(S)-Reutericyclin: Susceptibility Testing and In Vivo Effect on Murine Fecal Microbiome and Volatile Organic Compounds
Abstract
1. Introduction
2. Results
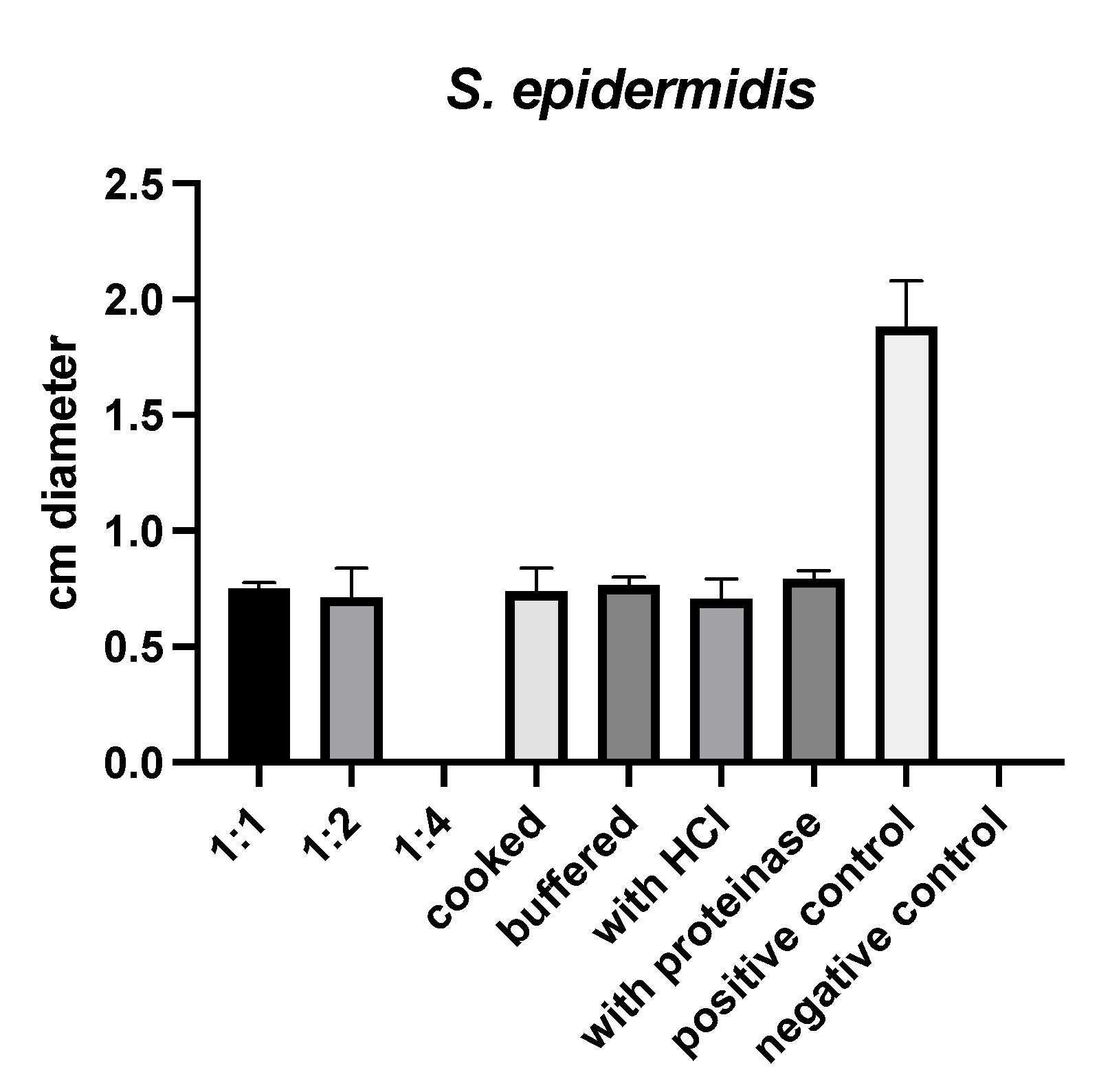
2.1. Susceptibility Testing
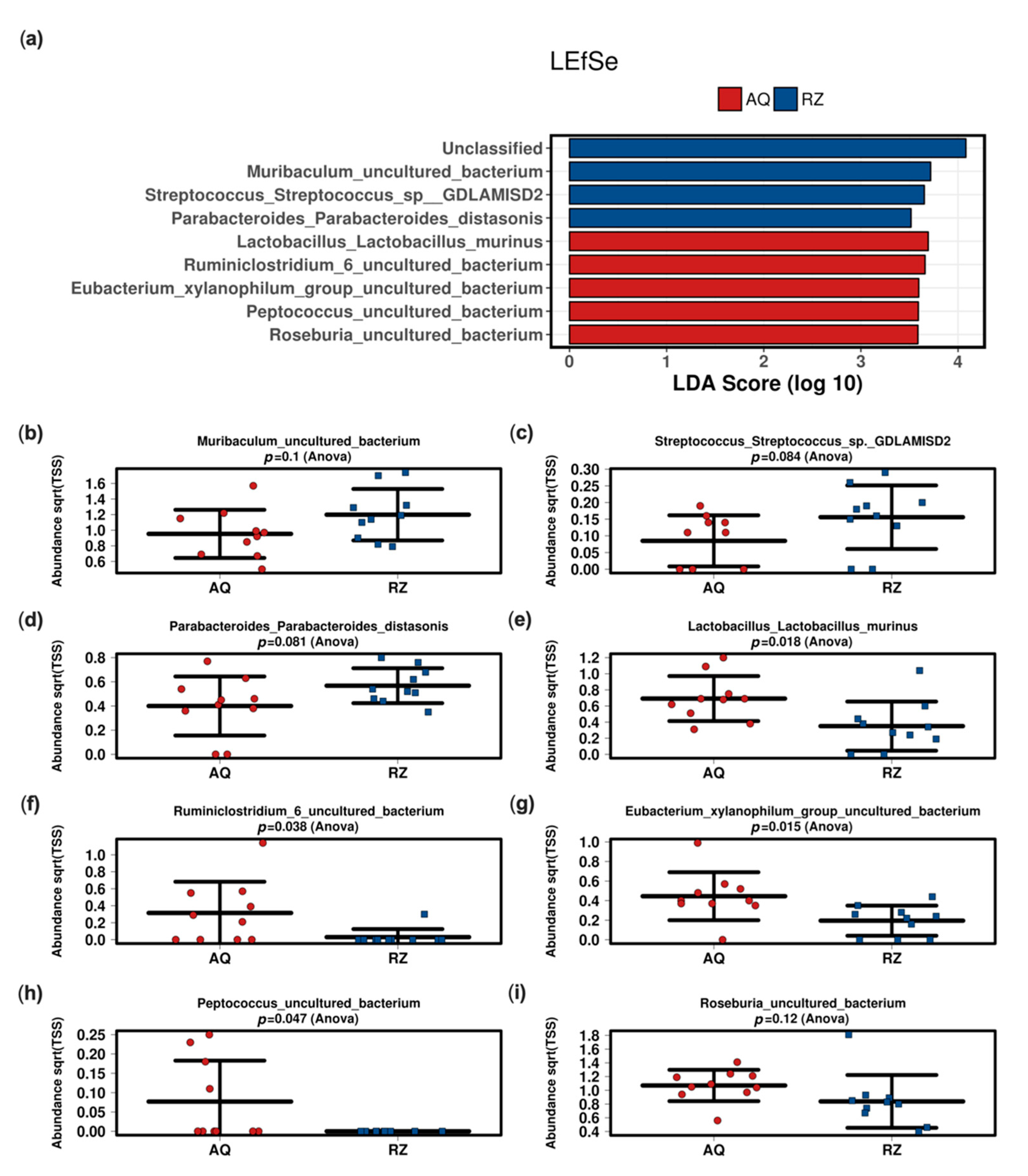
2.2. In Vivo Microbiome Analysis
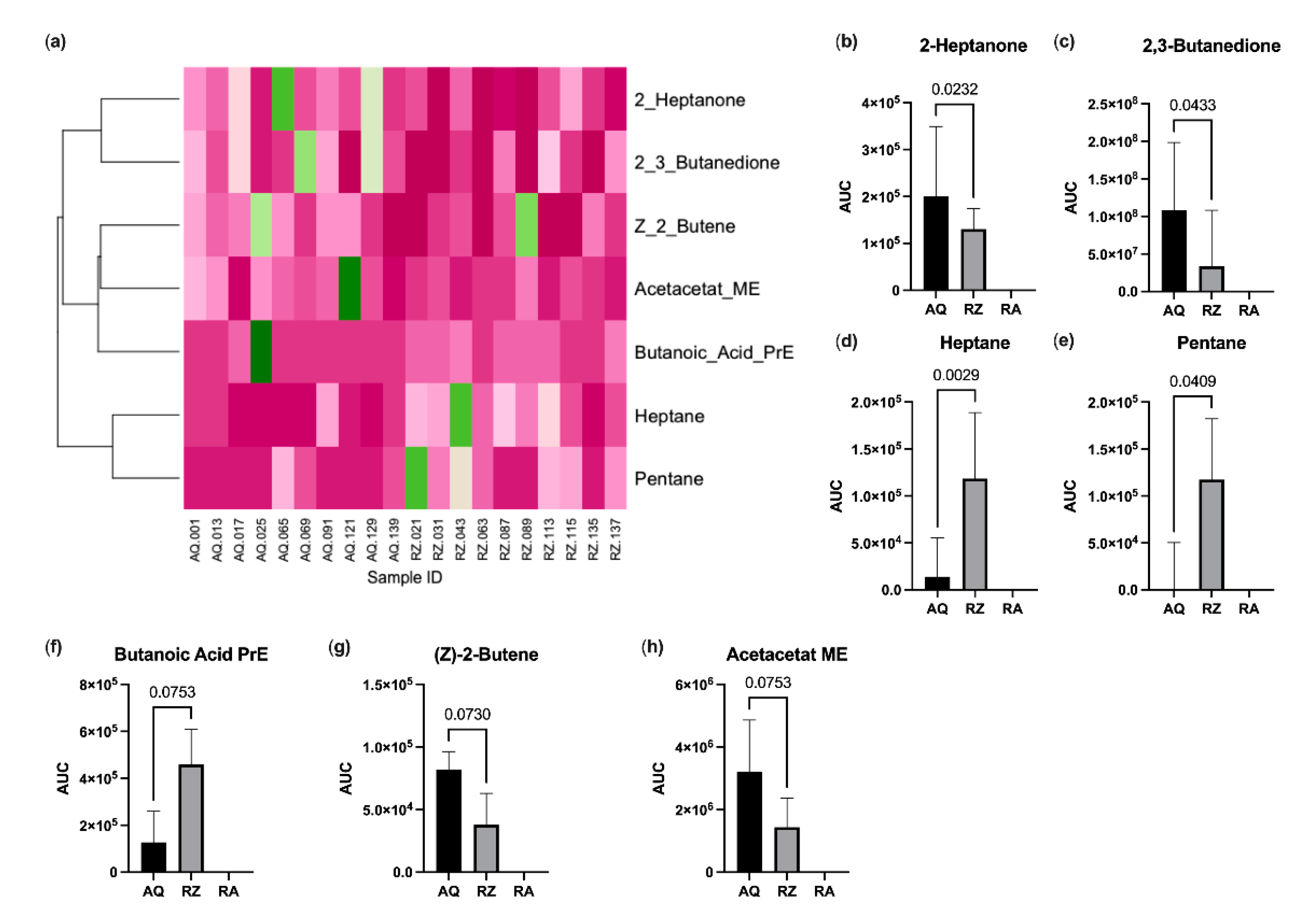
2.3. Fecal Volatile Organic Compound Analysis
2.4. Correlation Analysis
3. Discussion
4. Materials and Methods
4.1. Susceptibility Testing
4.2. Animal Model
4.3. 16S Based Microbiome Analysis
4.4. Stool VOC Analysis
4.5. Statistics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Angelakis, E.; Merhej, V.; Raoult, D. Related actions of probiotics and antibiotics on gut microbiota and weight modification. Lancet Infect. Dis. 2013, 13, 889–899. [Google Scholar] [CrossRef]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef]
- Spencer, M.D.; Hamp, T.J.; Reid, R.W.; Fischer, L.M.; Zeisel, S.H.; Fodor, A.A. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 2011, 140, 976–986. [Google Scholar] [CrossRef]
- Redman, M.G.; Ward, E.J.; Phillips, R.S. The efficacy and safety of probiotics in people with cancer: A systematic review. Ann. Oncol. 2014, 25, 1919–1929. [Google Scholar] [CrossRef]
- De Weirdt, R.; Crabbe, A.; Roos, S.; Vollenweider, S.; Lacroix, C.; van Pijkeren, J.P.; Britton, R.A.; Sarker, S.; Van de Wiele, T.; Nickerson, C.A. Glycerol supplementation enhances L. reuteri’s protective effect against S. Typhimurium colonization in a 3-D model of colonic epithelium. PLoS ONE 2012, 7, e37116. [Google Scholar] [CrossRef]
- Montiel, R.; Martin-Cabrejas, I.; Langa, S.; El Aouad, N.; Arques, J.L.; Reyes, F.; Medina, M. Antimicrobial activity of reuterin produced by Lactobacillus reuteri on Listeria monocytogenes in cold-smoked salmon. Food Microbiol. 2014, 44, 1–5. [Google Scholar] [CrossRef]
- Ganzle, M.G. Reutericyclin: Biological activity, mode of action, and potential applications. Appl. Microbiol. Biotechnol. 2004, 64, 326–332. [Google Scholar] [CrossRef]
- Ganzle, M.G.; Holtzel, A.; Walter, J.; Jung, G.; Hammes, W.P. Characterization of reutericyclin produced by Lactobacillus reuteri LTH2584. Appl. Environ. Microbiol. 2000, 66, 4325–4333. [Google Scholar] [CrossRef]
- Lin, X.B.; Lohans, C.T.; Duar, R.; Zheng, J.; Vederas, J.C.; Walter, J.; Ganzle, M. Genetic determinants of reutericyclin biosynthesis in Lactobacillus reuteri. Appl. Environ. Microbiol. 2015, 81, 2032–2041. [Google Scholar] [CrossRef]
- Holtzel, A.; Ganzle, M.G.; Nicholson, G.J.; Hammes, W.P.; Jung, G. The First Low Molecular Weight Antibiotic from Lactic Acid Bacteria: Reutericyclin, a New Tetramic Acid. Angew. Chem. Int. Ed. Engl. 2000, 39, 2766–2768. [Google Scholar] [CrossRef]
- Schobert, R. Domino syntheses of bioactive tetronic and tetramic acids. Naturwissenschaften 2007, 94, 1–11. [Google Scholar] [CrossRef]
- Messens, W.; De Vuyst, L. Inhibitory substances produced by Lactobacilli isolated from sourdoughs—A review. Int. J. Food Microbiol. 2002, 72, 31–43. [Google Scholar] [CrossRef]
- Ganzle, M.G.; Vogel, R.F. Studies on the mode of action of reutericyclin. Appl. Environ. Microbiol. 2003, 69, 1305–1307. [Google Scholar] [CrossRef]
- Cherian, P.T.; Wu, X.; Maddox, M.M.; Singh, A.P.; Lee, R.E.; Hurdle, J.G. Chemical modulation of the biological activity of reutericyclin: A membrane-active antibiotic from Lactobacillus reuteri. Sci. Rep. 2014, 4, 4721. [Google Scholar] [CrossRef]
- Hurdle, J.G.; Heathcott, A.E.; Yang, L.; Yan, B.; Lee, R.E. Reutericyclin and related analogues kill stationary phase Clostridium difficile at achievable colonic concentrations. J. Antimicrob. Chemother. 2011, 66, 1773–1776. [Google Scholar] [CrossRef]
- Yendapally, R.; Hurdle, J.G.; Carson, E.I.; Lee, R.B.; Lee, R.E. N-substituted 3-acetyltetramic acid derivatives as antibacterial agents. J. Med. Chem. 2008, 51, 1487–1491. [Google Scholar] [CrossRef]
- Casas, I.A.; Dobrogosz, W.J. Lactobacillus reuteri: An overview of a new probiotic for humans and animals. Microecol. Ther. 1997, 25, 221–231. [Google Scholar]
- Wang, W.; Zijlstra, R.T.; Ganzle, M.G. Feeding Limosilactobacillus fermentum K9-2 and Lacticaseibacillus casei K9-1, or Limosi Lactobacillus reuteri TMW1.656 Reduces Pathogen Load in Weanling Pigs. Front. Microbiol. 2020, 11, 608293. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, W.; Blaine, A.; Kane, S.T.; Zijlstra, R.T.; Ganzle, M.G. Impact of probiotic Lactobacillus sp. on autochthonous lactobacilli in weaned piglets. J. Appl. Microbiol. 2019, 126, 242–254. [Google Scholar] [CrossRef]
- Wu, X.; Cherian, P.T.; Lee, R.E.; Hurdle, J.G. The membrane as a target for controlling hypervirulent Clostridium difficile infections. J. Antimicrob. Chemother. 2013, 68, 806–815. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, X.; Le, M.H.; Zijlstra, R.T.; Ganzle, M.G. Reutericyclin producing Lactobacillus reuteri modulates development of fecal microbiota in weanling pigs. Front. Microbiol. 2015, 6, 762. [Google Scholar] [CrossRef]
- Riviere, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef] [PubMed]
- Imhann, F.; Bonder, M.J.; Vich Vila, A.; Fu, J.; Mujagic, Z.; Vork, L.; Tigchelaar, E.F.; Jankipersadsing, S.A.; Cenit, M.C.; Harmsen, H.J.; et al. Proton pump inhibitors affect the gut microbiome. Gut 2016, 65, 740–748. [Google Scholar] [CrossRef]
- Freedberg, D.E.; Toussaint, N.C.; Chen, S.P.; Ratner, A.J.; Whittier, S.; Wang, T.C.; Wang, H.H.; Abrams, J.A. Proton Pump Inhibitors Alter Specific Taxa in the Human Gastrointestinal Microbiome: A Crossover Trial. Gastroenterology 2015, 149, 883–885.e889. [Google Scholar] [CrossRef]
- Dobranowski, P.A.; Tang, C.; Sauve, J.P.; Menzies, S.C.; Sly, L.M. Compositional changes to the ileal microbiome precede the onset of spontaneous ileitis in SHIP deficient mice. Gut Microbes 2019, 10, 578–598. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Greenwood, R.; Costello Bde, L.; Ratcliffe, N.M.; Probert, C.S. An investigation of fecal volatile organic metabolites in irritable bowel syndrome. PLoS ONE 2013, 8, e58204. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Dickschat, J.S. Bacterial volatiles: The smell of small organisms. Nat. Prod. Rep. 2007, 24, 814–842. [Google Scholar] [CrossRef]
- Maddula, S.; Blank, L.M.; Schmid, A.; Baumbach, J.I. Detection of volatile metabolites of Escherichia coli by multi capillary column coupled ion mobility spectrometry. Anal. Bioanal. Chem. 2009, 394, 791–800. [Google Scholar] [CrossRef]
- Syhre, M.; Chambers, S.T. The scent of Mycobacterium tuberculosis. Tuberculosis 2008, 88, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Syhre, M.; Scotter, J.M.; Chambers, S.T. Investigation into the production of 2-Pentylfuran by Aspergillus fumigatus and other respiratory pathogens in vitro and human breath samples. Med. Mycol. 2008, 46, 209–215. [Google Scholar] [CrossRef]
- Trefz, P.; Koehler, H.; Klepik, K.; Moebius, P.; Reinhold, P.; Schubert, J.K.; Miekisch, W. Volatile emissions from Mycobacterium avium subsp. paratuberculosis mirror bacterial growth and enable distinction of different strains. PLoS ONE 2013, 8, e76868. [Google Scholar] [CrossRef]
- Lv, D.; Lu, S.; Tan, X.; Shao, M.; Xie, S.; Wang, L. Source profiles, emission factors and associated contributions to secondary pollution of volatile organic compounds (VOCs) emitted from a local petroleum refinery in Shandong. Environ. Pollut. 2021, 274, 116589. [Google Scholar] [CrossRef] [PubMed]
- Pitiriciu, M.; Tansel, B. Volatile organic contaminants (VOCs) emitted from sewer networks during wastewater collection and transport. J. Environ. Manag. 2021, 285, 112136. [Google Scholar] [CrossRef] [PubMed]
- Rath, M.; Mitchell, T.R.; Gold, S.E. Volatiles produced by Bacillus mojavensis RRC101 act as plant growth modulators and are strongly culture-dependent. Microbiol. Res. 2018, 208, 76–84. [Google Scholar] [CrossRef]
- Park, H.B.; Lee, B.; Kloepper, J.W.; Ryu, C.M. One shot-two pathogens blocked: Exposure of Arabidopsis to hexadecane, a long chain volatile organic compound, confers induced resistance against both Pectobacterium carotovorum and Pseudomonas syringae. Plant Signal. Behav. 2013, 8, e24619. [Google Scholar] [CrossRef]
- Yasmin, H.; Rashid, U.; Hassan, M.N.; Nosheen, A.; Naz, R.; Ilyas, N.; Sajjad, M.; Azmat, A.; Alyemeni, M.N. Volatile organic compounds produced by Pseudomonas pseudoalcaligenes alleviated drought stress by modulating defense system in maize (Zea mays L.). Physiol. Plant 2020. [Google Scholar] [CrossRef]
- Vitense, P.; Kasbohm, E.; Klassen, A.; Gierschner, P.; Trefz, P.; Weber, M.; Miekisch, W.; Schubert, J.K.; Mobius, P.; Reinhold, P.; et al. Detection of Mycobacterium avium ssp. paratuberculosis in Cultures from Fecal and Tissue Samples Using VOC Analysis and Machine Learning Tools. Front Vet. Sci. 2021, 8, 620327. [Google Scholar] [CrossRef]
- Mochalski, P.; Unterkofler, K. Quantification of selected volatile organic compounds in human urine by gas chromatography selective reagent ionization time of flight mass spectrometry (GC-SRI-TOF-MS) coupled with head-space solid-phase microextraction (HS-SPME). Analyst 2016, 141, 4796–4803. [Google Scholar] [CrossRef]
- Harshman, S.W.; Geier, B.A.; Fan, M.; Rinehardt, S.; Watts, B.S.; Drummond, L.A.; Preti, G.; Phillips, J.B.; Ott, D.K.; Grigsby, C.C. The identification of hypoxia biomarkers from exhaled breath under normobaric conditions. J. Breath Res. 2015, 9, 047103. [Google Scholar] [CrossRef]
- Klymiuk, I.; Singer, G.; Castellani, C.; Trajanoski, S.; Obermuller, B.; Till, H. Characterization of the Luminal and Mucosa-Associated Microbiome along the Gastrointestinal Tract: Results from Surgically Treated Preterm Infants and a Murine Model. Nutrients 2021, 13, 1030. [Google Scholar] [CrossRef]
- Klymiuk, I.; Bilgilier, C.; Stadlmann, A.; Thannesberger, J.; Kastner, M.T.; Hogenauer, C.; Puspok, A.; Biowski-Frotz, S.; Schrutka-Kolbl, C.; Thallinger, G.G.; et al. The Human Gastric Microbiome Is Predicated upon Infection with Helicobacter pylori. Front. Microbiol. 2017, 8, 2508. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewski, M.; Proietti, C.; Ellis, J.J.; Hasan, S.; Brion, M.J.; Berger, B.; Krause, L. Calypso: A user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics 2017, 33, 782–783. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, A.; Trefz, P.; Fischer, S.; Klepik, K.; Walter, G.; Steffens, M.; Ziller, M.; Schubert, J.K.; Reinhold, P.; Kohler, H.; et al. In Vivo Volatile Organic Compound Signatures of Mycobacterium avium subsp. paratuberculosis. PLoS ONE 2015, 10, e0123980. [Google Scholar] [CrossRef]
- Miekisch, W.; Trefz, P.; Bergmann, A.; Schubert, J.K. Microextraction techniques in breath biomarker analysis. Bioanalysis 2014, 6, 1275–1291. [Google Scholar] [CrossRef]
- Obermuller, B.; Singer, G.; Kienesberger, B.; Klymiuk, I.; Sperl, D.; Stadlbauer, V.; Horvath, A.; Miekisch, W.; Gierschner, P.; Grabherr, R.; et al. The Effects of Prebiotic Supplementation with OMNi-LOGiC® FIBRE on Fecal Microbiome, Fecal Volatile Organic Compounds, and Gut Permeability in Murine Neuroblastoma-Induced Tumor-Associated Cachexia. Nutrients 2020, 12, 2029. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kienesberger, B.; Obermüller, B.; Singer, G.; Mittl, B.; Grabherr, R.; Mayrhofer, S.; Heinl, S.; Stadlbauer, V.; Horvath, A.; Miekisch, W.; et al. (S)-Reutericyclin: Susceptibility Testing and In Vivo Effect on Murine Fecal Microbiome and Volatile Organic Compounds. Int. J. Mol. Sci. 2021, 22, 6424. https://doi.org/10.3390/ijms22126424
Kienesberger B, Obermüller B, Singer G, Mittl B, Grabherr R, Mayrhofer S, Heinl S, Stadlbauer V, Horvath A, Miekisch W, et al. (S)-Reutericyclin: Susceptibility Testing and In Vivo Effect on Murine Fecal Microbiome and Volatile Organic Compounds. International Journal of Molecular Sciences. 2021; 22(12):6424. https://doi.org/10.3390/ijms22126424
Chicago/Turabian StyleKienesberger, Bernhard, Beate Obermüller, Georg Singer, Barbara Mittl, Reingard Grabherr, Sigrid Mayrhofer, Stefan Heinl, Vanessa Stadlbauer, Angela Horvath, Wolfram Miekisch, and et al. 2021. "(S)-Reutericyclin: Susceptibility Testing and In Vivo Effect on Murine Fecal Microbiome and Volatile Organic Compounds" International Journal of Molecular Sciences 22, no. 12: 6424. https://doi.org/10.3390/ijms22126424
APA StyleKienesberger, B., Obermüller, B., Singer, G., Mittl, B., Grabherr, R., Mayrhofer, S., Heinl, S., Stadlbauer, V., Horvath, A., Miekisch, W., Fuchs, P., Klymiuk, I., Till, H., & Castellani, C. (2021). (S)-Reutericyclin: Susceptibility Testing and In Vivo Effect on Murine Fecal Microbiome and Volatile Organic Compounds. International Journal of Molecular Sciences, 22(12), 6424. https://doi.org/10.3390/ijms22126424







