Transcriptome-Wide Identification and Quantification of Caffeoylquinic Acid Biosynthesis Pathway and Prediction of Its Putative BAHDs Gene Complex in A. spathulifolius
Abstract
1. Introduction
2. Results
2.1. Assembly and Gene Annotation
2.2. Functional Characterization and DEGs of PP Biosynthesis
2.3. Prediction of BAHDs Superfamily
2.4. BAHDs Protein Phylogenies
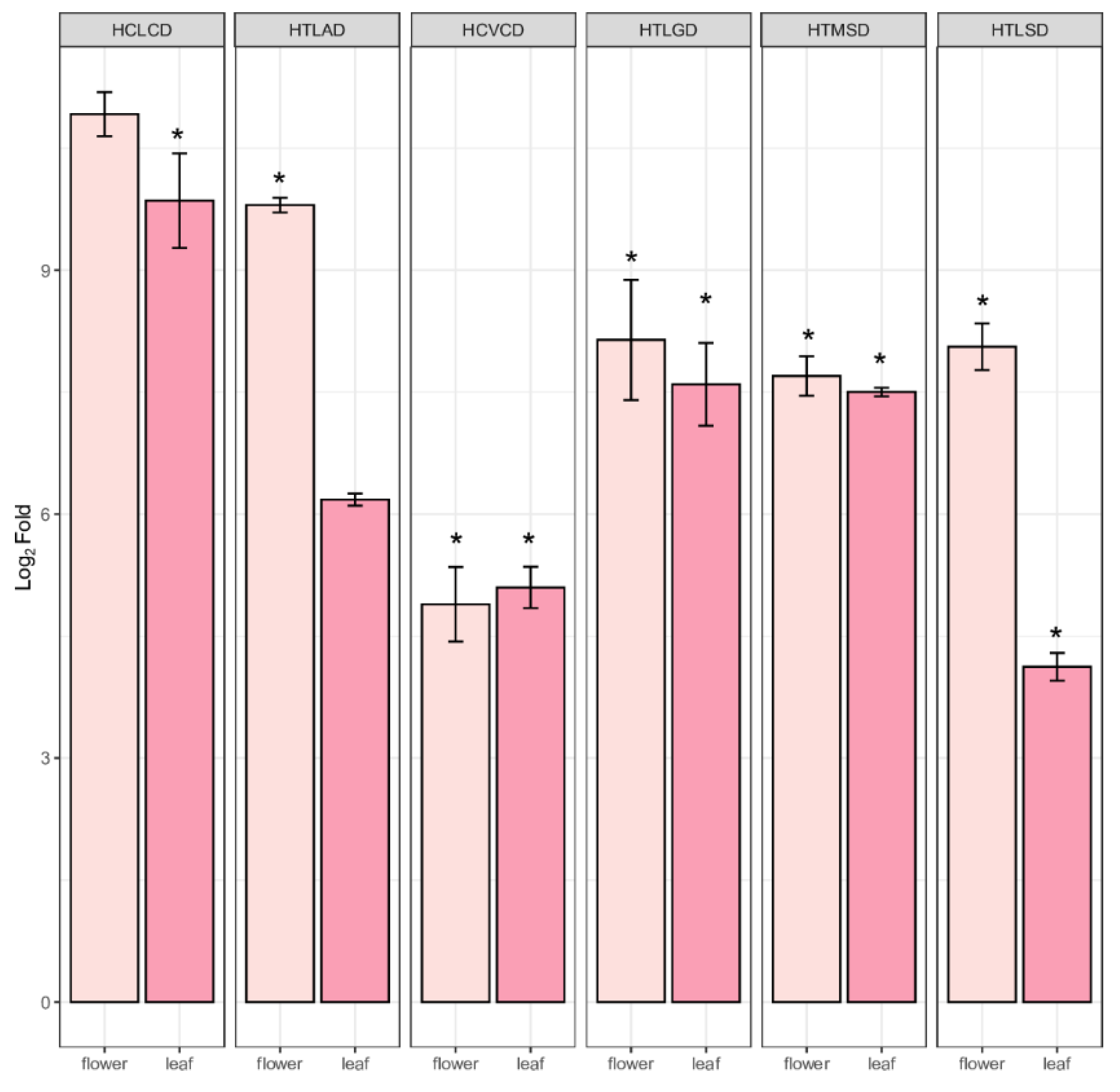
2.5. Quantification of PPP Unigenes
3. Discussion
4. Materials and Methods
4.1. RNA Isolation, cDNA Library Construction, and Illumina Sequencing
4.2. Denovo Assembly and Functional Annotation
4.3. Identification of PPP Unigenes
4.4. Differentially Expressed Genes (DEGs) Analysis
4.5. Structure of BAHD Family Member Unigenes
4.6. qRT-PCR Expression Studies
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arnold, P.A.; Kruuk, L.E.B.; Nicotra, A.B. How to analyse plant phenotypic plasticity in response to a changing climate. New Phytol. 2019, 222, 1235–1241. [Google Scholar] [CrossRef]
- Austen, N.; Walker, H.J.; Lake, J.A.; Phoenix, G.K.; Cameron, D.D. The Regulation of Plant Secondary Metabolism in Response to Abiotic Stress: Interactions between Heat Shock and Elevated CO2. Front. Plant Sci. 2019, 10, 1463. [Google Scholar] [CrossRef]
- Berini, J.L.; Brockman, S.A.; Hegeman, A.D.; Reich, P.B.; Muthukrishnan, R.; Montgomery, R.A.; Forester, J.D. Combinations of Abiotic Factors Differentially Alter Production of Plant Secondary Metabolites in Five Woody Plant Species in the Boreal-Temperate Transition Zone. Front. Plant Sci. 2018, 9, 1257. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wen, K.S.; Ruan, X.; Zhao, Y.X.; Wei, F.; Wang, Q. Response of Plant Secondary Metabolites to Environmental Factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef] [PubMed]
- Carrington, Y.; Guo, J.; Le, C.H.; Fillo, A.; Kwon, J.; Tran, L.T.; Ehlting, J. Evolution of a secondary metabolic pathway from primary metabolism: Shikimate and quinate biosynthesis in plants. Plant J. 2018, 95, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52. [Google Scholar] [CrossRef]
- Biala, W.; Jasinski, M. The Phenylpropanoid Case—It Is Transport That Matters. Front. Plant Sci. 2018, 9, 1610. [Google Scholar] [CrossRef]
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biotechnol. 2015, 148, 63–106. [Google Scholar]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino Acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Yoo, H.; Widhalm, J.R.; Qian, Y.; Maeda, H.; Cooper, B.R.; Jannasch, A.S.; Gonda, I.; Lewinsohn, E.; Rhodes, D.; Dudareva, N. An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine:phenylpyruvate aminotransferase. Nat. Commun. 2013, 4, 2833. [Google Scholar] [CrossRef] [PubMed]
- Tzin, V.; Galili, G. The Biosynthetic Pathways for Shikimate and Aromatic Amino Acids in Arabidopsis thaliana. Arab. Book 2010, 8, e0132. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Sood, P.; Citovsky, V. The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol. Plant Pathol. 2010, 11, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, K.M.; Weaver, L.M. The Shikimate Pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 473–503. [Google Scholar] [CrossRef]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.J.; Reddy, M.S.; Wang, L. The phenylpropanoid pathway and plant defence-a genomics perspective. Mol. Plant Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef]
- Cle, C.; Hill, L.M.; Niggeweg, R.; Martin, C.R.; Guisez, Y.; Prinsen, E.; Jansen, M.A. Modulation of chlorogenic acid biosynthesis in Solanum lycopersicum; consequences for phenolic accumulation and UV-tolerance. Phytochemistry 2008, 69, 2149–2156. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.C.; Dai, F.; Zhou, B.; Yang, L.; Liu, Z.L. Antioxidant activity of hydroxycinnamic acid derivatives in human low density lipoprotein: Mechanism and structure-activity relationship. Food Chem. 2007, 104, 132–139. [Google Scholar] [CrossRef]
- Rahman, M.A.; Abdullah, N.; Aminudin, N. Antioxidative Effects and Inhibition of Human Low Density Lipoprotein Oxidation In Vitro of Polyphenolic Compounds in Flammulina velutipes (Golden Needle Mushroom). Oxid. Med. Cell Longev. 2015, 2015, 403023. [Google Scholar] [CrossRef]
- Clifford, M.N. Chlorogenic acids and other cinnamates—Nature, occurrence, dietary burden, absorption and metabolism. J. Sci. Food Agric. 2000, 80, 1033–1043. [Google Scholar] [CrossRef]
- Olthof, M.R.; Hollman, P.C.; Katan, M.B. Chlorogenic acid and caffeic acid are absorbed in humans. J. Nutr. 2001, 131, 66–71. [Google Scholar] [CrossRef]
- Cho, A.S.; Jeon, S.M.; Kim, M.J.; Yeo, J.; Seo, K.I.; Choi, M.S.; Lee, M.K. Chlorogenic acid exhibits anti-obesity property and improves lipid metabolism in high-fat diet-induced-obese mice. Food Chem. Toxicol. 2010, 48, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Gonthier, M.P.; Verny, M.A.; Besson, C.; Remesy, C.; Scalbert, A. Chlorogenic acid bioavailability largely depends on its metabolism by the gut microflora in rats. J. Nutr. 2003, 133, 1853–1859. [Google Scholar] [CrossRef] [PubMed]
- Azuma, K.; Ippoushi, K.; Nakayama, M.; Ito, H.; Higashio, H.; Terao, J. Absorption of chlorogenic acid and caffeic acid in rats after oral administration. J. Agric. Food Chem. 2000, 48, 5496–5500. [Google Scholar] [CrossRef] [PubMed]
- Uranga, J.G.; Podio, N.S.; Wunderlin, D.A.; Santiago, A.N. Theoretical and Experimental Study of the Antioxidant Behaviors of 5-O-Caffeoylquinic, Quinic and Caffeic Acids Based on Electronic and Structural Properties. Chemistryselect 2016, 1, 4113–4120. [Google Scholar] [CrossRef]
- Awad, M.A.; de Jager, A.; van Westing, L.M. Flavonoid and chlorogenic acid levels in apple fruit: Characterisation of variation. Sci. Hortic-Amst. 2000, 83, 249–263. [Google Scholar] [CrossRef]
- Xi, Y.; Cheng, D.; Zeng, X.; Cao, J.; Jiang, W. Evidences for Chlorogenic Acid--A Major Endogenous Polyphenol Involved in Regulation of Ripening and Senescence of Apple Fruit. PLoS ONE 2016, 11, e0146940. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.; Nakamura, K.; Ma, L.; Li, J.Z.; Kayahara, H. Analyses of arbutin and chlorogenic acid, the major phenolic constituents in Oriental pear. J. Agric. Food Chem. 2005, 53, 3882–3887. [Google Scholar] [CrossRef]
- Jeon, J.S.; Kim, H.T.; Jeong, I.H.; Hong, S.R.; Oh, M.S.; Yoon, M.H.; Shim, J.H.; Jeong, J.H.; Abd El-Aty, A.M. Contents of chlorogenic acids and caffeine in various coffee-related products. J. Adv. Res. 2019, 17, 85–94. [Google Scholar] [CrossRef]
- Comino, C.; Hehn, A.; Moglia, A.; Menin, B.; Bourgaud, F.; Lanteri, S.; Portis, E. The isolation and mapping of a novel hydroxycinnamoyltransferase in the globe artichoke chlorogenic acid pathway. BMC Plant Biol. 2009, 9, 30. [Google Scholar] [CrossRef]
- Schutz, K.; Kammerer, D.; Carle, R.; Schieber, A. Identification and quantification of caffeoylquinic acids and flavonoids from artichoke (Cynara scolymus L.) heads, juice, and pomace by HPLC-DAD-ESI/MS(n). J. Agric. Food Chem. 2004, 52, 4090–4096. [Google Scholar] [CrossRef]
- Gonzalez-Perez, S.; Merck, K.B.; Vereijken, J.M.; van Koningsveld, G.A.; Gruppen, H.; Voragen, A.G. Isolation and characterization of undenatured chlorogenic acid free sunflower (Helianthus annuus) proteins. J. Agric. Food Chem. 2002, 50, 1713–1719. [Google Scholar] [CrossRef]
- Cheevarungnapakul, K.; Khaksar, G.; Panpetch, P.; Boonjing, P.; Sirikantaramas, S. Identification and Functional Characterization of Genes Involved in the Biosynthesis of Caffeoylquinic Acids in Sunflower (Helianthus annuus L.). Front. Plant Sci. 2019, 10, 968. [Google Scholar] [CrossRef]
- Ulbrich, B.; Zenk, M.H. Partial purification and properties of p-hydroxycinnamoyl-CoA: Shikimate-p-hydroxycinnamoyl transferase from higher plants. Phytochemistry 1980, 19, 1625–1629. [Google Scholar] [CrossRef]
- Stockigt, J.; Zenk, M.H. Enzymatic synthesis of chlorogenic acid from caffeoyl coenzyme A and quinic acid. FEBS Lett. 1974, 42, 131–134. [Google Scholar] [CrossRef]
- Mølgaard, P.; Ravn, H. Evolutionary aspects of caffeoyl ester distribution In Dicotyledons. Phytochemistry 1988, 27, 2411–2421. [Google Scholar] [CrossRef]
- Okuda, T.; Yoshida, T.; Hatano, T.; Iwasaki, M.; Kubo, M.; Orime, T.; Yoshizaki, M.; Naruhashi, N. Hydrolysable tannins as chemotaxonomic markers in the rosaceae. Phytochemistry 1992, 31, 3091–3096. [Google Scholar] [CrossRef]
- Whitaker, B.D.; Stommel, J.R. Distribution of hydroxycinnamic acid conjugates in fruit of commercial eggplant (Solanum melongena L.) cultivars. J. Agric. Food Chem. 2003, 51, 3448–3454. [Google Scholar] [CrossRef]
- D’Auria, J.C. Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 2006, 9, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Kitts, D.D. Role of Chlorogenic Acids in Controlling Oxidative and Inflammatory Stress Conditions. Nutrients 2015, 8, 16. [Google Scholar] [CrossRef]
- Schroder, J. Probing plant polyketide biosynthesis. Nat. Struct. Biol. 1999, 6, 714–716. [Google Scholar] [CrossRef]
- Santana-Galvez, J.; Cisneros-Zevallos, L.; Jacobo-Velazquez, D.A. Chlorogenic Acid: Recent Advances on Its Dual Role as a Food Additive and a Nutraceutical against Metabolic Syndrome. Molecules 2017, 22, 358. [Google Scholar] [CrossRef]
- Abrankó, L.; Clifford, M.N. An Unambiguous Nomenclature for the Acyl-quinic Acids Commonly Known as Chlorogenic Acids. J. Agric. Food Chem. 2017, 65, 3602–3608. [Google Scholar] [CrossRef]
- Miyamae, Y.; Kurisu, M.; Han, J.; Isoda, H.; Shigemori, H. Structure-activity relationship of caffeoylquinic acids on the accelerating activity on ATP production. Chem. Pharm. Bull. (Tokyo) 2011, 59, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, H.; Seki, E.; Aitani, M. Inhibitory effect of green coffee bean extract on fat accumulation and body weight gain in mice. BMC Complement. Altern. Med. 2006, 6, 9. [Google Scholar] [CrossRef]
- Sun, J.; Song, Y.L.; Zhang, J.; Huang, Z.; Huo, H.X.; Zheng, J.; Zhang, Q.; Zhao, Y.F.; Li, J.; Tu, P.F. Characterization and quantitative analysis of phenylpropanoid amides in eggplant (Solanum melongena L.) by high performance liquid chromatography coupled with diode array detection and hybrid ion trap time-of-flight mass spectrometry. J. Agric. Food Chem. 2015, 63, 3426–3436. [Google Scholar] [CrossRef] [PubMed]
- Rigano, M.M.; Raiola, A.; Docimo, T.; Ruggieri, V.; Calafiore, R.; Vitaglione, P.; Ferracane, R.; Frusciante, L.; Barone, A. Metabolic and Molecular Changes of the Phenylpropanoid Pathway in Tomato (Solanum lycopersicum) Lines Carrying Different Solanum pennellii Wild Chromosomal Regions. Front. Plant Sci. 2016, 7, 1484. [Google Scholar] [CrossRef] [PubMed]
- Docimo, T.; Francese, G.; Ruggiero, A.; Batelli, G.; De Palma, M.; Bassolino, L.; Toppino, L.; Rotino, G.L.; Mennella, G.; Tucci, M. Phenylpropanoids Accumulation in Eggplant Fruit: Characterization of Biosynthetic Genes and Regulation by a MYB Transcription Factor. Front. Plant Sci. 2015, 6, 1233. [Google Scholar] [CrossRef]
- Hoffmann, L.; Maury, S.; Martz, F.; Geoffroy, P.; Legrand, M. Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J. Biol. Chem 2003, 278, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Cao, J.; Feng, Q.; Peng, J.; Hu, Y. Roles of chlorogenic Acid on regulating glucose and lipids metabolism: A review. Evid. Based Complement. Altern. Med. 2013, 2013, 801457. [Google Scholar] [CrossRef]
- Niggeweg, R.; Michael, A.J.; Martin, C. Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 2004, 22, 746–754. [Google Scholar] [CrossRef]
- El-Seedi, H.R.; El-Said, A.M.; Khalifa, S.A.; Goransson, U.; Bohlin, L.; Borg-Karlson, A.K.; Verpoorte, R. Biosynthesis, natural sources, dietary intake, pharmacokinetic properties, and biological activities of hydroxycinnamic acids. J. Agric. Food Chem. 2012, 60, 10877–10895. [Google Scholar] [CrossRef]
- Clifford, M.N.; Jaganath, I.B.; Ludwig, I.A.; Crozier, A. Chlorogenic acids and the acyl-quinic acids: Discovery, biosynthesis, bioavailability and bioactivity. Nat. Prod. Rep. 2017, 34, 1391–1421. [Google Scholar] [CrossRef]
- Jean Claude, S.; Park, S. Aster spathulifolius Maxim. A leaf transcriptome provides an overall functional characterization, discovery of SSR marker and phylogeny analysis. PLoS ONE 2020, 15, e0244132. [Google Scholar] [CrossRef]
- Xu, H.; Park, N.I.; Li, X.; Kim, Y.K.; Lee, S.Y.; Park, S.U. Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in Scutellaria baicalensis. Bioresour. Technol. 2010, 101, 9715–9722. [Google Scholar] [CrossRef]
- Mahesh, V.; Million-Rousseau, R.; Ullmann, P.; Chabrillange, N.; Bustamante, J.; Mondolot, L.; Morant, M.; Noirot, M.; Hamon, S.; de Kochko, A.; et al. Functional characterization of two p-coumaroyl ester 3’-hydroxylase genes from coffee tree: Evidence of a candidate for chlorogenic acid biosynthesis. Plant Mol. Biol. 2007, 64, 145–159. [Google Scholar] [CrossRef]
- Liu, C.J. Deciphering the enigma of lignification: Precursor transport, oxidation, and the topochemistry of lignin assembly. Mol. Plant 2012, 5, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.Q. Phenylalanine ammonia-lyase, a key component used for phenylpropanoids production by metabolic engineering. RSC Adv. 2015, 5, 62587–62603. [Google Scholar] [CrossRef]
- Tsai, C.J.; Harding, S.A.; Tschaplinski, T.J.; Lindroth, R.L.; Yuan, Y. Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol. 2006, 172, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Weitzel, C.; Petersen, M. Enzymes of phenylpropanoid metabolism in the important medicinal plant Melissa officinalis L. Planta 2010, 232, 731–742. [Google Scholar] [CrossRef]
- De Paolis, A.; Pignone, D.; Morgese, A.; Sonnante, G. Characterization and differential expression analysis of artichoke phenylalanine ammonia-lyase-coding sequences. Physiol. Plant 2008, 132, 33–43. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C.J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 2015, 8, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Watanabe, M.; Hoefgen, R.; Fernie, A.R. The evolution of phenylpropanoid metabolism in the green lineage. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 123–152. [Google Scholar] [CrossRef]
- Hamberger, B.; Hahlbrock, K. The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc. Natl. Acad. Sci. USA 2004, 101, 2209–2214. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kim, J.I.; Pysh, L.; Chapple, C. Four Isoforms of Arabidopsis 4-Coumarate: CoA Ligase Have Overlapping yet Distinct Roles in Phenylpropanoid Metabolism. Plant Physiol. 2015, 169, 2409–2421. [Google Scholar] [CrossRef] [PubMed]
- Schoch, G.; Goepfert, S.; Morant, M.; Hehn, A.; Meyer, D.; Ullmann, P.; Werck-Reichhart, D. CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 2001, 276, 36566–36574. [Google Scholar] [CrossRef] [PubMed]
- Unno, H.; Ichimaida, F.; Suzuki, H.; Takahashi, S.; Tanaka, Y.; Saito, A.; Nishino, T.; Kusunoki, M.; Nakayama, T. Structural and mutational studies of anthocyanin malonyltransferases establish the features of BAHD enzyme catalysis. J. Biol. Chem. 2007, 282, 15812–15822. [Google Scholar] [CrossRef] [PubMed]
- Haslam, T.M.; Manas-Fernandez, A.; Zhao, L.F.; Kunst, L. Arabidopsis ECERIFERUM2 Is a Component of the Fatty Acid Elongation Machinery Required for Fatty Acid Extension to Exceptional Lengths. Plant Physiol. 2012, 160, 1164–1174. [Google Scholar] [CrossRef]
- Legrand, G.; Delporte, M.; Khelifi, C.; Harant, A.; Vuylsteker, C.; Morchen, M.; Hance, P.; Hilbert, J.L.; Gagneul, D. Identification and Characterization of Five BAHD Acyltransferases Involved in Hydroxycinnamoyl Ester Metabolism in Chicory. Front. Plant Sci. 2016, 7, 741. [Google Scholar] [CrossRef]
- Menin, B.; Comino, C.; Moglia, A.; Dolzhenko, Y.; Portis, E.; Lanteri, S. Identification and mapping of genes related to caffeoylquinic acid synthesis in Cynara cardunculus L. Plant Sci. 2010, 179, 338–347. [Google Scholar] [CrossRef]
- Yu, X.H.; Gou, J.Y.; Liu, C.J. BAHD superfamily of acyl-CoA dependent acyltransferases in Populus and Arabidopsis: Bioinformatics and gene expression. Plant Mol. Biol. 2009, 70, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Breitler, J.C.; Campa, C.; Georget, F.; Bertrand, B.; Etienne, H. A single-step method for RNA isolation from tropical crops in the field. Sci. Rep. 2016, 6, 38368. [Google Scholar] [CrossRef]
- Roser, L.G.; Aguero, F.; Sanchez, D.O. FastqCleaner: An interactive Bioconductor application for quality-control, filtering and trimming of FASTQ files. BMC Bioinform. 2019, 20, 361. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.H.; Davis, F.G.; et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Huala, E.; Dickerman, A.W.; Garcia-Hernandez, M.; Weems, D.; Reiser, L.; LaFond, F.; Hanley, D.; Kiphart, D.; Zhuang, M.; Huang, W.; et al. The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001, 29, 102–105. [Google Scholar] [CrossRef]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008, 36, D1009-14. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.C.; Yao, J.; Ho, K.S.; Lambowitz, A.M.; Wilke, C.O. Limitations of alignment-free tools in total RNA-seq quantification. BMC Genom. 2018, 19, 510. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Comino, C.; Lanteri, S.; Portis, E.; Acquadro, A.; Romani, A.; Hehn, A.; Larbat, R.; Bourgaud, F. Isolation and functional characterization of a cDNA coding a hydroxycinnamoyltransferase involved in phenylpropanoid biosynthesis in Cynara cardunculus L. BMC Plant Biol. 2007, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Scaglione, D.; Reyes-Chin-Wo, S.; Acquadro, A.; Froenicke, L.; Portis, E.; Beitel, C.; Tirone, M.; Mauro, R.; Lo Monaco, A.; Mauromicale, G.; et al. The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci. Rep. 2016, 6, 19427. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
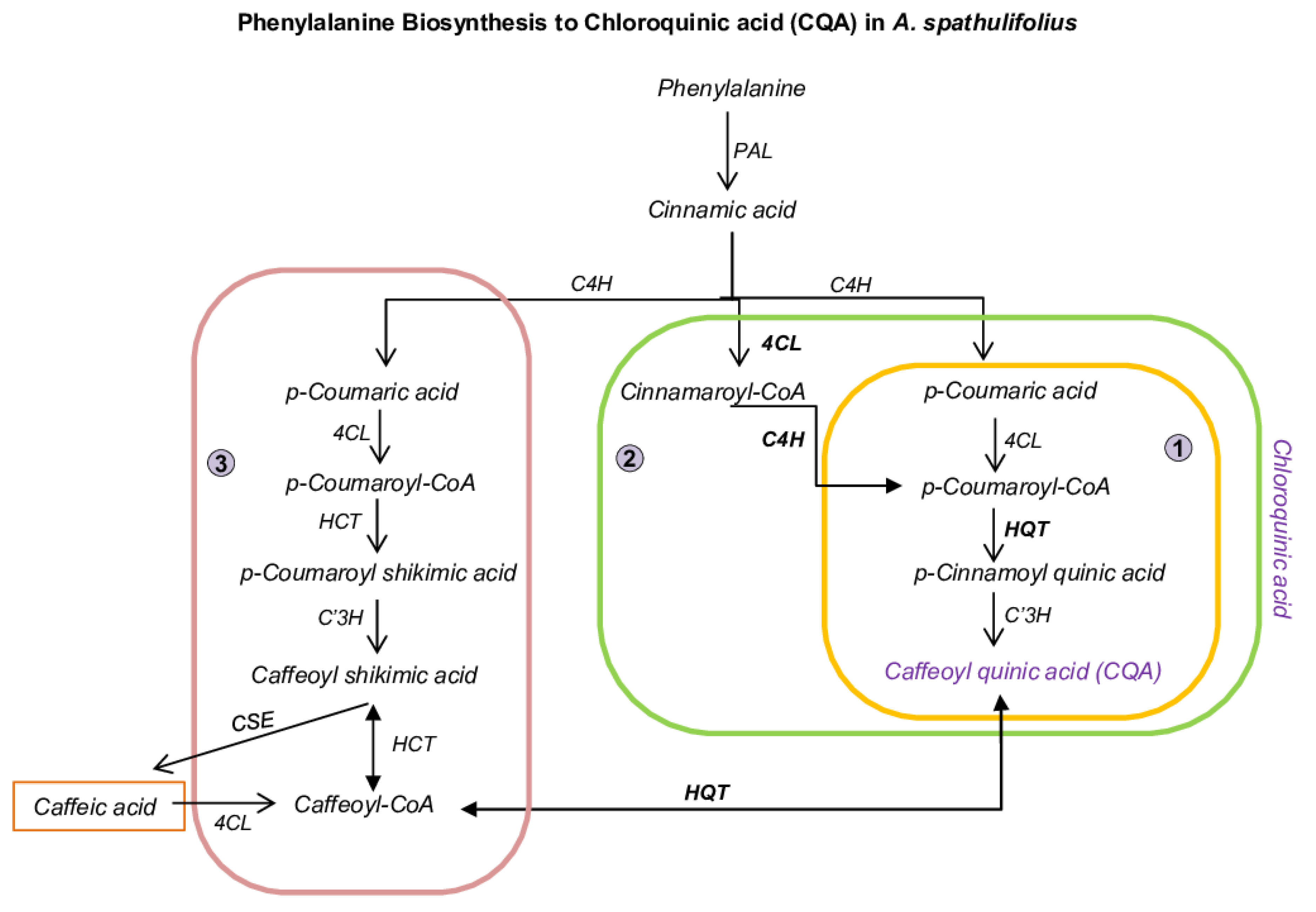
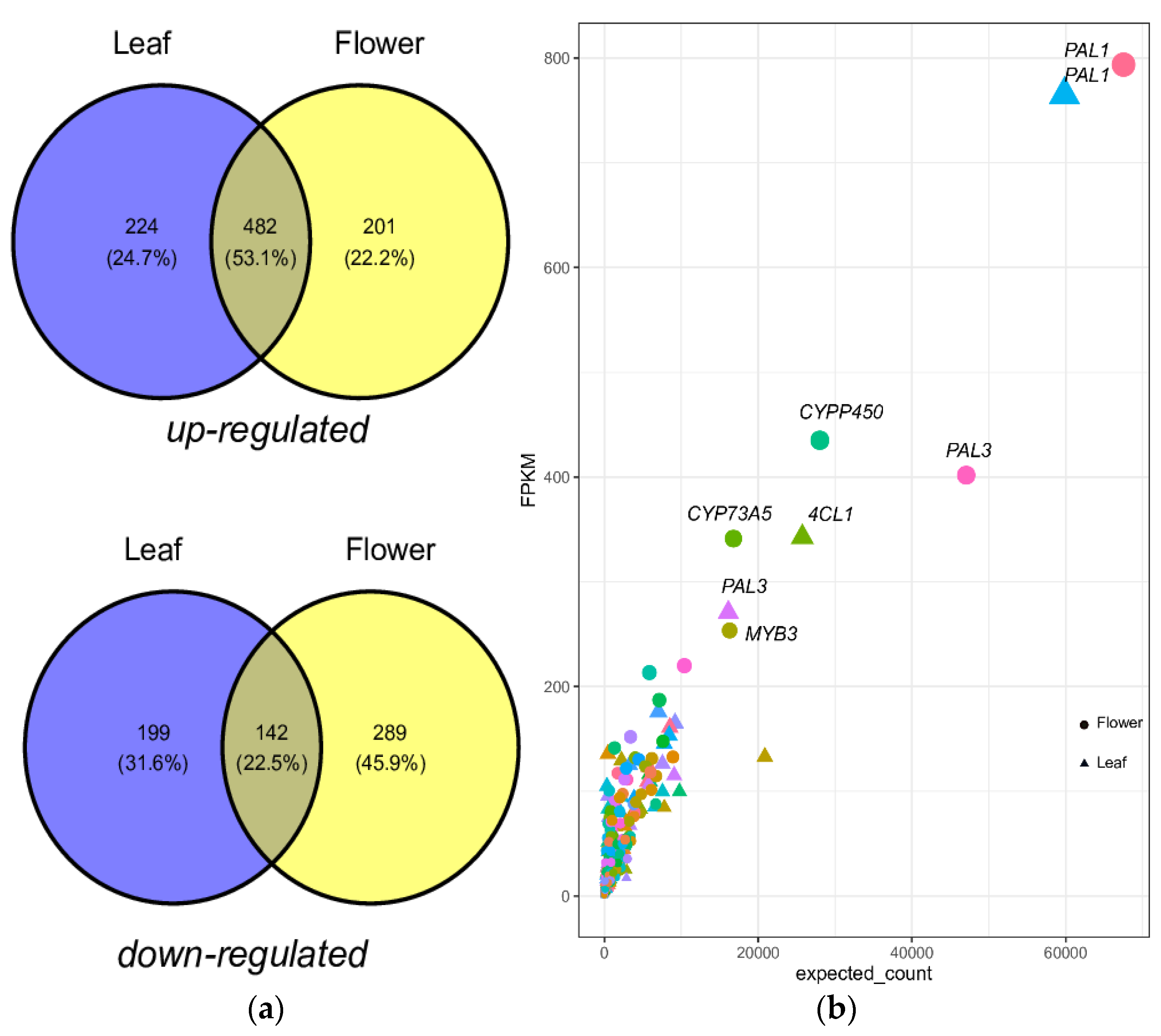

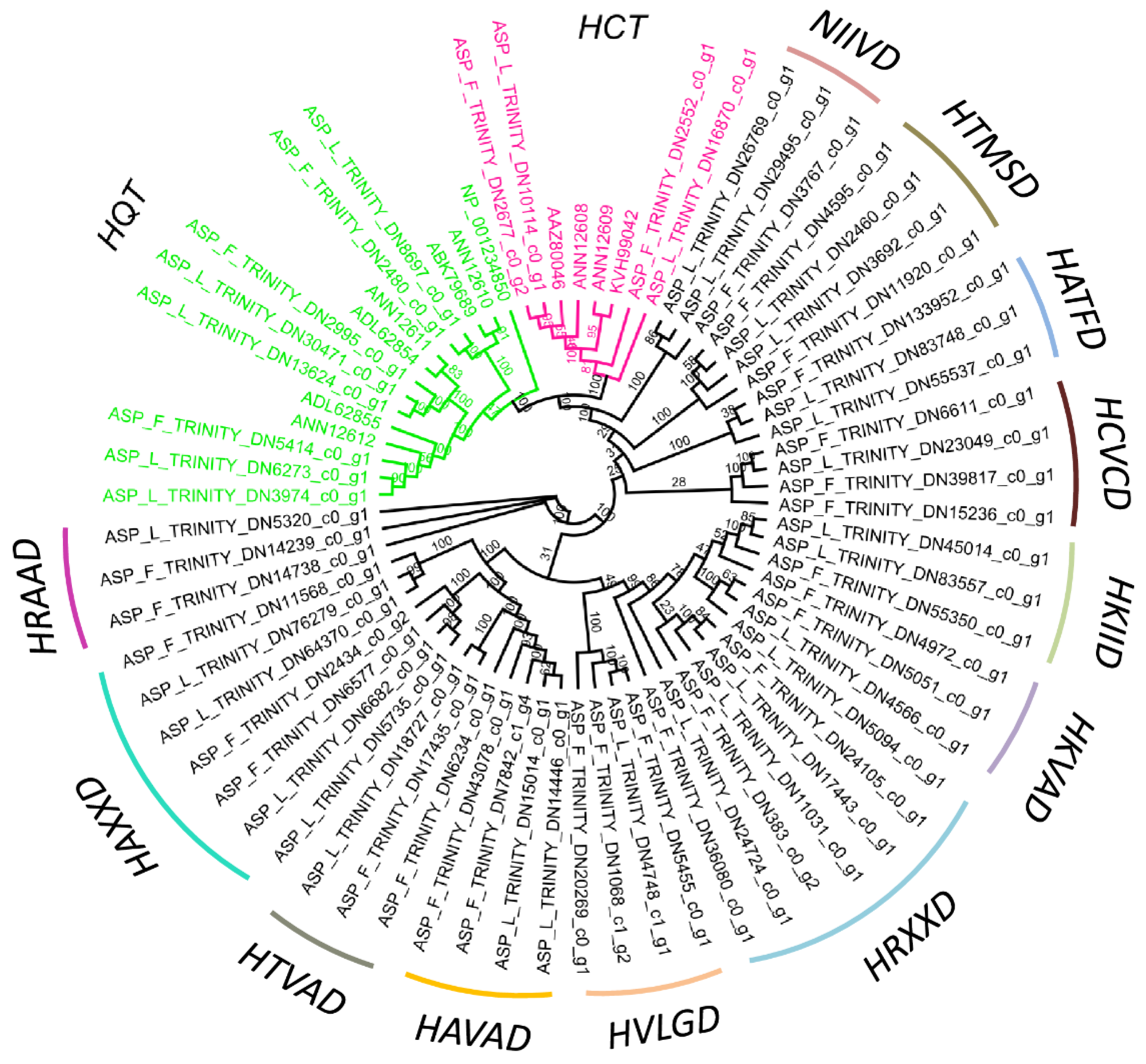
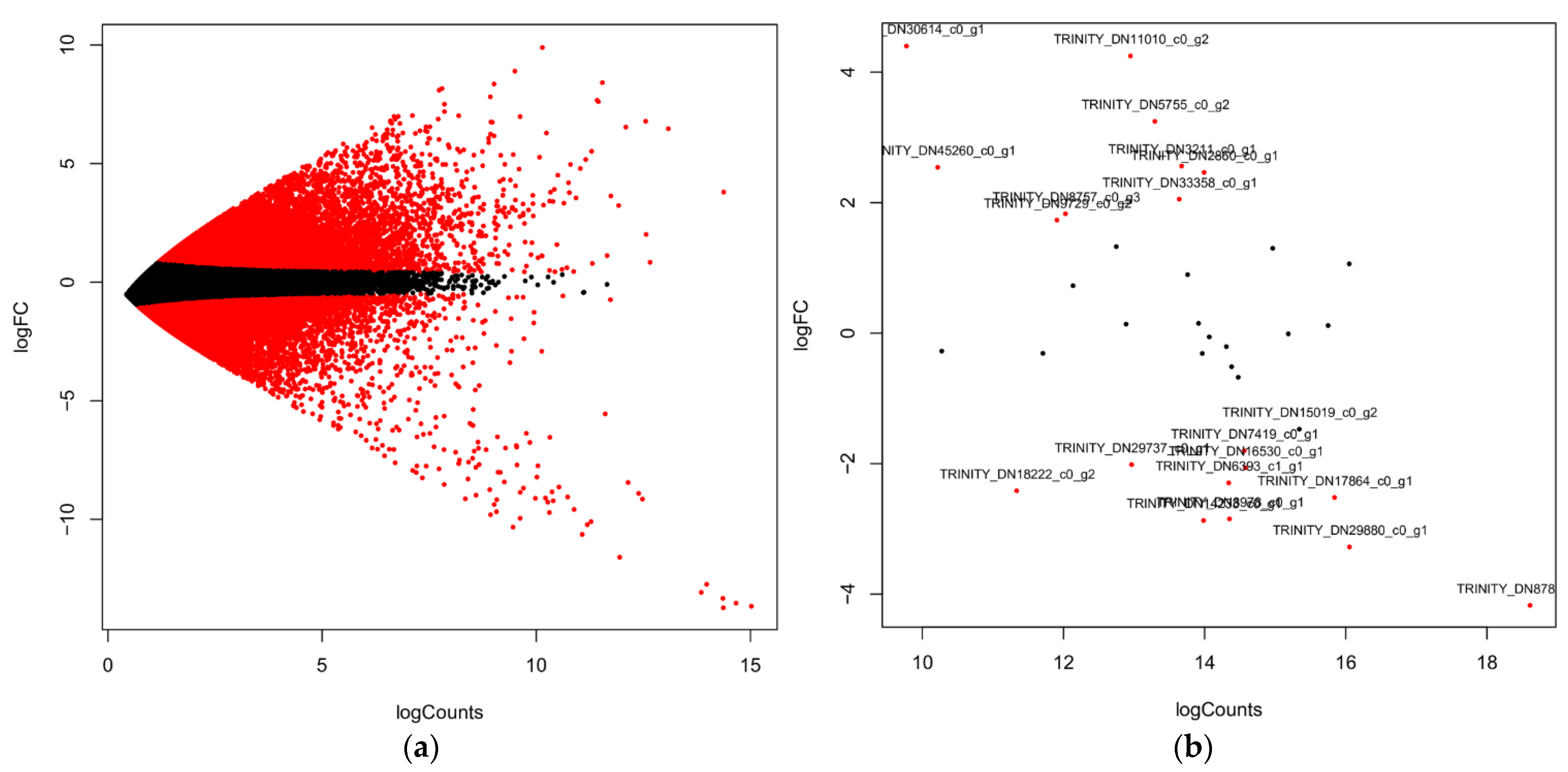

| S. No. | Sample | Total Base | Unigenes | Uniprot | KEGG | PPP | BAHD |
|---|---|---|---|---|---|---|---|
| 1 | Flower | 98,467,589 | 146,337 | 65,129 | 70,019 | 1128 | 82 |
| 2 | leaf | 71,660,029 | 98,860 | 48,896 | 39,890 | 1287 | 72 |
| GO ID | BP TERM | % | p-Value |
|---|---|---|---|
| GO:0009698 | phenylpropanoid metabolic process | 40.9 | 9.00 × 10−21 |
| GO:0009800 | cinnamic acid biosynthetic process | 18.2 | 1.10 × 10−8 |
| GO:2000762 | regulation of phenylpropanoid metabolic process | 18.2 | 3.80 × 10−8 |
| GO:0009611 | response to wounding | 31.8 | 4.90 × 10−8 |
| GO:0009699 | phenylpropanoid biosynthetic process | 18.2 | 6.90 × 10−6 |
| GO:0006559 | L-phenylalanine catabolic process | 13.6 | 3.10 × 10−5 |
| GO:0009809 | lignin biosynthetic process | 22.7 | 2.80 × 10−4 |
| GO:0008152 | metabolic process | 13.6 | 3.80 × 10−4 |
| GO:0009411 | response to UV | 13.6 | 2.40 × 10−3 |
| GO:0080167 | response to karrikin | 13.6 | 8.30 × 10−3 |
| GO:0009813 | flavonoid biosynthetic process | 13.6 | 1.10 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Claude, S.-J.; Park, S.; Park, S.-J. Transcriptome-Wide Identification and Quantification of Caffeoylquinic Acid Biosynthesis Pathway and Prediction of Its Putative BAHDs Gene Complex in A. spathulifolius. Int. J. Mol. Sci. 2021, 22, 6333. https://doi.org/10.3390/ijms22126333
Claude S-J, Park S, Park S-J. Transcriptome-Wide Identification and Quantification of Caffeoylquinic Acid Biosynthesis Pathway and Prediction of Its Putative BAHDs Gene Complex in A. spathulifolius. International Journal of Molecular Sciences. 2021; 22(12):6333. https://doi.org/10.3390/ijms22126333
Chicago/Turabian StyleClaude, Sivagami-Jean, Sunmi Park, and Seon-Joo Park. 2021. "Transcriptome-Wide Identification and Quantification of Caffeoylquinic Acid Biosynthesis Pathway and Prediction of Its Putative BAHDs Gene Complex in A. spathulifolius" International Journal of Molecular Sciences 22, no. 12: 6333. https://doi.org/10.3390/ijms22126333
APA StyleClaude, S.-J., Park, S., & Park, S.-J. (2021). Transcriptome-Wide Identification and Quantification of Caffeoylquinic Acid Biosynthesis Pathway and Prediction of Its Putative BAHDs Gene Complex in A. spathulifolius. International Journal of Molecular Sciences, 22(12), 6333. https://doi.org/10.3390/ijms22126333







