The Role of Anthocyanins, Deoxyanthocyanins and Pyranoanthocyanins on the Modulation of Tyrosinase Activity: An In Vitro and In Silico Approach
Abstract
1. Introduction
2. Results and Discussion
2.1. Effect of Anthocyanins and Related Compounds on Tyrosinase Activity
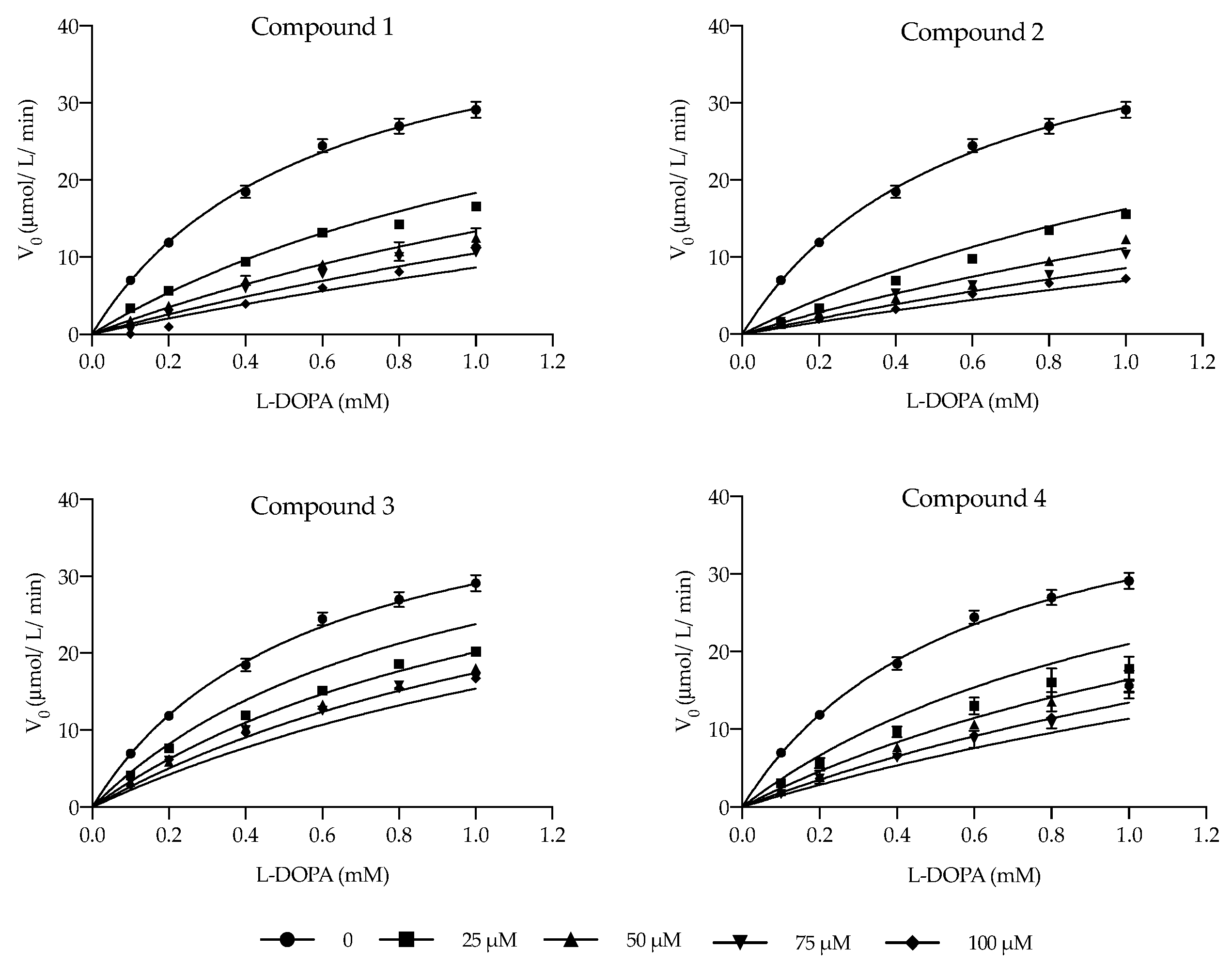
2.2. Kinetic Analysis and Determination of the Inhibition Type
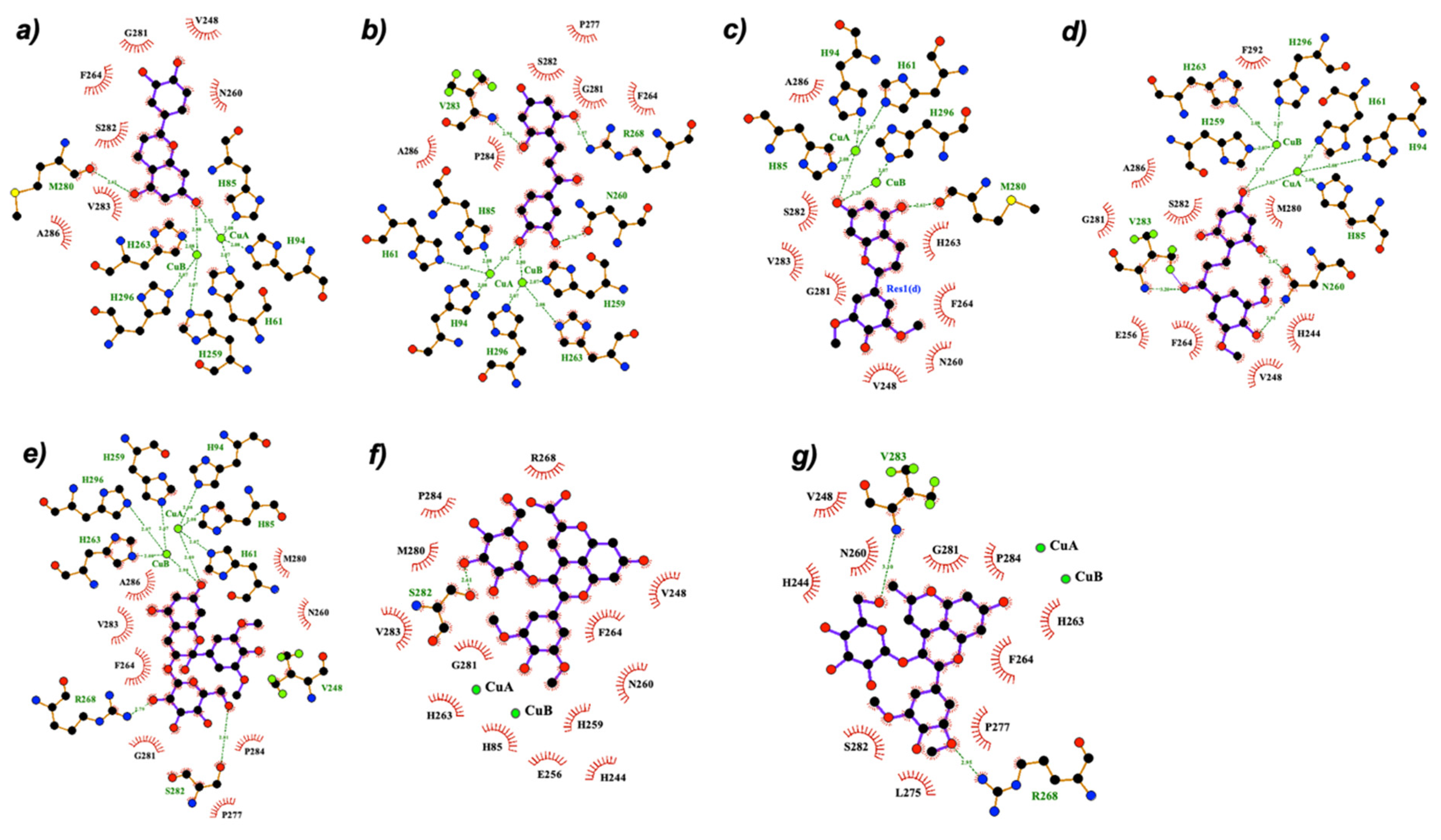
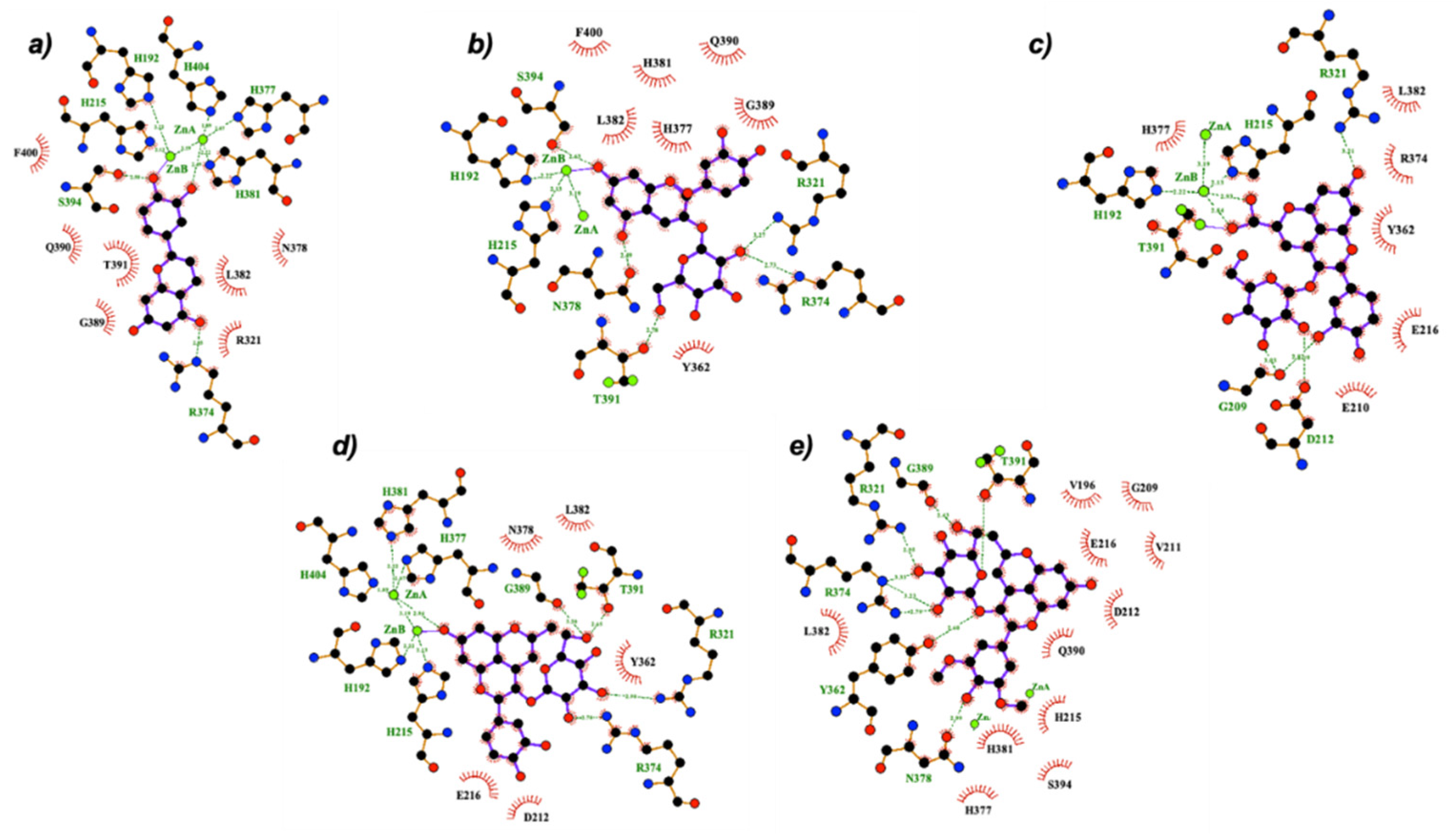
2.3. Molecular Docking
3. Materials and Methods
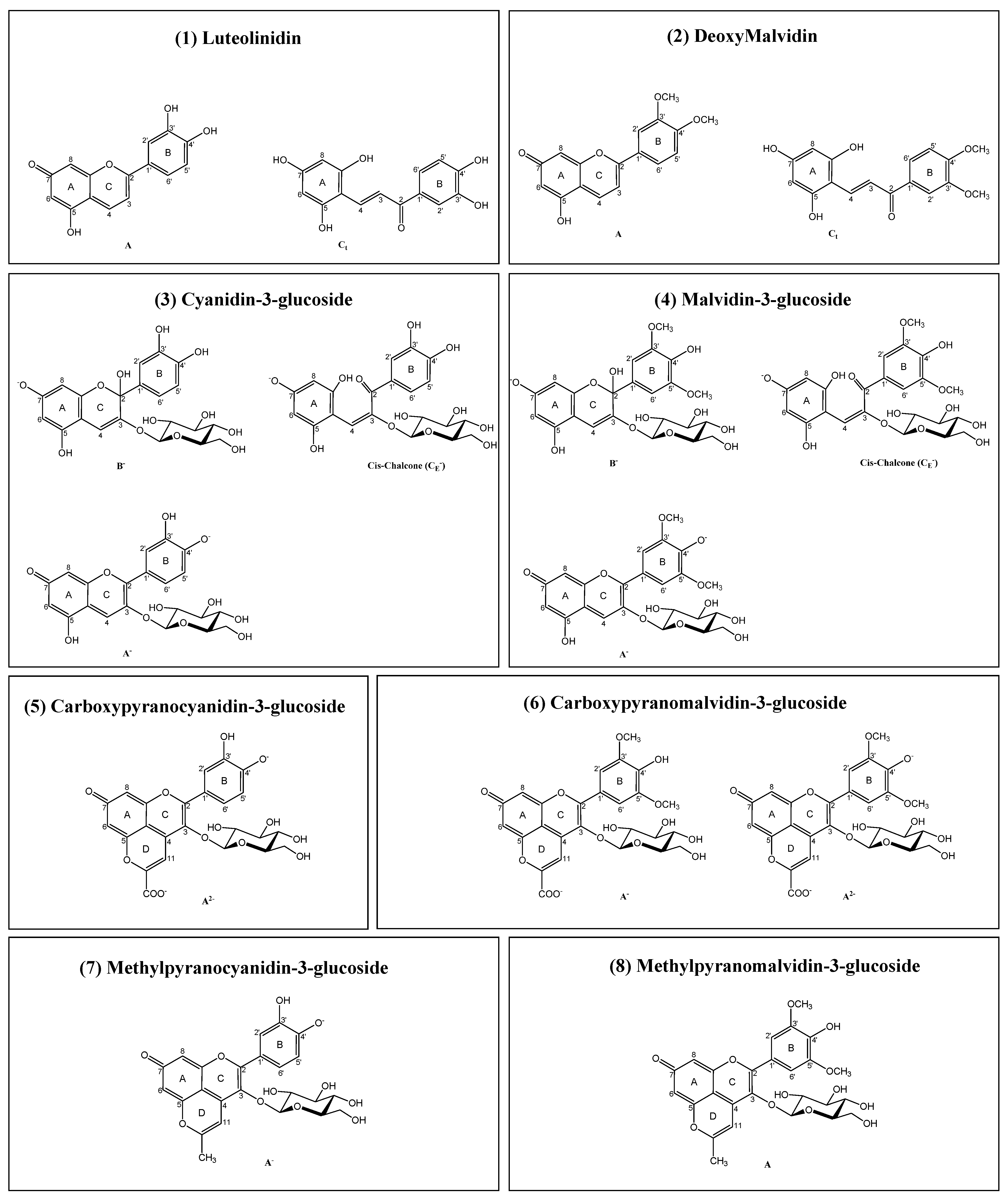
3.1. Isolation and Synthesis of the Different Compounds
3.2. HPLC-DAD and LC-MS Analysis
3.3. Mushroom Tyrosinase Inhibition Assay
3.4. Kinetic Analysis of Tyrosinase Inhibition
3.5. Molecular Modeling and Molecular Docking
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments

Conflicts of Interest
References
- Lin, J.Y.; Fisher, D.E. Melanocyte biology and skin pigmentation. Nat. Cell Biol. 2007, 445, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Hebert, D.N. Tyrosinase maturation through the mammalian secretory pathway: Bringing color to life. Pigment. Cell Res. 2006, 19, 3–18. [Google Scholar] [CrossRef]
- D’Mello, S.A.N.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling Pathways in Melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Niki, Y.; Ito, M.; Akiyama, K.; Matsui, M.S.; Yarosh, D.B.; Ichihashi, M. Melanosomes Are Transferred from Melanocytes to Keratinocytes through the Processes of Packaging, Release, Uptake, and Dispersion. J. Investig. Dermatol. 2012, 132, 1222–1229. [Google Scholar] [CrossRef] [PubMed]
- Serre, C.; Busuttil, V.; Botto, J. Intrinsic and extrinsic regulation of human skin melanogenesis and pigmentation. Int. J. Cosmet. Sci. 2018, 40, 328–347. [Google Scholar] [CrossRef] [PubMed]
- Kanteev, M.; Goldfeder, M.; Fishman, A. Structure-function correlations in tyrosinases. Protein Sci. 2015, 24, 1360–1369. [Google Scholar] [CrossRef]
- Olivares, C.; Solano, F. New insights into the active site structure and catalytic mechanism of tyrosinase and its related proteins. Pigment. Cell Melanoma Res. 2009, 22, 750–760. [Google Scholar] [CrossRef]
- Deri, B.; Kanteev, M.; Goldfeder, M.; Lecina, D.; Guallar, V.; Adir, N.; Fishman, A. The unravelling of the complex pattern of tyrosinase inhibition. Sci. Rep. 2016, 6, 34993. [Google Scholar] [CrossRef]
- Nordlund, J.J.; Grimes, P.E.; Ortonne, J.P. The safety of hydroquinone. J. Eur. Acad. Dermatol. Venereol. 2006, 20, 781–787. [Google Scholar] [CrossRef]
- Zolghadri, S.; Bahrami, A.; Khan, M.T.H.; Munoz-Munoz, J.; Garcia-Molina, F.; Garcia-Canovas, F.; Saboury, A.A. A comprehensive review on tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2019, 34, 279–309. [Google Scholar] [CrossRef]
- Hwang, J.-M.; Kuo, H.-C.; Lin, C.-T.; Kao, E.-S. Inhibitory effect of liposome-encapsulated anthocyanin on melanogenesis in human melanocytes. Pharm. Biol. 2013, 51, 941–947. [Google Scholar] [CrossRef]
- Kubota, M.; Hosoya, T.; Fukumoto, S.; Miyagi, T.; Kumazawa, S. Anti-melanogenic compounds in Rubus croceacanthus. J. Berry Res. 2014, 4, 127–135. [Google Scholar] [CrossRef]
- Hemachandran, H.; Anantharaman, A.; Mohan, S.; Mohan, G.; Kumar, T.; Dey, D.; Dey, P.; Choudhury, A.; Doss, C.G.P.; Ramamoorthy, S. Unraveling the inhibition mechanism of cyanidin-3-sophoroside on polyphenol oxidase and its effect on enzymatic browning of apples. Food Chem. 2017, 227, 102–110. [Google Scholar] [CrossRef]
- Trouillas, P.; Sancho-Garcia, J.-C.; Freitas, V.; Gierschner, J.; Otyepka, M.; Dangles, O. Stabilizing and Modulating Color by Copigmentation: Insights from Theory and Experiment. Chem. Rev. 2016, 116, 4937–4982. [Google Scholar] [CrossRef]
- Aramwit, P.; Bang, N.; Srichana, T. The properties and stability of anthocyanins in mulberry fruits. Food Res. Int. 2010, 43, 1093–1097. [Google Scholar] [CrossRef]
- Cásedas, G.; Les, F.; Gómez-Serranillos, M.P.; Smith, C.; López, V. Anthocyanin profile, antioxidant activity and enzyme inhibiting properties of blueberry and cranberry juices: A comparative study. Food Funct. 2017, 8, 4187–4193. [Google Scholar] [CrossRef]
- Oliveira, J.; Mateus, N.; Freitas, V. Previous and recent advances in pyranoanthocyanins equilibria in aqueous solution. Dyes Pigment. 2014, 100, 190–200. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhang, P.; Warner, R.D.; Fang, Z. 3-Deoxyanthocyanidin Colorant: Nature, Health, Synthesis, and Food Applications. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1533–1549. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, J.; Basílio, N.; De Freitas, V.; Pina, F. New Procedure To Calculate All Equilibrium Constants in Flavylium Compounds: Application to the Copigmentation of Anthocyanins. ACS Omega 2019, 4, 12058–12070. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.L.; Oliveira, J.; Araújo, P.; Calogero, G.; Freitas, V.; Pina, F.; Parola, A.J.; Lima, J.C. Study of the multi-equilibria of red wine colorants pyranoanthocyanins and evaluation of their potential in dye-sensitized solar cells. Sol. Energy 2019, 191, 100–108. [Google Scholar] [CrossRef]
- Oliveira, J.; Mateus, N.; De Freitas, V. Network of carboxypyranomalvidin-3-O-glucoside (vitisin A) equilibrium forms in aqueous solution. Tetrahedron Lett. 2013, 54, 5106–5110. [Google Scholar] [CrossRef]
- Oliveira, J.; Petrov, V.; Parola, A.J.; Pina, F.; Azevedo, J.; Teixeira, N.; Braás, N.F.; Fernandes, P.A.; Mateus, N.; Ramos, M.J.; et al. Chemical Behavior of Methylpyranomalvidin-3-O-glucoside in Aqueous Solution Studied by NMR and UV−Visible Spectroscopy. J. Phys. Chem. B 2011, 115, 1538–1545. [Google Scholar] [CrossRef] [PubMed]
- Pina, F.; Melo, M.J.; Laia, C.; Parola, A.J.; Lima, J.C. Chemistry and applications of flavylium compounds: A handful of colours. Chem. Soc. Rev. 2012, 41, 869–908. [Google Scholar] [CrossRef]
- Mazza, G.; Brouillard, R. Color stability and structural transformations of cyanidin 3,5-diglucoside and four 3-deoxyanthocyanins in aqueous solutions. J. Agric. Food Chem. 1987, 35, 422–426. [Google Scholar] [CrossRef]
- Noh, H.; Lee, S.J.; Jo, H.-J.; Choi, H.W.; Hong, S.; Kong, K.-H. Histidine residues at the copper-binding site in human tyrosinase are essential for its catalytic activities. J. Enzym. Inhib. Med. Chem. 2020, 35, 726–732. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Biswas, R.; Sharma, A.; Banerjee, S.; Biswas, S.; Katiyar, C. Validation of medicinal herbs for anti-tyrosinase potential. J. Herb. Med. 2018, 14, 1–16. [Google Scholar] [CrossRef]
- Tsuda, T.; Osawa, T. Inhibition of Tyrosinase Activity by the Anthocyanin Pigments Isolated from Phaseolus vulgaris L. Food Sci. Technol. Int. Tokyo 1997, 3, 82–83. [Google Scholar] [CrossRef]
- Xue, Y.-L.; Miyakawa, T.; Hayashi, Y.; Okamoto, K.; Hu, F.; Mitani, N.; Furihata, K.; Sawano, Y.; Tanokura, M. Isolation and Tyrosinase Inhibitory Effects of Polyphenols from the Leaves of Persimmon, Diospyros kaki. J. Agric. Food Chem. 2011, 59, 6011–6017. [Google Scholar] [CrossRef]
- Somaatmadja, D.; Powers, J.J.; Hamdy, M.K. Anthocyanins. VI. Chelation Studies on Anthocyanins and Other Related Compounds. J. Food Sci. 1964, 29, 655–660. [Google Scholar] [CrossRef]
- Chen, Q.-X.; Kubo, I. Kinetics of Mushroom Tyrosinase Inhibition by Quercetin. J. Agric. Food Chem. 2002, 50, 4108–4112. [Google Scholar] [CrossRef] [PubMed]
- Copeland, R.A. Reversible Modes of Inhibitor Interactions with Enzymes. In Evaluation of Enzyme Inhibitors in Drug Discovery 2013; John Wiley & Sons, Inc.: Hoboken, NJ, USA; pp. 57–121. [CrossRef]
- Sun, W.; Wendt, M.; Klebe, G.; Röhm, K.-H. On the interpretation of tyrosinase inhibition kinetics. J. Enzym. Inhib. Med. Chem. 2013, 29, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-S.; Chen, H.-J.; Huang, J.-P.; Lee, P.-C.; Tsai, C.-R.; Hsu, T.-F.; Huang, W.-Y. Kinetics of Tyrosinase Inhibitory Activity Using Vitis vinifera Leaf Extracts. BioMed Res. Int. 2017, 2017, 5232680. [Google Scholar] [CrossRef]
- Fan, M.; Zhang, G.; Hu, X.; Xu, X.; Gong, D. Quercetin as a tyrosinase inhibitor: Inhibitory activity, conformational change and mechanism. Food Res. Int. 2017, 100, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Pina, F.; Oliveira, J.; Freitas, V. Anthocyanins and derivatives are more than flavylium cations. Tetrahedron 2015, 71, 3107–3114. [Google Scholar] [CrossRef]
- Flurkey, A.; Cooksey, J.; Reddy, A.; Spoonmore, K.; Rescigno, A.; Inlow, J.; Flurkey, W.H. Enzyme, Protein, Carbohydrate, and Phenolic Contaminants in Commercial Tyrosinase Preparations: Potential Problems Affecting Tyrosinase Activity and Inhibition Studies. J. Agric. Food Chem. 2008, 56, 4760–4768. [Google Scholar] [CrossRef]
- Cásedas, G.; Les, F.; González-Burgos, E.; Gómez-Serranillos, M.P.; Smith, C.; López, V. Cyanidin-3-O-glucoside inhibits different enzymes involved in central nervous system pathologies and type-2 diabetes. S. Afr. J. Bot. 2019, 120, 241–246. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- Karakaya, G.; Türe, A.; Ercan, A.; Öncül, S.; Aytemir, M.D. Synthesis, computational molecular docking analysis and effectiveness on tyrosinase inhibition of kojic acid derivatives. Bioorg. Chem. 2019, 88, 102950. [Google Scholar] [CrossRef] [PubMed]
- Ielo, L.; Deri, B.; Germanò, M.P.; Vittorio, S.; Mirabile, S.; Gitto, R.; Rapisarda, A.; Ronsisvalle, S.; Floris, S.; Pazy, Y.; et al. Exploiting the 1-(4-fluorobenzyl)piperazine fragment for the development of novel tyrosinase inhibitors as anti-melanogenic agents: Design, synthesis, structural insights and biological profile. Eur. J. Med. Chem. 2019, 178, 380–389. [Google Scholar] [CrossRef]
- Rafiq, M.; Nazir, Y.; Ashraf, Z.; Rafique, H.; Afzal, S.; Mumtaz, A.; Hassan, M.; Ali, A.; Afzal, K.; Yousuf, M.R.; et al. Synthesis, computational studies, tyrosinase inhibitory kinetics and antimelanogenic activity of hydroxy substituted 2-[(4-acetylphenyl)amino]-2-oxoethyl derivatives. J. Enzym. Inhib. Med. Chem. 2019, 34, 1562–1572. [Google Scholar] [CrossRef] [PubMed]
- Brasil, E.M.; Canavieira, L.M.; Cardoso, É.T.C.; Silva, E.O.; Lameira, J.; Nascimento, J.L.M.; Eifler-Lima, V.L.; Macchi, B.M.; Sriram, D.; Bernhardt, P.V.; et al. Inhibition of tyrosinase by 4H -chromene analogs: Synthesis, kinetic studies, and computational analysis. Chem. Biol. Drug Des. 2017, 90, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Cabanes, J.; Chazarra, S.; Garcia-Carmona, F. Kojic Acid, a Cosmetic Skin Whitening Agent, is a Slow-binding Inhibitor of Catecholase Activity of Tyrosinase. J. Pharm. Pharmacol. 2011, 46, 982–985. [Google Scholar] [CrossRef]
- Lai, X.; Wichers, H.; Soler-Lopez, M.; Dijkstra, B.W. Structure of Human Tyrosinase Related Protein 1 Reveals a Binuclear Zinc Active Site Important for Melanogenesis. Angew. Chem. Int. Ed. 2017, 56, 9812–9815. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.; Thornton, J. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Espín, J.C.; Varón, R.; Fenoll, L.G.; Gilabert, M.A.; García-Ruíz, P.A.; Tudela, J.; García-Cánovas, F. Kinetic characterization of the substrate specificity and mechanism of mushroom tyrosinase. JBIC J. Biol. Inorg. Chem. 2000, 267, 1270–1279. [Google Scholar] [CrossRef]
- Al Bittar, S.; Mora, N.; Loonis, M.; Dangles, O. A simple synthesis of 3-deoxyanthocyanidins and their O-glucosides. Tetrahedron 2016, 72, 4294–4302. [Google Scholar] [CrossRef]
- Fernandes, I.; Faria, A.; Azevedo, J.; Soares, S.; Calhau, C.; Freitas, V.; Mateus, N. Influence of Anthocyanins, Derivative Pigments and Other Catechol and Pyrogallol-Type Phenolics on Breast Cancer Cell Proliferation. J. Agric. Food Chem. 2010, 58, 3785–3792. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.; Fernandes, V.; Miranda, C.; Santos-Buelga, C.; Silva, A.; Freitas, V.; Mateus, N. Color Properties of Four Cyanidin−Pyruvic Acid Adducts. J. Agric. Food Chem. 2006, 54, 6894–6903. [Google Scholar] [CrossRef] [PubMed]
- Ismaya, W.T.; Rozeboom, H.J.; Weijn, A.; Mes, J.J.; Fusetti, F.; Wichers, H.J.; Dijkstra, B.W. Crystal Structure of Agaricus bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone. Biochemistry 2011, 50, 5477–5486. [Google Scholar] [CrossRef]
- Dennington, R.; Keith, T.A.; Millam, J.M. GaussView, Version 5; Semichem Inc.: Shawnee, KS, USA, 2009. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
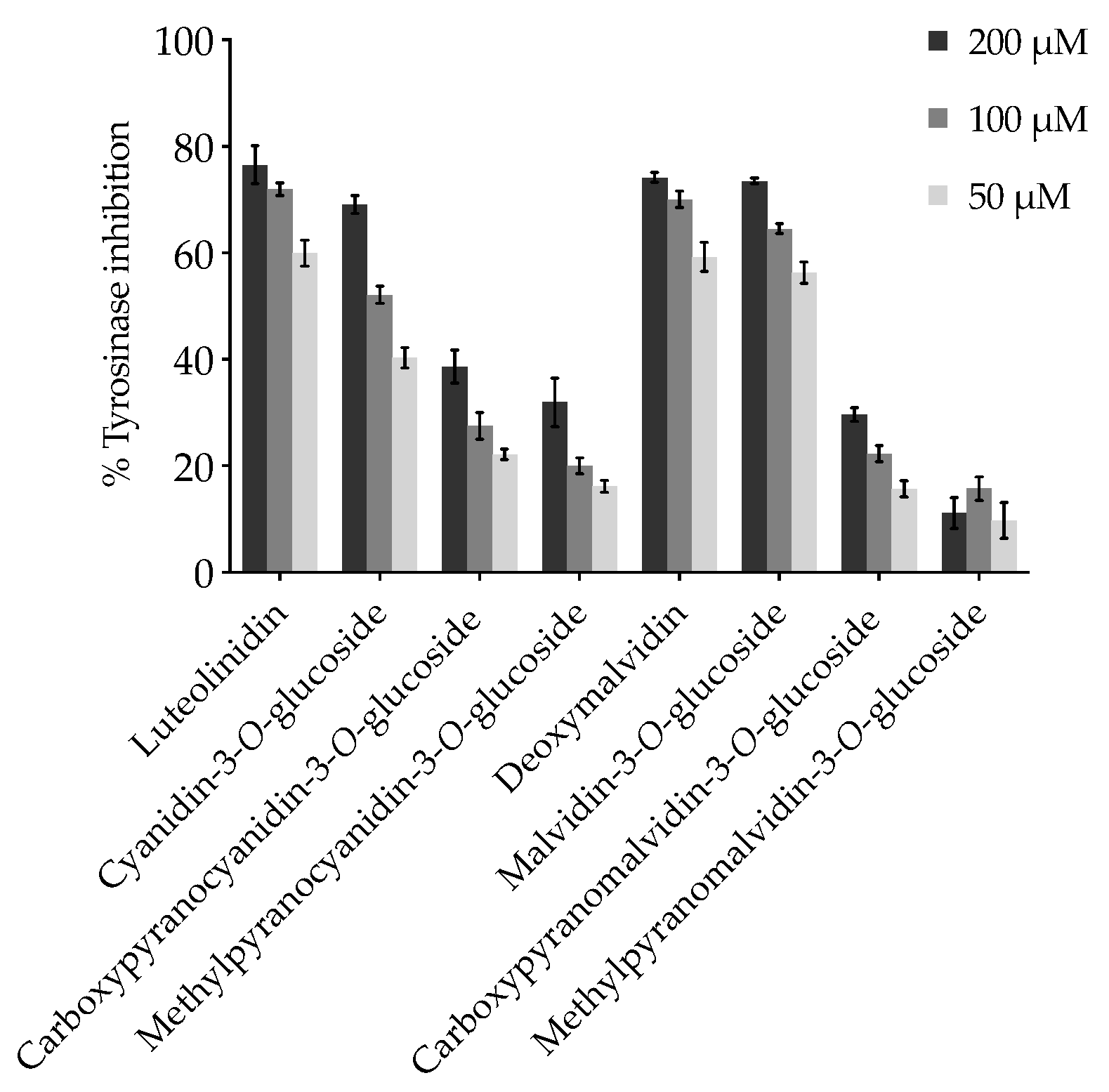
| Compound | % mTYR Inhibition at 50 µM |
|---|---|
| Kojic acid 1−8 | 76.94 ± 3.214 |
| (1) Luteolinidin 3,5−8 | 58.52 ± 2.082 |
| (3) Cyanidin-3-O-glucoside 2,5−8 | 40.39 ± 1.998 |
| (5) Carboxypyranocyanidin-3-O-glucoside 2,4 | 22.14 ± 0.9620 |
| (7) Methylpyranocyanidin-3-O-glucoside 2,4 | 16.19 ± 1.135 |
| (2) Deoxymalvidin 6,8 | 57.34 ± 2.381 |
| (4) Malvidin-3-O-glucoside 6,8 | 54.43 ± 4.055 |
| (6) Carboxypyranomalvidin-3-O-glucoside | 15.71 ± 1.483 |
| (8) Methylpyranomalvidin-3-O-glucoside | 9.742 ± 3.357 |
| IC50 (µM) | Ki (µM) | α | Inhibition Type | |
|---|---|---|---|---|
| Compound 1 | 39.21 ± 2.711 | 15.08 ± 1.182 | 21.00 | Competitive |
| Compound 2 | 20.31 ± 4.308 | 11.28 ± 0.985 | ∞ | Competitive |
| Compound 3 | 77.91 ± 4.804 | 40.31 ± 3.611 | 4.43 | Competitive |
| Compound 4 | 27.60 ± 4.202 | 23.22 ± 1.889 | ∞ | Competitive |
| Compound | ΔG binding (kcal/mol) | Average Distance to Cu (Å) | Coordinating Group to the Metal | Interactions | |
|---|---|---|---|---|---|
| H-Bonds | Hydrophobic | ||||
| Luteolinidin (1-A) | −6.0 | 2.9 | O-C7 (ring A) | N260, H263 | H263, F264, V283 |
| Luteolinidin (1-Ct) | −6.6 | 2.9 | HO-C4’ (ring B) | N260, R268 | H263, F264, V283 |
| Malvidin-3-O-glucoside (4-B−) | −8.1 | 2.9 | O-C7 (ring A) | N260, H263 | H244, V248, H263, F264, V283 |
| Cyanidin-3-O-glucoside (3-A−) | −8.5 | 2.9 | O-C7 (ring A) | H263, R268, S282 | V248, N260, H263, F264, V283 |
| Malvidin-3-O-glucoside (3-A−) | −8.4 | 2.9 | O-C7 (ring A) | H244, M280, G281, S282 | V248, H263, F264, V283, A286 |
| Deoxymalvidin (2-Ct) | −7.0 | 3.0 | O-C7 (ring A) | H244, H263, N260 | H263, F264, V283 |
| Deoxymalvidin (2-A) | −6.0 | 3.0 | O-C7 (ring A) | N260, H263 | H244, H263, F264, V283 |
| Cyanidin-3-O-glucoside (3-B−) | −9.0 | 3.1 | O-C7 (ring A) | H263, S282 | V248, H263, F264, V283 |
| Kojic acid (KA) | −4.5 | 3.3 | OH | N260 | H263, F264, V283 |
| Tropolone | −4.9 | 3.4 | O | - | H263, F264, V283 |
| Malvidin-3-O-glucoside (4-CE−) | −8.6 | 3.5 | O-C7 (ring A) | H263 R268, S282 | H244, V248, H263, F264, V283 |
| Cyanidin-3-O-glucoside (3-CE−) | −8.8 | 3.5 | O-C7 (ring A) | N260, R268 | H263, F264, V283 |
| Carboxypyranomalvidin-3-O-glucoside (6-A2−) | −8.3 | 3.6 | O-C4’ (ring B) | H244, R268, S282 | V248, F264, V283 |
| Methylpyranocyanidin-3-O-glucoside (7) | −8.6 | 3.6 | O-C4’ (ring B) | H244, R268, S282 | V248, F264, V283 |
| Carboxypyranomalvidin-3-O-glucoside (6-A−) | −7.9 | 3.6 | OH-C4’ (ring B) | H244, R268, S282 | V248, F264, V283 |
| Carboxypyranocyanidin-3-O-glucoside (5) | −6.9 | 3.8 | O-C4’ (ring B) | H244, N260, R268 | V248, F264, V283 |
| Methylpyranomalvidin-3-O-glucoside (8) | −8.8 | 4.5 | O-C7 (ring A) | R268, S282 | V248, F264, V283 |
| Compound | ΔG binding (kcal/mol) | Average Distance to Zn (Å) | Coordinating Group to the Metal | Interactions | |
|---|---|---|---|---|---|
| H-bonds | Hydrophobic | ||||
| Luteolinidin (1-Ct) | −7.1 | 1.9 | HO-C3’, HO-C4’ (ring B) | Y362, S394 | Y362, H381 |
| Tropolone | −6.0 | 2.0 | O | T391, S394 | L382, H381, T391 |
| Kojic acid | −5.2 | 2.0 | O | S394 | L382, H381, T391 |
| Luteolinidin (1-A) | −7.5 | 2.1 | HO-C3’, HO-C4’ (ring B) | R374, S394 | Y362, L382, H381 |
| Malvidin-3-O-glucoside (4-B−) | −7.6 | 2.3 | O-C7 (ring A) | R374, G389, T391 | Y362, L382, H381 |
| Deoxymalvidin (2-Ct) | −6.8 | 2.4 | HO-C4’ (ring B) | Y362, S394 | H381 |
| Methylpyranocyanidin-3-O-glucoside (7) | −8.6 | 2.4 | O-C7 (ring A) | Y362, T391, S394 | Y362, L382, H381 |
| Deoxymalvidin (2-A) | −6.1 | 2.5 | O-C7 (ring A) | N378, T391, S394 | L382, H381 |
| Cyanidin-3-O-glucoside (3-B−) | −10.8 | 2.6 | O-C7 (ring A) | Y362, R374, N378, T391, S394 | Y362, L382, H381 |
| Cyanidin-3-O-glucoside (3-A−) | −8.5 | 2.6 | O-C7 (ring A) | R321, R354, H377, N378, T391, S394 | L382, H381, T391, F400 |
| Malvidin-3-O-glucoside (3-A−) | −8.4 | 2.7 | O-C7 (ring A) | R321, Y362, R374, G388, G389, T391 | L382, H381, Q390 |
| Carboxypyranomalvidin-3-O-glucoside (6-A2−) | −9.6 | 2.7 | COO- (ring D) | D212, E216, Y362, R374, T391 | L382 |
| Carboxypyranocyanidin-3-O-glucoside (5) | −9.4 | 2.9 | COO- (ring D) | N378, T391, S394 | Y362, L382 |
| Carboxypyranomalvidin-3-O-glucoside (6-A−) | −10.0 | 2.9 | COO- (ring D) | D212, E216, R374, T391 | Y362 |
| Methylpyranomalvidin-3-O-glucoside (8) | −8.7 | 3.2 | OMe-C3’ (ring B) | Y362, R374, N378, T391 | L382 |
| Cyanidin-3-O-glucoside (3-Cc−) | −7.3 | 4.2 | O-C7 (ring A) | R374, G389, T391 | L382 |
| Malvidin-3-O-glucoside (4-Cc−) | −9.4 | 5.6 | O-C7 (ring A) | R374, T391 | L382 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Correia, P.; Oliveira, H.; Araújo, P.; Brás, N.F.; Pereira, A.R.; Moreira, J.; de Freitas, V.; Mateus, N.; Oliveira, J.; Fernandes, I. The Role of Anthocyanins, Deoxyanthocyanins and Pyranoanthocyanins on the Modulation of Tyrosinase Activity: An In Vitro and In Silico Approach. Int. J. Mol. Sci. 2021, 22, 6192. https://doi.org/10.3390/ijms22126192
Correia P, Oliveira H, Araújo P, Brás NF, Pereira AR, Moreira J, de Freitas V, Mateus N, Oliveira J, Fernandes I. The Role of Anthocyanins, Deoxyanthocyanins and Pyranoanthocyanins on the Modulation of Tyrosinase Activity: An In Vitro and In Silico Approach. International Journal of Molecular Sciences. 2021; 22(12):6192. https://doi.org/10.3390/ijms22126192
Chicago/Turabian StyleCorreia, Patrícia, Hélder Oliveira, Paula Araújo, Natércia F. Brás, Ana Rita Pereira, Joana Moreira, Victor de Freitas, Nuno Mateus, Joana Oliveira, and Iva Fernandes. 2021. "The Role of Anthocyanins, Deoxyanthocyanins and Pyranoanthocyanins on the Modulation of Tyrosinase Activity: An In Vitro and In Silico Approach" International Journal of Molecular Sciences 22, no. 12: 6192. https://doi.org/10.3390/ijms22126192
APA StyleCorreia, P., Oliveira, H., Araújo, P., Brás, N. F., Pereira, A. R., Moreira, J., de Freitas, V., Mateus, N., Oliveira, J., & Fernandes, I. (2021). The Role of Anthocyanins, Deoxyanthocyanins and Pyranoanthocyanins on the Modulation of Tyrosinase Activity: An In Vitro and In Silico Approach. International Journal of Molecular Sciences, 22(12), 6192. https://doi.org/10.3390/ijms22126192











