LncRSPH9-4 Facilitates Meningitic Escherichia coli-Caused Blood–Brain Barrier Disruption via miR-17-5p/MMP3 Axis
Abstract
1. Introduction
2. Results
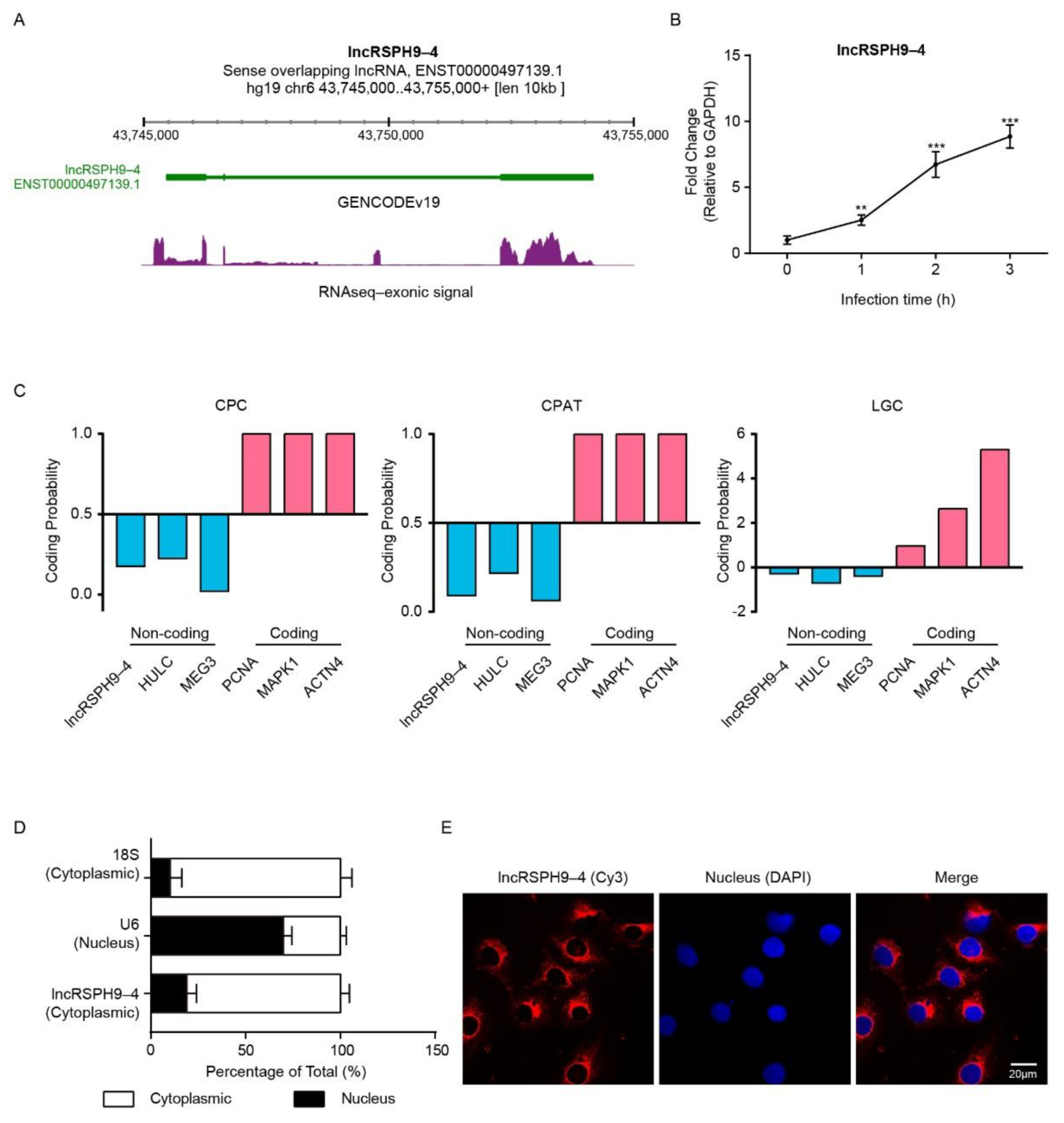
2.1. LncRSPH9-4 Was Significantly Up-Regulated and Cytoplasmically Distributed in Meningitic E. coli-Challenged hBMECs
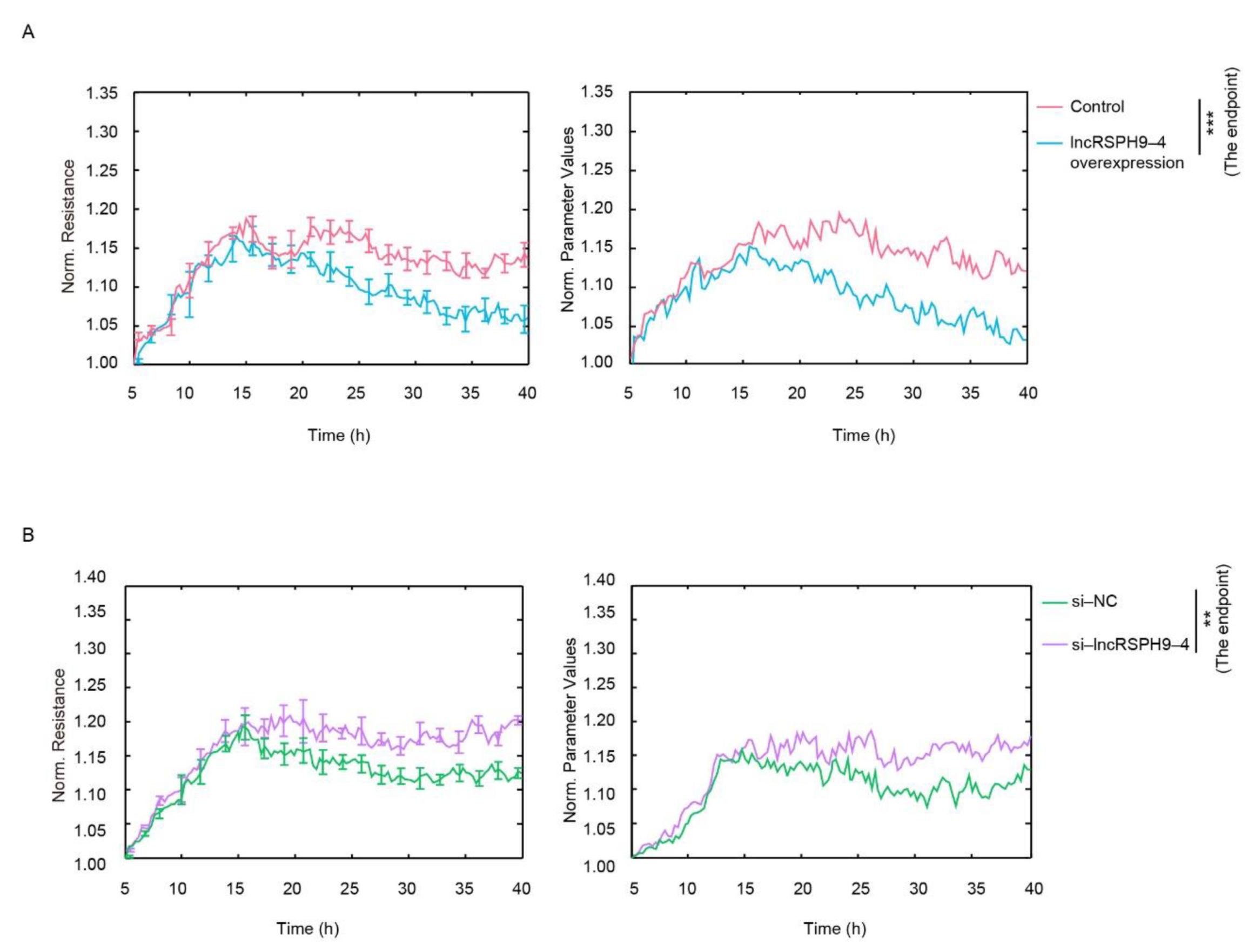
2.2. LncRSPH9-4 Negatively Affected the Barrier Permeability of hBMECs
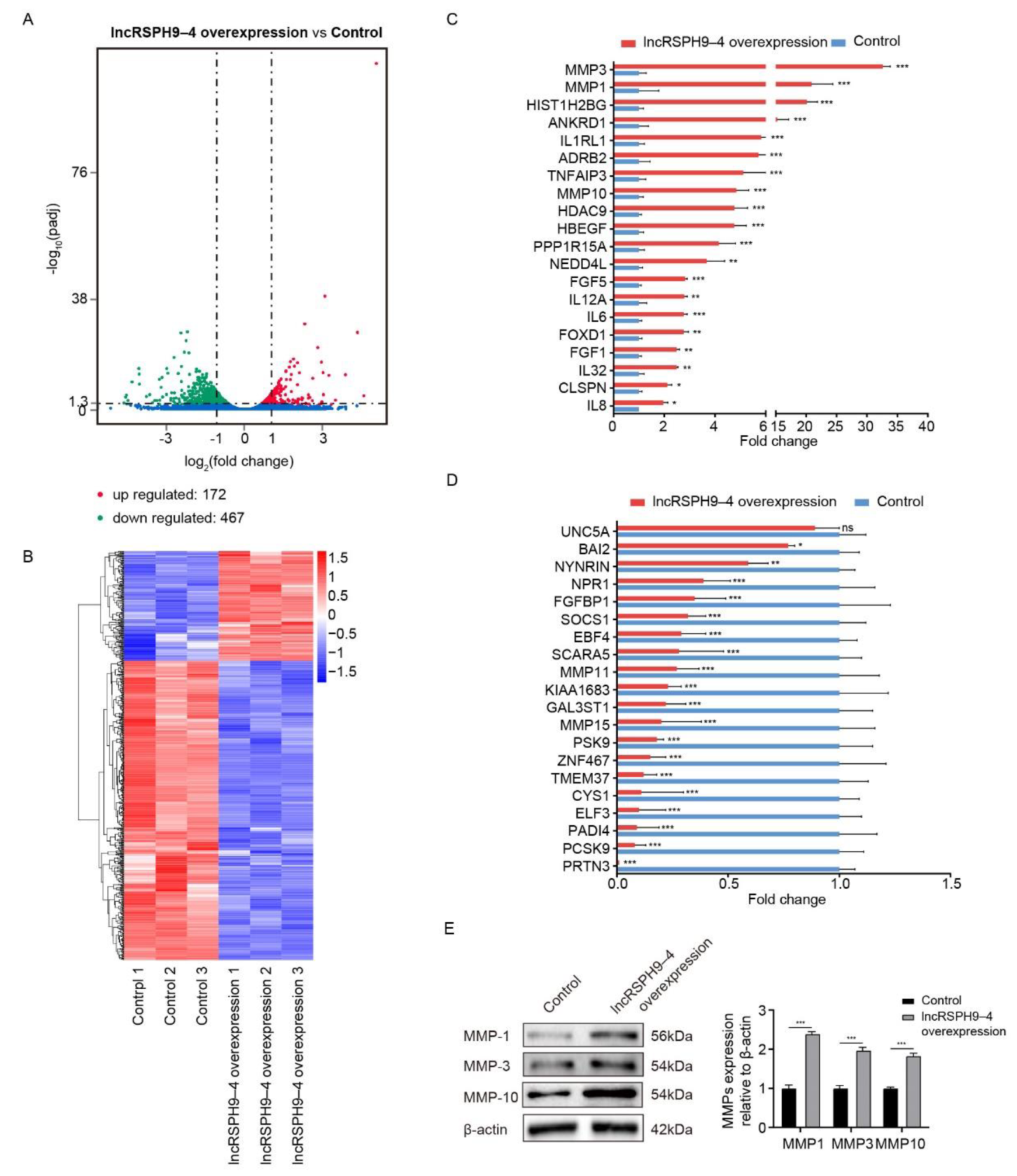
2.3. Differential mRNAs Transcription in hBMECs with LncRSPH9-4 Overexpression
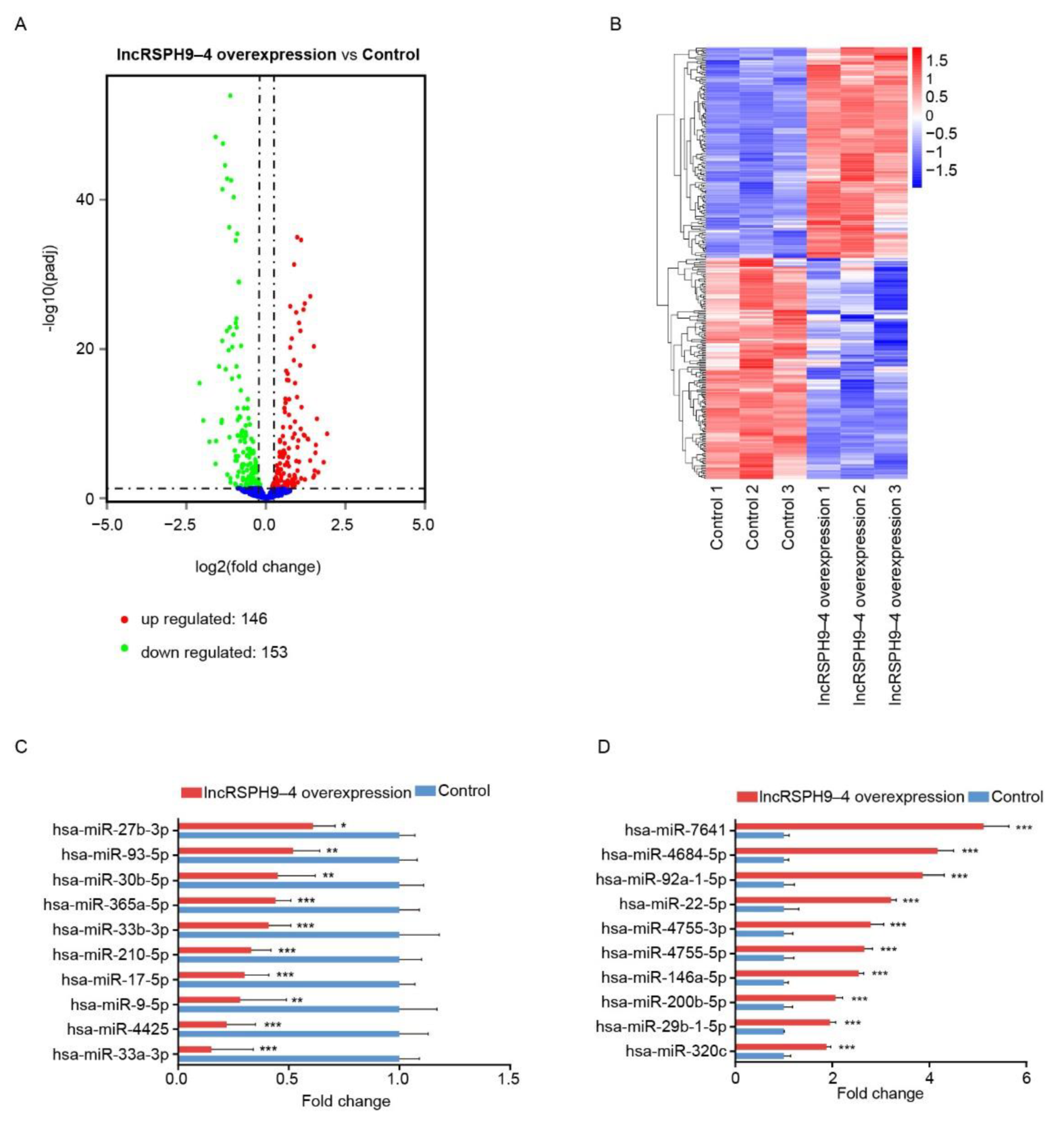
2.4. Altered miRNAs Profiles in hBMECs with LncRSPH9-4 Overexpression
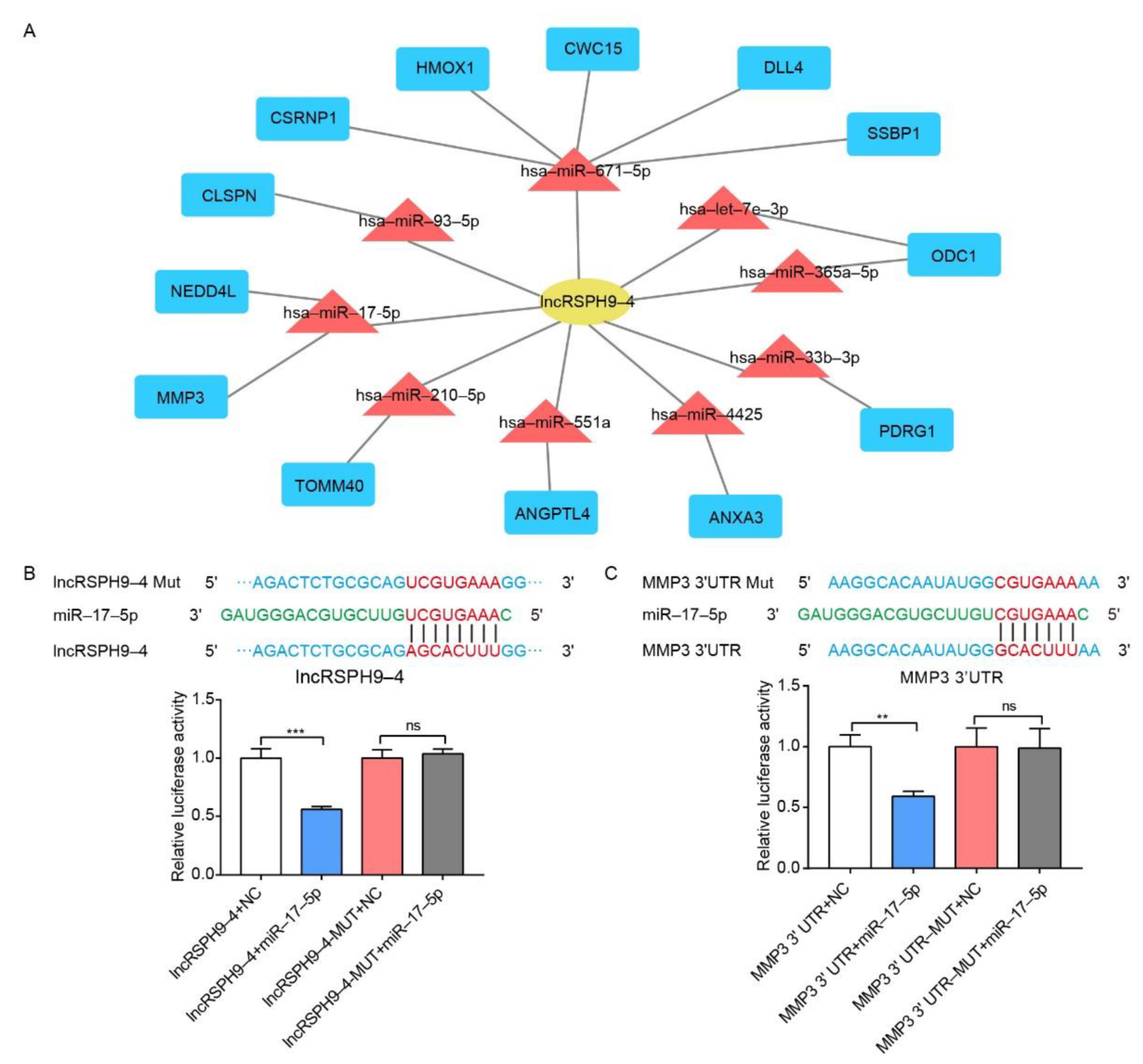
2.5. The Competitive Endogenous RNA Network Analysis of LncRSPH9-4
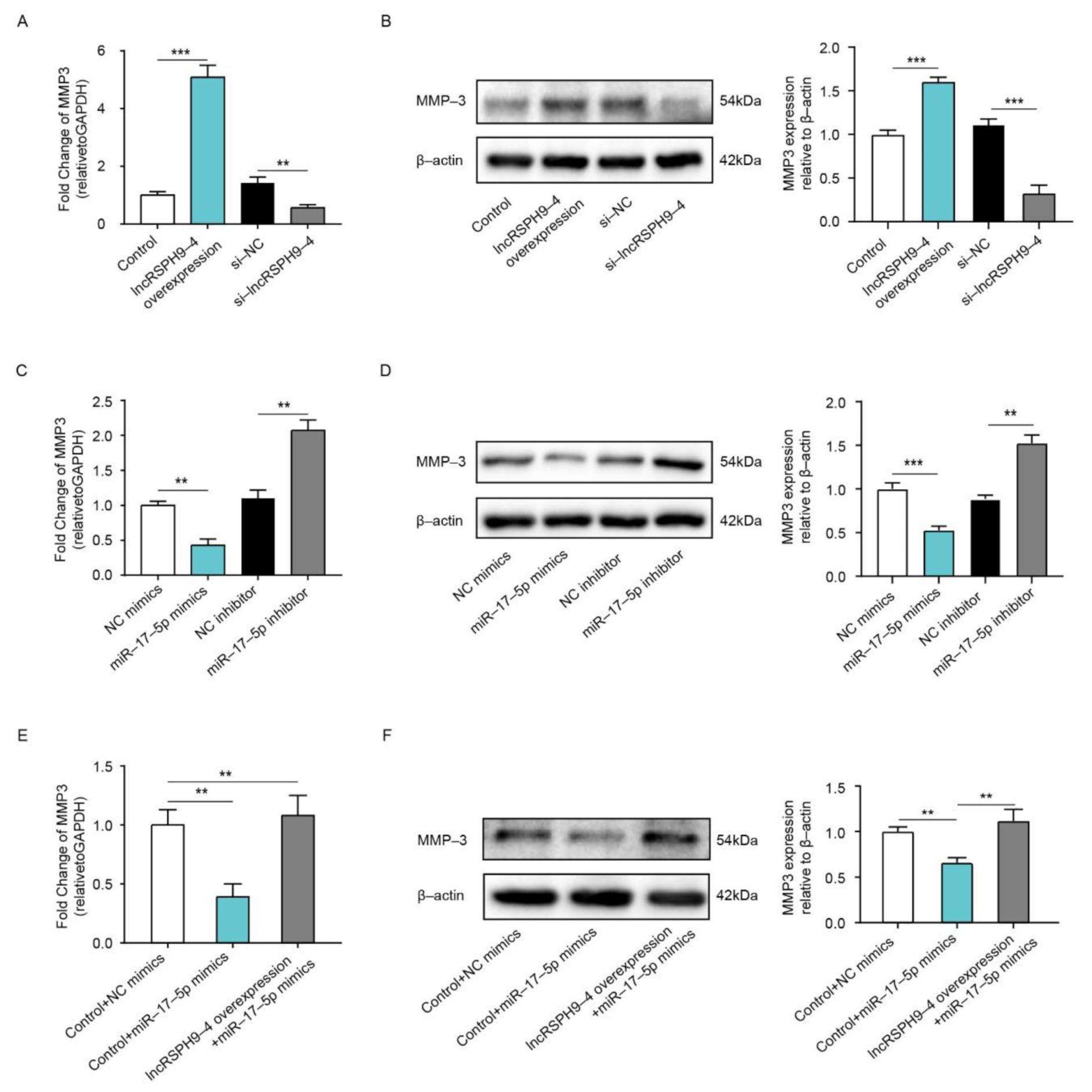
2.6. LncRSPH9-4 Positively Regulated MMP3 Expression
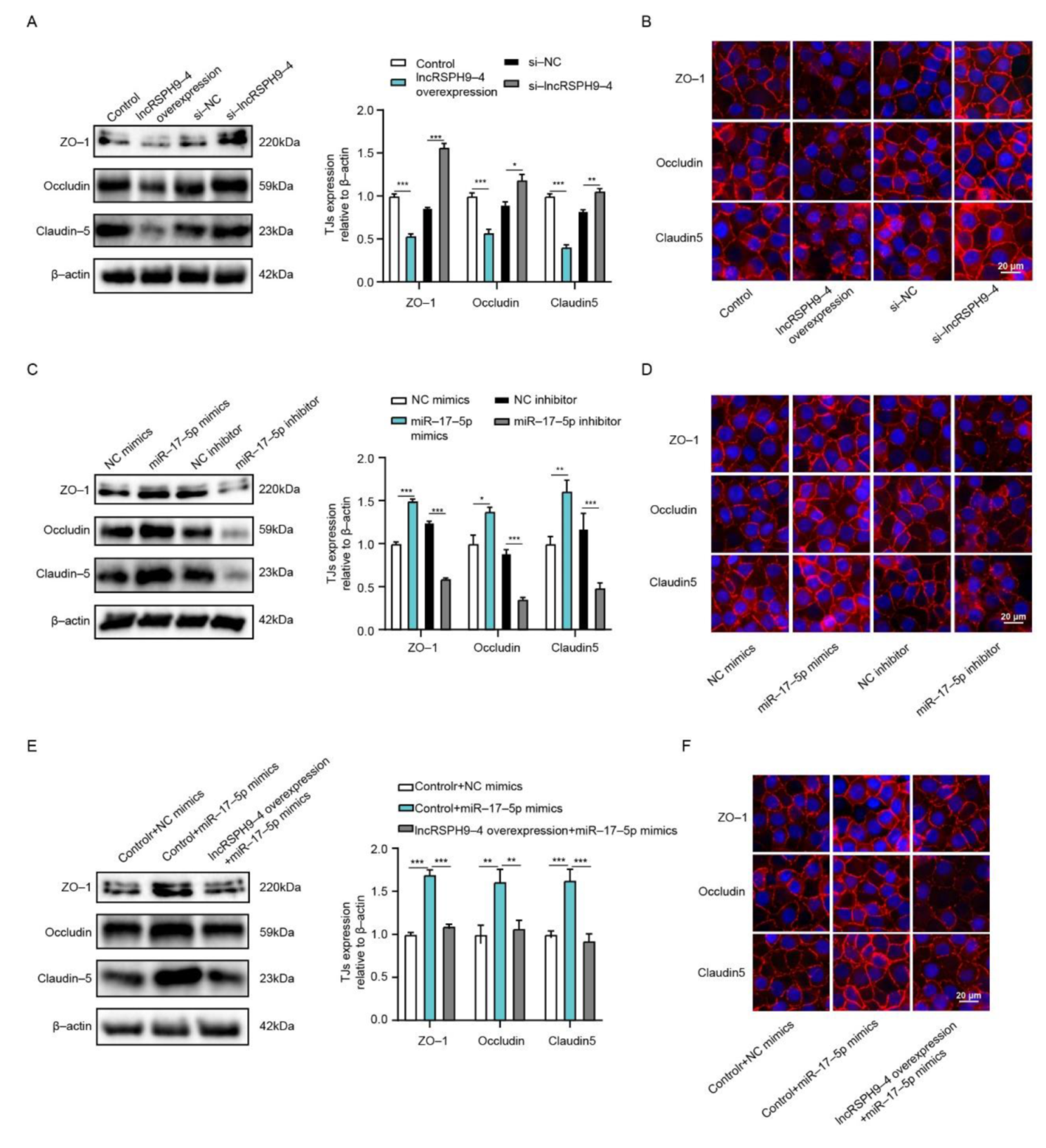
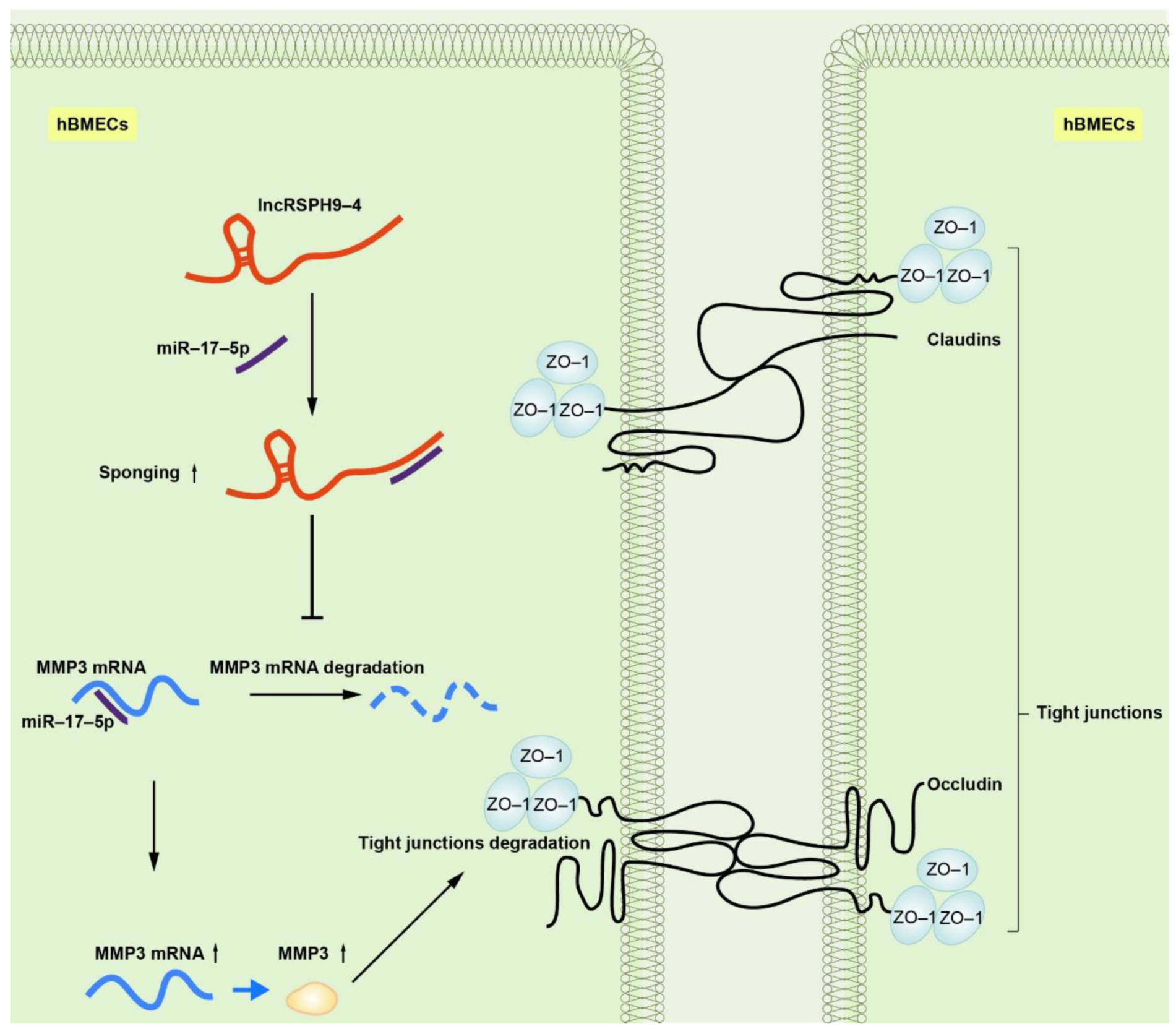
2.7. LncRSPH9-4 Disrupted BBB Junction Protein through miR-17-5p/MMP3 Axis
3. Discussion
4. Materials and Methods
4.1. Cell Line and Cell Culture
4.2. Meningitic E. coli Infection of hBMECs
4.3. Transfection
4.4. Electrical Cell-Substrate Impedance Sensing
4.5. RNA Isolation and Quantitative Real-Time PCR Analysis
4.6. mRNA Library Preparation and Sequencing
4.7. Small RNA Library Preparation and Sequencing
4.8. Construction of the lncRNA-miRNA-mRNA ceRNA Network
4.9. Dual-Luciferase Reporter Assays
4.10. FISH
4.11. Western Blotting
4.12. Immunofluorescence Assay
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, K.S. Mechanisms of microbial traversal of the blood-brain barrier. Nat. Rev. Microbiol. 2008, 6, 625–634. [Google Scholar] [CrossRef]
- Kim, K.S. Pathogenesis of bacterial meningitis: From bacteraemia to neuronal injury. Nat. Rev. Neurosci. 2003, 4, 376–385. [Google Scholar] [CrossRef]
- Sandoval, K.E.; Witt, K.A. Blood-brain barrier tight junction permeability and ischemic stroke. Neurobiol. Dis. 2008, 32, 200–219. [Google Scholar] [CrossRef] [PubMed]
- Dejana, E.; Tournier-Lasserve, E.; Weinstein, B.M. The control of vascular integrity by endothelial cell junctions: Molecular basis and pathological implications. Dev. Cell 2009, 16, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Edwards, V.L.; Wang, L.C.; Dawson, V.; Stein, D.C.; Song, W. Neisseria gonorrhoeae breaches the apical junction of polarized epithelial cells for transmigration by activating EGFR. Cell. Microbiol. 2013, 15, 1042–1057. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hawkins, B.T.; Davis, T.P. The blood-brain barrier/neurovascular unit in health and disease. Pharmacol. Rev. 2005, 57, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Chang, H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef]
- Lee, J.T.; Bartolomei, M.S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2013, 152, 1308–1323. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Rinn, J.L. Large non-coding RNAs: Missing links in cancer? Hum. Mol. Genet. 2010, 19, R152–R161. [Google Scholar] [CrossRef]
- Huang, W.; Thomas, B.; Flynn, R.A.; Gavzy, S.J.; Wu, L.; Kim, S.V.; Hall, J.A.; Miraldi, E.R.; Ng, C.P.; Rigo, F.; et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 2015, 528, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xu, J.; Wang, Y.; Cao, X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 2017, 358, 1051–1055. [Google Scholar] [CrossRef]
- Zhang, L.; Luo, X.; Chen, F.; Yuan, W.; Xiao, X.; Zhang, X.; Dong, Y.; Zhang, Y.; Liu, Y. LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J. Cell. Biochem. 2018, 119, 5460–5472. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Chen, L.; Wang, Y.; Jiang, X.; Xia, H.; Zhuang, Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J. Neurooncol. 2015, 121, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Yu, Z.; Fang, X.; Liu, M.; Pu, Y.; Shao, Q.; Wang, D.; Zhao, X.; Huang, A.; Xiang, Z.; et al. LncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Rep. 2017, 18, 1801–1816. [Google Scholar] [CrossRef]
- Liu, W.Q.; Wang, Y.J.; Zheng, Y.; Chen, X. Effects of long non-coding RNA NEAT1 on sepsis-induced brain injury in mice via NF-kappaB. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3933–3939. [Google Scholar] [CrossRef]
- Li, Z.; Li, J.; Tang, N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience 2017, 354, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Huang, F.; Fu, J.; Dou, B.; Xu, B.; Miao, L.; Liu, W.; Yang, X.; Tan, C.; Chen, H.; et al. Differential transcription profiles of long non-coding RNAs in primary human brain microvascular endothelial cells in response to meningitic Escherichia coli. Sci. Rep. 2016, 6, 38903. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zi, X.H. LncRNA SNHG1 alleviates OGD induced injury in BMEC via miR-338/HIF-1alpha axis. Brain Res. 2019, 1714, 174–181. [Google Scholar] [CrossRef]
- Zhang, T.; Tian, C.; Wu, J.; Zhang, Y.; Wang, J.; Kong, Q.; Mu, L.; Sun, B.; Ai, T.; Wang, Y.; et al. MicroRNA-182 exacerbates blood-brain barrier (BBB) disruption by downregulating the mTOR/FOXO1 pathway in cerebral ischemia. FASEB J. 2020. [Google Scholar] [CrossRef]
- Bobbili, M.R.; Mader, R.M.; Grillari, J.; Dellago, H. OncomiR-17-5p: Alarm signal in cancer? Oncotarget 2017, 8, 71206–71222. [Google Scholar] [CrossRef] [PubMed]
- Dellago, H.; Bobbili, M.R.; Grillari, J. MicroRNA-17-5p: At the Crossroads of Cancer and Aging—A Mini-Review. Gerontology 2017, 63, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Gurney, K.J.; Estrada, E.Y.; Rosenberg, G.A. Blood-brain barrier disruption by stromelysin-1 facilitates neutrophil infiltration in neuroinflammation. Neurobiol. Dis. 2006, 23, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.; Qian, Y.; Xu, G.; Luo, Q.; Zhang, Z. Long-term characterization of activated microglia/macrophages facilitating the development of experimental brain metastasis through intravital microscopic imaging. J. Neuroinflamm. 2019, 16, 4. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol. Dis. 2012, 46, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.H.; Szeto, V.; Yang, B.B.; Zhu, S.Z.; Sun, H.S.; Feng, Z.P. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018, 9, 281. [Google Scholar] [CrossRef]
- Ruan, W.; Li, J.; Xu, Y.; Wang, Y.; Zhao, F.; Yang, X.; Jiang, H.; Zhang, L.; Saavedra, J.M.; Shi, L.; et al. MALAT1 Up-Regulator Polydatin Protects Brain Microvascular Integrity and Ameliorates Stroke Through C/EBPbeta/MALAT1/CREB/PGC-1alpha/PPARgamma Pathway. Cell. Mol. Neurobiol. 2019, 39, 265–286. [Google Scholar] [CrossRef]
- Mus, E.; Hof, P.R.; Tiedge, H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2007, 104, 10679–10684. [Google Scholar] [CrossRef]
- Arisi, I.; D’Onofrio, M.; Brandi, R.; Felsani, A.; Capsoni, S.; Drovandi, G.; Felici, G.; Weitschek, E.; Bertolazzi, P.; Cattaneo, A. Gene expression biomarkers in the brain of a mouse model for Alzheimer’s disease: Mining of microarray data by logic classification and feature selection. J. Alzheimers Dis. 2011, 24, 721–738. [Google Scholar] [CrossRef]
- Yue, X.H.; Guo, L.; Wang, Z.Y.; Jia, T.H. Inhibition of miR-17-5p promotes mesenchymal stem cells to repair spinal cord injury. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3899–3907. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yang, F.; Cao, H.; Liang, Z. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015, 29, 3054–3064. [Google Scholar] [CrossRef]
- Li, S.; Zhu, A.; Ren, K.; Li, S.; Chen, L. IFNbeta-induced exosomal linc-EPHA6-1 promotes cytotoxicity of NK cells by acting as a ceRNA for hsa-miR-4485-5p to up-regulate NKp46 expression. Life Sci. 2020, 257, 118064. [Google Scholar] [CrossRef]
- Hermann, D.M.; ElAli, A. The abluminal endothelial membrane in neurovascular remodeling in health and disease. Sci. Signal 2012, 5, re4. [Google Scholar] [CrossRef]
- Song, J.; Hu, Y.; Li, H.; Huang, X.; Zheng, H.; Hu, Y.; Wang, J.; Jiang, X.; Li, J.; Yang, Z.; et al. miR-1303 regulates BBB permeability and promotes CNS lesions following CA16 infections by directly targeting MMP9. Emerg. Microbes Infect. 2018, 7, 155. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Zhong, Y.; Gu, L.; Shen, W.; Guo, J. MiR-21 involve in ERK-mediated upregulation of MMP9 in the rat hippocampus following cerebral ischemia. Brain Res. Bull. 2013, 94, 56–62. [Google Scholar] [CrossRef]
- Kalani, A.; Kamat, P.K.; Familtseva, A.; Chaturvedi, P.; Muradashvili, N.; Narayanan, N.; Tyagi, S.C.; Tyagi, N. Role of microRNA29b in blood-brain barrier dysfunction during hyperhomocysteinemia: An epigenetic mechanism. J. Cereb. Blood Flow Metab. 2014, 34, 1212–1222. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Zhang, Y.; Du, X.F.; Li, J.; Zi, H.X.; Bu, J.W.; Yan, Y.; Han, H.; Du, J.L. Neurons secrete miR-132-containing exosomes to regulate brain vascular integrity. Cell. Res. 2017, 27, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Kimura-Ohba, S.; Thompson, J.F.; Salayandia, V.M.; Cosse, M.; Raz, L.; Jalal, F.Y.; Rosenberg, G.A. Vascular tight junction disruption and angiogenesis in spontaneously hypertensive rat with neuroinflammatory white matter injury. Neurobiol. Dis. 2018, 114, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Stins, M.F.; Badger, J.; Sik Kim, K. Bacterial invasion and transcytosis in transfected human brain microvascular endothelial cells. Microb. Pathog. 2001, 30, 19–28. [Google Scholar] [CrossRef]
- Stins, M.F.; Gilles, F.; Kim, K.S. Selective expression of adhesion molecules on human brain microvascular endothelial cells. J. Neuroimmunol. 1997, 76, 81–90. [Google Scholar] [CrossRef]
- Kim, B.J.; Hancock, B.M.; Bermudez, A.; Del Cid, N.; Reyes, E.; van Sorge, N.M.; Lauth, X.; Smurthwaite, C.A.; Hilton, B.J.; Stotland, A.; et al. Bacterial induction of Snail1 contributes to blood-brain barrier disruption. J. Clin. Investig. 2015, 125, 2473–2483. [Google Scholar] [CrossRef]
- Liu, T.; Miao, Z.; Jiang, J.; Yuan, S.; Fang, W.; Li, B.; Chen, Y. Visfatin Mediates SCLC Cells Migration across Brain Endothelial Cells through Upregulation of CCL2. Int. J. Mol. Sci. 2015, 16, 11439–11451. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Chen, J.; Xu, B.; Yang, B.; Fu, J.; Xiao, S.; Tan, C.; Chen, H.; Wang, X. circ_2858 Helps Blood-Brain Barrier Disruption by Increasing VEGFA via Sponging miR-93-5p during Escherichia coli Meningitis. Mol. Ther. Nucleic Acids 2020, 22, 708–721. [Google Scholar] [CrossRef]
- Lerner, A.; Neidhofer, S.; Reuter, S.; Matthias, T. MMP3 is a reliable marker for disease activity, radiological monitoring, disease outcome predictability, and therapeutic response in rheumatoid arthritis. Best Pract. Res. Clin. Rheumatol. 2018, 32, 550–562. [Google Scholar] [CrossRef]
- Zhang, Q.; Zheng, M.; Betancourt, C.E.; Liu, L.; Sitikov, A.; Sladojevic, N.; Zhao, Q.; Zhang, J.H.; Liao, J.K.; Wu, R. Increase in Blood-Brain Barrier (BBB) Permeability Is Regulated by MMP3 via the ERK Signaling Pathway. Oxid. Med. Cell Longev. 2021, 2021, 6655122. [Google Scholar] [CrossRef]
- Rosenberg, G.A.; Yang, Y. Vasogenic edema due to tight junction disruption by matrix metalloproteinases in cerebral ischemia. Neurosurg. Focus 2007, 22, E4. [Google Scholar] [CrossRef]
- Von Wedel-Parlow, M.; Schrot, S.; Lemmen, J.; Treeratanapiboon, L.; Wegener, J.; Galla, H.J. Neutrophils cross the BBB primarily on transcellular pathways: An in vitro study. Brain Res. 2011, 1367, 62–76. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, B.; Yang, R.; Fu, J.; Yang, B.; Chen, J.; Tan, C.; Chen, H.; Wang, X. LncRSPH9-4 Facilitates Meningitic Escherichia coli-Caused Blood–Brain Barrier Disruption via miR-17-5p/MMP3 Axis. Int. J. Mol. Sci. 2021, 22, 6343. https://doi.org/10.3390/ijms22126343
Xu B, Yang R, Fu J, Yang B, Chen J, Tan C, Chen H, Wang X. LncRSPH9-4 Facilitates Meningitic Escherichia coli-Caused Blood–Brain Barrier Disruption via miR-17-5p/MMP3 Axis. International Journal of Molecular Sciences. 2021; 22(12):6343. https://doi.org/10.3390/ijms22126343
Chicago/Turabian StyleXu, Bojie, Ruicheng Yang, Jiyang Fu, Bo Yang, Jiaqi Chen, Chen Tan, Huanchun Chen, and Xiangru Wang. 2021. "LncRSPH9-4 Facilitates Meningitic Escherichia coli-Caused Blood–Brain Barrier Disruption via miR-17-5p/MMP3 Axis" International Journal of Molecular Sciences 22, no. 12: 6343. https://doi.org/10.3390/ijms22126343
APA StyleXu, B., Yang, R., Fu, J., Yang, B., Chen, J., Tan, C., Chen, H., & Wang, X. (2021). LncRSPH9-4 Facilitates Meningitic Escherichia coli-Caused Blood–Brain Barrier Disruption via miR-17-5p/MMP3 Axis. International Journal of Molecular Sciences, 22(12), 6343. https://doi.org/10.3390/ijms22126343






