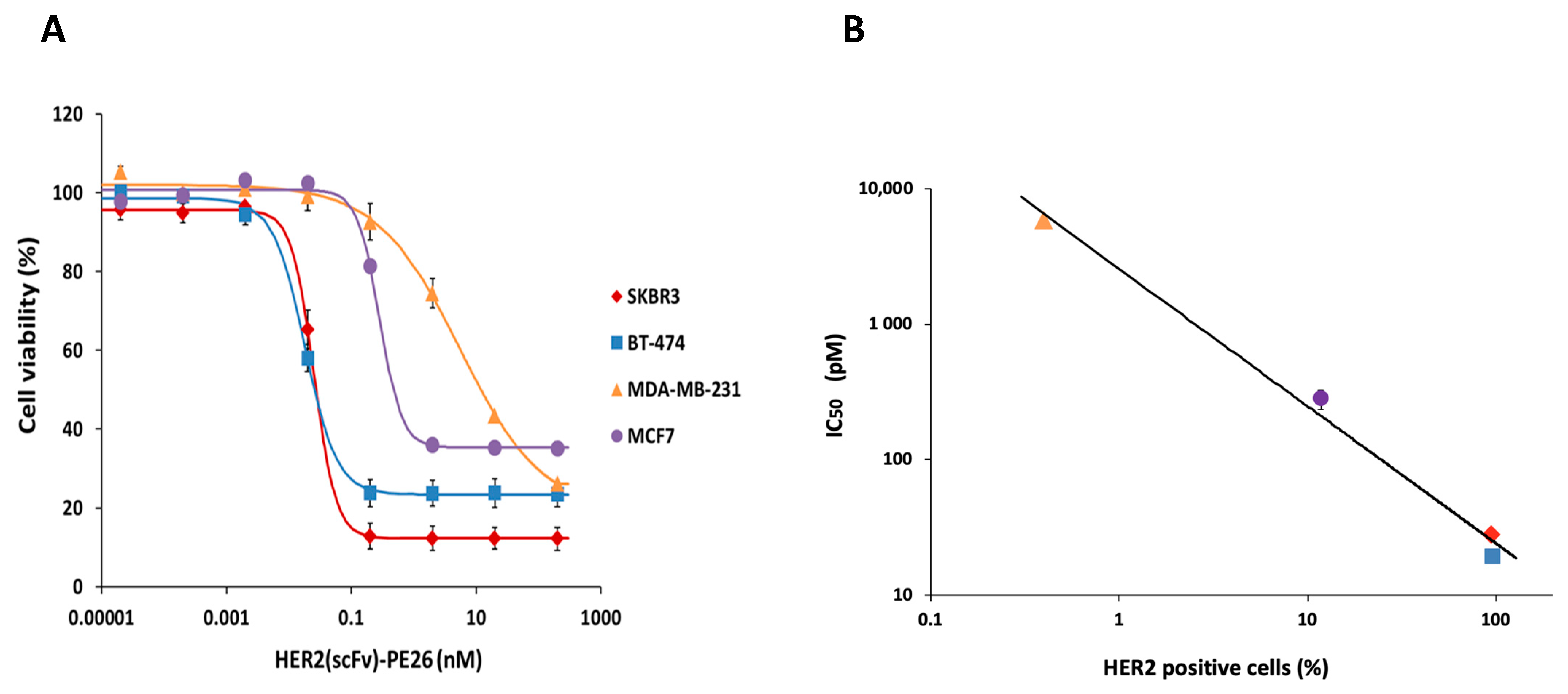
2.8. Correlation between HER2 Levels and the IC50 of HER2(scFv)-PE24B
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Construction of Plasmids
4.3. Expression and Solubility Test of MBP-HER2(scFv)-PE24B in E. coli
4.4. Purification and Tag Removal of HER2(scFv)-PE24B
4.5. Purification of MBP-HER2(scFv)-GFP and Tag Removal to Generate HER2(scFv)-GFP
4.6. Purification of HER2(scFv) and PE24B
4.7. Electrophoresis and Quantification of Protein Expression and the Solubility Level
4.8. Determination of the HER2(scFv)-PE24B Purity by HPLC and Silver Staining
4.9. Endotoxin Removal and Endotoxin Assay
4.10. Mass Spectrometry Analysis of the Purified HER2(scFv)-PE24B
4.11. Flow Cytometric Analysis
4.12. MTT Assay
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, Y.; Zhu, X.; Wei, X.; Tang, C.; Zhang, W. HER2-targeted therapies in gastric cancer. Biochim. Biophys. Acta (BBA) Bioenerg. 2021, 1876, 188549. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thurlimann, B.; Senn, H.J.; Panel, M. Strategies for subtypes--dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, C.; Perrone, F.; Gallo, C.; De Laurentiis, M.; Lauria, R.; Morabito, A.; Pettinato, G.; Panico, L.; D’Antonio, A.; Bianco, A.R.; et al. C-erb B2 overexpression decreases the benefit of adjuvant tamoxifen in early-stage breast cancer without axillary lymph node metastases. J. Clin. Oncol 1996, 14, 2702–2708. [Google Scholar] [CrossRef]
- Olayioye, M.A.; Neve, R.M.; Lane, H.A.; Hynes, N.E. NEW EMBO MEMBERS’ REVIEW: The ErbB signaling network: Receptor heterodimerization in development and cancer. EMBO J. 2000, 19, 3159–3167. [Google Scholar] [CrossRef]
- Gutierrez, C.; Schiff, R. HER2: Biology detection and clinical implications. Arch. Pathol. Lab. Med. 2011, 135, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Li, S.G.; Li, L. Targeted therapy in HER2-positive breast cancer. Biomed. Rep. 2013, 1, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Hudis, C.A. Trastuzumab—Mechanism of Action and Use in Clinical Practice. N. Engl. J. Med. 2007, 357, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.; Claret, F.X. Trastuzumab: Updated mechanisms of action and resistance in breast cancer. Front. Oncol. 2012, 2, 62. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, C.L.; Sliwkowski, M.X.; Osborne, C.K.; Perez, E.A.; Puglisi, F.; Gianni, L. Treatment of HER2-positive breast cancer: Current status and future perspectives. Nat. Rev. Clin. Oncol. 2011, 9, 16–32. [Google Scholar] [CrossRef]
- Mathew, M.; Verma, R.S. Humanized immunotoxins: A new generation of immunotoxins for targeted cancer therapy. Cancer Sci. 2009, 100, 1359–1365. [Google Scholar] [CrossRef]
- Yamaizumi, M.; Mekada, E.; Uchida, T.; Okada, Y. One molecule of diphtheria toxin fragment a introduced into a cell can kill the cell. Cell 1978, 15, 245–250. [Google Scholar] [CrossRef]
- Antignani, A.; Fitzgerald, D. Immunotoxins: The Role of the Toxin. Toxins 2013, 5, 1486–1502. [Google Scholar] [CrossRef]
- Alewine, C.; Hassan, R.; Pastan, I. Advances in Anticancer Immunotoxin Therapy. Oncologist 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Wolf, P.; Elsässer-Beile, U. Pseudomonas exotoxin A: From virulence factor to anti-cancer agent. Int. J. Med. Microbiol. 2009, 299, 161–176. [Google Scholar] [CrossRef] [PubMed]
- Seetharam, S.; Chaudhary, V.K.; FitzGerald, D.; Pastan, I. Increased cytotoxic activity of Pseudomonas exotoxin and two chimeric toxins ending in KDEL. J. Biol. Chem. 1991, 266, 17376–17381. [Google Scholar] [CrossRef]
- Weldon, J.E.; Xiang, L.; Chertov, O.; Margulies, I.; Kreitman, R.J.; Fitzgerald, D.J.; Pastan, I. A protease-resistant immunotoxin against CD22 with greatly increased activity against CLL and diminished animal toxicity. Blood 2009, 113, 3792–3800. [Google Scholar] [CrossRef] [PubMed]
- Onda, M.; Beers, R.; Xiang, L.; Lee, B.; Weldon, J.; Kreitman, R.J.; Pastan, I. Recombinant immunotoxin against B-cell malignancies with no immunogenicity in mice by removal of B-cell epitopes. Proc. Natl. Acad. Sci. USA 2011, 108, 5742–5747. [Google Scholar] [CrossRef]
- Mazor, R.; Vassall, A.N.; Eberle, J.A.; Beers, R.; Weldon, J.; Venzon, D.J.; Tsang, K.Y.; Benhar, I.; Pastan, I. Identification and elimination of an immunodominant T-cell epitope in recombinant immunotoxins based on Pseudomonas exotoxin A. Proc. Natl. Acad. Sci. USA 2012, 109, E3597–E3603. [Google Scholar] [CrossRef]
- Mazor, R.; Onda, M.; Park, D.; Addissie, S.; Xiang, L.; Zhang, J.; Hassan, R.; Pastan, I. Dual B- and T-cell de-immunization of recombinant immunotoxin targeting mesothelin with high cytotoxic activity. Oncotarget 2016, 7, 29916–29926. [Google Scholar] [CrossRef]
- Pastan, I.; Beers, R.; Bera, T.K.; Lo, B.K.C. Recombinant Immunotoxins in the Treatment of Cancer. Antib. Eng. 2004, 248, 503–518. [Google Scholar] [CrossRef]
- Barkhordari, F.; Sohrabi, N.; Davami, F.; Mahboudi, F.; Garoosi, Y.T. Cloning, expression and characterization of a HER2-alpha luffin fusion protein in Escherichia coli. Prep. Biochem. Biotechnol. 2019, 49, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Goleij, Z.; Hosseini, H.M.; Sedighian, H.; Behzadi, E.; Halabian, R.; Sorouri, R.; Fooladi, A.A.I. Breast cancer targeted/therapeutic with double and triple fusion Immunotoxins. J. Steroid Biochem. Mol. Biol. 2020, 200, 105651. [Google Scholar] [CrossRef]
- Lee, S.; Park, S.; Nguyen, M.T.; Lee, E.; Kim, J.; Baek, S.; Kim, C.J.; Jang, Y.J.; Choe, H. A chemical conjugate between HER2-targeting antibody fragment and Pseudomonas exotoxin A fragment demonstrates cytotoxic effects on HER2-expressing breast cancer cells. BMB Rep. 2019, 52, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Allahyari, H.; Heidari, S.; Ghamgosha, M.; Saffarian, P.; Amani, J. Immunotoxin: A new tool for cancer therapy. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef]
- Akbari, B.; Farajnia, S.; Khosroshahi, S.A.; Safari, F.; Yousefi, M.; Dariushnejad, H.; Rahbarnia, L. Immunotoxins in cancer therapy: Review and update. Int. Rev. Immunol. 2017, 36, 207–219. [Google Scholar] [CrossRef]
- Khirehgesh, M.R.; Sharifi, J.; Safari, F.; Akbari, B. Immunotoxins and nanobody-based immunotoxins: Review and update. J. Drug Target. 2021, 1–15. [Google Scholar] [CrossRef]
- Batra, J.K.; Kasprzyk, P.G.; Bird, R.E.; Pastan, I.; King, C.R. Recombinant anti-erbB2 immunotoxins containing Pseudomonas exotoxin. Proc. Natl. Acad. Sci. USA 1992, 89, 5867–5871. [Google Scholar] [CrossRef] [PubMed]
- Riccio, G.; D’Avino, C.; Raines, R.T.; De Lorenzo, C. A novel fully human antitumor ImmunoRNase resistant to the RNase inhibitor. Protein Eng. Des. Sel. 2012, 26, 243–248. [Google Scholar] [CrossRef]
- Vu, T.T.T.; Jeong, B.; Krupa, M.; Kwon, U.; Song, J.-A.; Do, B.H.; Nguyen, M.T.; Seo, T.; Nguyen, A.N.; Joo, C.H.; et al. Soluble Prokaryotic Expression and Purification of Human Interferon Alpha-2b Using a Maltose-Binding Protein Tag. J. Mol. Microbiol. Biotechnol. 2016, 26, 359–368. [Google Scholar] [CrossRef]
- Do, B.H.; Nguyen, M.T.; Song, J.-A.; Park, S.; Yoo, J.; Jang, J.; Lee, S.; So, S.; Yoon, Y.; Kim, I.; et al. Soluble Prokaryotic Expression and Purification of Bioactive Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand. J. Microbiol. Biotechnol. 2017, 27, 2156–2164. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Ke, N.; Berkmen, M. Use of the SHuffle Strains in Production of Proteins. Curr. Protoc. Protein Sci. 2016, 85, 5261–5262. [Google Scholar] [CrossRef]
- Nguyen, T.; Vu, T.; Nguyen, M.; Ta, H.; Park, K.; Kim, S.; Kim, C.; Jang, Y.; Choe, H. Soluble Prokaryotic Overexpression and Purification of Human GM-CSF Using the Protein Disulfide Isomerase b′a′ Domain. Int. J. Mol. Sci. 2021, 22, 5267. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.T.T.; Jeong, B.; Yu, J.; Koo, B.-K.; Jo, S.-H.; Robinson, R.C.; Choe, H. Soluble prokaryotic expression and purification of crotamine using an N-terminal maltose-binding protein tag. Toxicon 2014, 92, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.; Koo, B.-K.; Vu, T.T.T.; Song, J.-A.; Chong, S.-H.; Jeong, B.; Ryu, H.-B.; Moh, S.-H.; Choe, H. Prokaryotic Soluble Overexpression and Purification of Bioactive Human Growth Hormone by Fusion to Thioredoxin, Maltose Binding Protein, and Protein Disulfide Isomerase. PLoS ONE 2014, 9, e89038. [Google Scholar] [CrossRef]
- Do, B.H.; Ryu, H.-B.; Hoang, P.; Koo, B.-K.; Choe, H. Soluble Prokaryotic Overexpression and Purification of Bioactive Human Granulocyte Colony-Stimulating Factor by Maltose Binding Protein and Protein Disulfide Isomerase. PLoS ONE 2014, 9, e89906. [Google Scholar] [CrossRef]
- Song, J.-A.; Koo, B.-K.; Chong, S.H.; Kwak, J.; Ryu, H.-B.; Nguyen, M.T.; Vu, T.T.T.; Jeong, B.; Kim, S.W.; Choe, H. Expression and Purification of Biologically Active Human FGF2 Containing the b′a′ Domains of Human PDI in Escherichia coli. Appl. Biochem. Biotechnol. 2013, 170, 67–80. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Krupa, M.; Koo, B.K.; Song, J.A.; Vu, T.T.; Do, B.H.; Nguyen, A.N.; Seo, T.; Yoo, J.; Jeong, B.; et al. Prokaryotic Soluble Overexpression and Purification of Human VEGF165 by Fusion to a Maltose Binding Protein Tag. PLoS ONE 2016, 11, 0156296. [Google Scholar] [CrossRef]
- Vu, T.T.T.; Koo, B.-K.; Song, J.-A.; Chong, S.-H.; Park, C.R.; Nguyen, M.T.; Jeong, B.; Ryu, H.-B.; Seong, J.Y.; Jang, Y.J.; et al. Soluble overexpression and purification of bioactive human CCL2 in E. coli by maltose-binding protein. Mol. Biol. Rep. 2014, 42, 651–663. [Google Scholar] [CrossRef]
- Nguyen, A.N.; Song, J.-A.; Nguyen, M.T.; Do, B.H.; Kwon, G.G.; Park, S.S.; Yoo, J.; Jang, J.; Jin, J.; Osborn, M.J.; et al. Prokaryotic soluble expression and purification of bioactive human fibroblast growth factor 21 using maltose-binding protein. Sci. Rep. 2017, 7, 16139. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Prima, M.J.; Song, J.-A.; Kim, J.; Do, B.H.; Yoo, J.; Park, S.; Jang, J.; Lee, S.; Lee, E.; et al. Prokaryotic soluble overexpression and purification of oncostatin M using a fusion approach and genetically engineered E. coli strains. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Kapust, R.B.; Waugh, D.S. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein. Sci. 1999, 8, 1668–1674. [Google Scholar] [CrossRef]
- Yu, L.; Wang, Y.; Yao, Y.; Li, W.; Lai, Q.; Li, J.; Zhou, Y.; Kang, T.; Xie, Y.; Wu, Y.; et al. Eradication of Growth of HER2-Positive Ovarian Cancer With Trastuzumab-DM1, an Antibody-Cytotoxic Drug Conjugate in Mouse Xenograft Model. Int. J. Gynecol. Cancer 2014, 24, 1158–1164. [Google Scholar] [CrossRef]
- Half, E.; Broaddus, R.; Danenberg, K.D.; Danenberg, P.V.; Ayers, G.D.; Sinicrope, F.A. HER-2 receptor expression, localization, and activation in colorectal cancer cell lines and human tumors. Int. J. Cancer 2004, 108, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.-W.; Chien, S.-Y.; Kuo, S.-J.; Tseng, L.-M.; Lin, H.-Y.; Chi, C.-W.; Chen, D.-R. The Potential Utility of Curcumin in the Treatment of HER-2-Overexpressed Breast Cancer: AnIn VitroandIn VivoComparison Study with Herceptin. Evid.-Based Complement. Altern. Med. 2011, 2012, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kunisue, H.; Kurebayashi, J.; Otsuki, T.; Tang, C.K.; Kurosumi, M.; Yamamoto, S.; Tanaka, K.; Doihara, H.; Shimizu, N.; Sonoo, H. Anti-HER2 antibody enhances the growth inhibitory effect of anti-oestrogen on breast cancer cells expressing both oestrogen receptors and HER2. Br. J. Cancer 2000, 82, 46–51. [Google Scholar] [CrossRef]
- Magnani, E.; Bartling, L.; Hake, S. From Gateway to MultiSite Gateway in one recombination event. BMC Mol. Biol. 2006, 7. [Google Scholar] [CrossRef]
- Petersen, L.K.; Stowers, R.S. A Gateway MultiSite recombination cloning toolkit. PLoS ONE 2011, 6, e24531. [Google Scholar] [CrossRef]
- Chen, W.; Yuan, Y.; Jiang, X. Antibody and antibody fragments for cancer immunotherapy. J. Control. Release 2020, 328, 395–406. [Google Scholar] [CrossRef]
- Busso, D.; Delagoutte-Busso, B.; Moras, D. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 2005, 343, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Pe’delacq, J.-D.; Cabantous, S.; Tran, T.; Terwilliger, T.; Waldo, G.S. Correction: Corrigendum: Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006, 24, 1170. [Google Scholar] [CrossRef]
- Song, J.A.; Koo, B.K.; Chong, S.H.; Kim, K.; Choi, D.K.; Thi Vu, T.T.; Nguyen, M.T.; Jeong, B.; Ryu, H.B.; Kim, I.; et al. Soluble expression of human leukemia inhibitory factor with protein disulfide isomerase in Escherichia coli and its simple purification. PLoS ONE 2013, 8, e83781. [Google Scholar] [CrossRef]


| Purification Step | Total Protein (mg) | Purity (%) | HER2(scFv)-PE24B (mg) | Yield (%) |
|---|---|---|---|---|
| Bacterial culture (500 mL) | 1400 | |||
| Supernatant | 103.2 | 41.07 | 23.23 | 100 |
| First cation exchange | 12.07 | 68.18 | 4.51 | 19.41 |
| Second IMAC | 4.96 | 28.72 | 1.42 | 6.11 |
| Third GPC | 0.2 | 100 | 0.25 | 1.07 |
| Cell Lines | Recombinant HER2(scFv)-PE24B, IC50 (pM, n ≥ 3) | Hill Coefficient | Conjugated HER2(scFv)-PE24B, IC50 (pM, n ≥ 3) 1 | HER2-Positive Cells (%) |
|---|---|---|---|---|
| BT-474 | 19 ± 1.4 | 1.87 ± 0.17 | 6.7 ± 3 | 97.5 |
| SKBR3 | 28.1 ± 2.5 | 2.24 ± 0.16 | 43 ± 8 | 93.8 |
| MCF7 | 280 ± 46 | 3.22 ± 1.87 | 1010 ± 380 | 11.8 |
| MDA-MB-231 | 5800 ± 280 | 0.65 ± 0.21 | 9440 ± 300 | 0.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Nguyen, M.Q.; Ta, H.K.K.; Nguyen, M.T.; Lee, G.; Kim, C.J.; Jang, Y.J.; Choe, H. Soluble Cytoplasmic Expression and Purification of Immunotoxin HER2(scFv)-PE24B as a Maltose Binding Protein Fusion. Int. J. Mol. Sci. 2021, 22, 6483. https://doi.org/10.3390/ijms22126483
Park S, Nguyen MQ, Ta HKK, Nguyen MT, Lee G, Kim CJ, Jang YJ, Choe H. Soluble Cytoplasmic Expression and Purification of Immunotoxin HER2(scFv)-PE24B as a Maltose Binding Protein Fusion. International Journal of Molecular Sciences. 2021; 22(12):6483. https://doi.org/10.3390/ijms22126483
Chicago/Turabian StylePark, Sangsu, Minh Quan Nguyen, Huynh Kim Khanh Ta, Minh Tan Nguyen, Gunsup Lee, Chong Jai Kim, Yeon Jin Jang, and Han Choe. 2021. "Soluble Cytoplasmic Expression and Purification of Immunotoxin HER2(scFv)-PE24B as a Maltose Binding Protein Fusion" International Journal of Molecular Sciences 22, no. 12: 6483. https://doi.org/10.3390/ijms22126483
APA StylePark, S., Nguyen, M. Q., Ta, H. K. K., Nguyen, M. T., Lee, G., Kim, C. J., Jang, Y. J., & Choe, H. (2021). Soluble Cytoplasmic Expression and Purification of Immunotoxin HER2(scFv)-PE24B as a Maltose Binding Protein Fusion. International Journal of Molecular Sciences, 22(12), 6483. https://doi.org/10.3390/ijms22126483








