SREBP-1c Deficiency Affects Hippocampal Micromorphometry and Hippocampus-Dependent Memory Ability in Mice
Abstract
1. Introduction
2. Results
2.1. SREBP-1c KO Mice Demonstrate Significantly Decreased Hippocampus-Dependent Memory Retention
2.2. SREBP-1c KO Mice Display Altered Dendritic Morphology in the Hippocampus
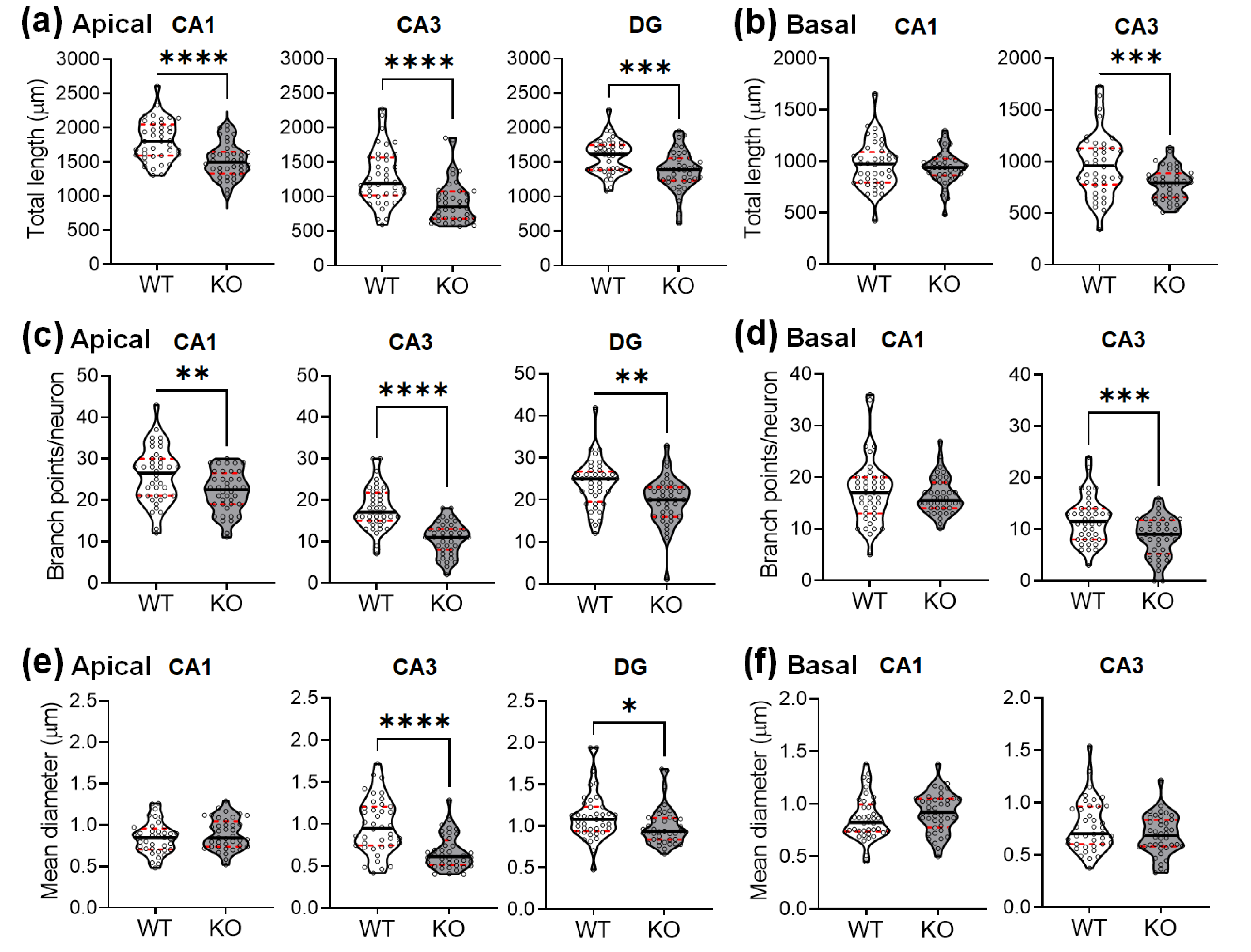
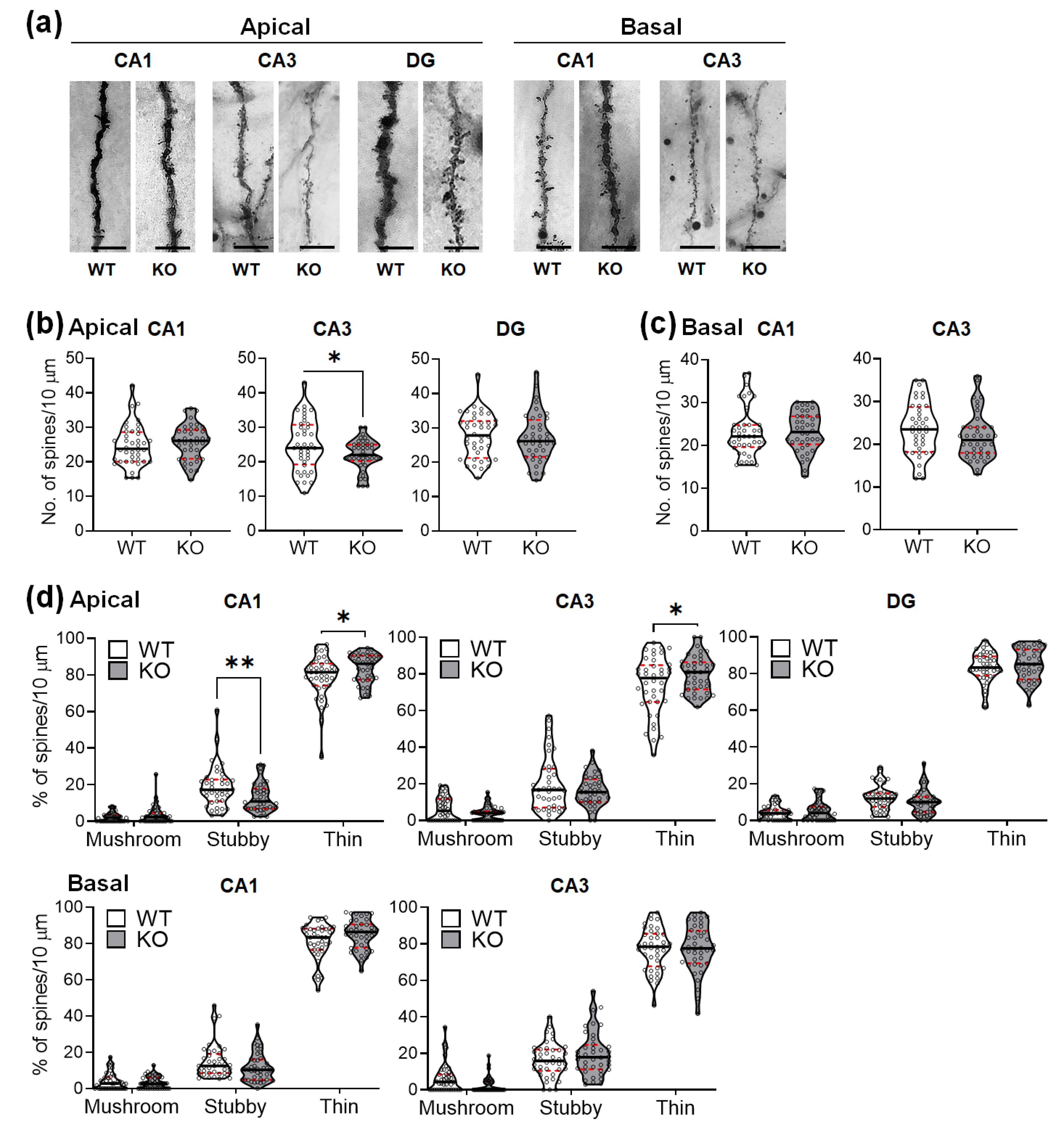
2.3. SREBP-1c KO Mice Show Altered Dendritic Spine Density and Morphology in the Hippocampus
2.4. The CA3 Subregion of SREBP-1c KO Mice Shows Significantly Greater Decreases in Dendritic and Spine Morphological Parameters Compared to Those of Other Subregions
2.5. SREBP-1c KO Mice Show Significantly Reduced SREBP-1 Protein Expression in the CA3 Subregion of the Hippocampus
3. Discussion
4. Materials and Methods
4.1. Animals and Treatment
4.2. Passive Avoidance
4.3. Golgi Staining
4.4. Sholl Analysis
4.5. Analysis of Dendritic Diameter
4.6. Analysis of Dendritic Spine Density and Morphology
4.7. Immunofluorescence of Free-Floating Sections
4.8. Analysis of Differences in the Degree of Morphological Alterations in Each Hippocampal Subregion
4.9. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dhuriya, Y.K.; Sharma, D. Neuronal plasticity: Neuronal organization is associated with neurological disorders. J. Mol. Neurosci. 2020, 70, 1684–1701. [Google Scholar] [CrossRef]
- Gonçalves, J.T.; Schafer, S.T.; Gage, F.H. Adult neurogenesis in the hippocampus: From stem cells to behavior. Cell. 2016, 167, 897–914. [Google Scholar] [CrossRef]
- França, T.F.A. Plasticity and redundancy in the integration of adult born neurons in the hippocampus. Neurobiol. Learn. Mem. 2018, 155, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, K.J. The hippocampus: From memory, to map, to memory map. Trends Neurosci. 2018, 41, 64–66. [Google Scholar] [CrossRef]
- Haukvik, U.K.; Tamnes, C.K.; Söderman, E.; Agartz, I. Neuroimaging hippocampal subfields in schizophrenia and bipolar disorder: A systematic review and meta-analysis. J. Psychiatr. Res. 2018, 104, 217–226. [Google Scholar] [CrossRef]
- Heckers, S. Neuroimaging studies of the hippocampus in schizophrenia. Hippocampus 2001, 11, 520–528. [Google Scholar] [CrossRef]
- Harrison, P.J. The hippocampus in schizophrenia: A review of the neuropathological evidence and its pathophysiological implications. Psychopharmacology 2004, 174, 151–162. [Google Scholar] [CrossRef]
- Gothelf, D.; Soreni, N.; Nachman, R.P.; Tyano, S.; Hiss, Y.; Reiner, O.; Weizman, A. Evidence for the involvement of the hippocampus in the pathophysiology of schizophrenia. Eur. Neuropsychopharmacol. 2000, 10, 389–395. [Google Scholar] [CrossRef]
- Heckers, S.; Konradi, C. Hippocampal pathology in schizophrenia. Curr. Top. Behav. Neurosci. 2010, 4, 529–553. [Google Scholar] [CrossRef]
- Bazan, N.G. Lipid signaling in neural plasticity, brain repair, and neuroprotection. Mol. Neurobiol. 2005, 32, 89–103. [Google Scholar] [CrossRef]
- Dietschy, J.M.; Turley, S.D. Thematic review series: Brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J. Lipid Res. 2004, 45, 1375–1397. [Google Scholar] [CrossRef] [PubMed]
- Edmond, J.; Higa, T.A.; Korsak, R.A.; Bergner, E.A.; Lee, W.N. Fatty acid transport and utilization for the developing brain. J. Neurochem. 1998, 70, 1227–1234. [Google Scholar] [CrossRef]
- Shimano, H. SREBPs: Physiology and pathophysiology of the SREBP family. FEBS J. 2009, 276, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Eberle, D.; Hegarty, B.; Bossard, P.; Ferre, P.; Foufelle, F. SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie 2004, 86, 839–848. [Google Scholar] [CrossRef]
- Hua, X.; Wu, J.; Goldstein, J.L.; Brown, M.S.; Hobbs, H.H. Structure of the human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics 1995, 25, 667–673. [Google Scholar] [CrossRef]
- Miserez, A.R.; Cao, G.; Probst, L.C.; Hobbs, H.H. Structure of the human gene encoding sterol regulatory element binding protein 2 (SREBF2). Genomics 1997, 40, 31–40. [Google Scholar] [CrossRef]
- Shimomura, I.; Shimano, H.; Horton, J.D.; Goldstein, J.L.; Brown, M.S. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Investig. 1997, 99, 838–845. [Google Scholar] [CrossRef]
- Brown, M.S.; Goldstein, J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef]
- Lee, S.; Kang, S.; Ang, M.J.; Kim, J.; Kim, J.C.; Kim, S.H.; Jeon, T.I.; Jung, C.; Im, S.S.; Moon, C. Deficiency of sterol regulatory element-binding protein-1c induces schizophrenia-like behavior in mice. Genes Brain Behav. 2019, 18, e12540. [Google Scholar] [CrossRef]
- Ang, M.J.; Kim, J.; Lee, S.; Kim, S.-H.; Kim, J.-C.; Jeon, T.-I.; Im, S.-S.; Moon, C. Transcriptome profiling reveals novel candidate genes related to hippocampal dysfunction in SREBP-1c knockout mice. Int. J. Mol. Sci. 2020, 21, 4131. [Google Scholar] [CrossRef]
- Carbon, M.; Correll, C.U. Thinking and acting beyond the positive: The role of the cognitive and negative symptoms in schizophrenia. CNS Spectr. 2014, 19 (Suppl. 1), 38–52. [Google Scholar] [CrossRef]
- Rosoklija, G.; Toomayan, G.; Ellis, S.P.; Keilp, J.; Mann, J.J.; Latov, N.; Hays, A.P.; Dwork, A.J. Structural abnormalities of subicular dendrites in subjects with schizophrenia and mood disorders: Preliminary findings. Arch. Gen. Psychiatry 2000, 57, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Moyer, C.E.; Shelton, M.A.; Sweet, R.A. Dendritic spine alterations in schizophrenia. Neurosci. Lett. 2015, 601, 46–53. [Google Scholar] [CrossRef]
- Akum, B.F.; Chen, M.; Gunderson, S.I.; Riefler, G.M.; Scerri-Hansen, M.M.; Firestein, B.L. Cypin regulates dendrite patterning in hippocampal neurons by promoting microtubule assembly. Nat. Neurosci. 2004, 7, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Van Aelst, L.; Cline, H.T. Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat. Neurosci. 2000, 3, 217–225. [Google Scholar] [CrossRef]
- Hoogenraad, C.C.; Milstein, A.D.; Ethell, I.M.; Henkemeyer, M.; Sheng, M. GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat. Neurosci. 2005, 8, 906–915. [Google Scholar] [CrossRef]
- Camargo, N.; Smit, A.B.; Verheijen, M.H. SREBPs: SREBP function in glia–neuron interactions. FEBS J. 2009, 276, 628–636. [Google Scholar] [CrossRef]
- Nieweg, K.; Schaller, H.; Pfrieger, F.W. Marked differences in cholesterol synthesis between neurons and glial cells from postnatal rats. J. Neurochem. 2009, 109, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rodríguez, R.A.; Tabernero, A.; Velasco, A.; Lavado, E.M.; Medina, J.M. The neurotrophic effect of oleic acid includes dendritic differentiation and the expression of the neuronal basic helix-loop-helix transcription factor NeuroD2. J. Neurochem. 2004, 88, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Lee, S.; Kim, J.; Kim, J.-C.; Kim, S.-H.; Son, Y.; Shin, T.; Youn, B.; Kim, J.-S.; Wang, H. Chronic treatment with combined chemotherapeutic agents affects hippocampal micromorphometry and function in mice, independently of neuroinflammation. Exp. Neurobiol. 2018, 27, 419. [Google Scholar] [CrossRef] [PubMed]
- Penzes, P.; Cahill, M.E.; Jones, K.A.; VanLeeuwen, J.-E.; Woolfrey, K.M. Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 2011, 14, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Glantz, L.A.; Lewis, D.A. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch. Gen. Psychiatry 2000, 57, 65–73. [Google Scholar] [CrossRef]
- Sweet, R.A.; Henteleff, R.A.; Zhang, W.; Sampson, A.R.; Lewis, D.A. Reduced dendritic spine density in auditory cortex of subjects with schizophrenia. Neuropsychopharmacology 2009, 34, 374–389. [Google Scholar] [CrossRef]
- Kolomeets, N.S.; Orlovskaya, D.D.; Rachmanova, V.I.; Uranova, N.A. Ultrastructural alterations in hippocampal mossy fiber synapses in schizophrenia: A postmortem morphometric study. Synapse 2005, 57, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Koyama, Y.; Hattori, T.; Nishida, T.; Hori, O.; Tohyama, M. Alterations in dendrite and spine morphology of cortical pyramidal neurons in DISC1-binding zinc finger protein (DBZ) knockout mice. Front. Neuroanat. 2015, 9, 52. [Google Scholar] [CrossRef]
- Konur, S.; Rabinowitz, D.; Fenstermaker, V.L.; Yuste, R. Systematic regulation of spine sizes and densities in pyramidal neurons. J. Neurobiol. 2003, 56, 95–112. [Google Scholar] [CrossRef]
- Toni, N.; Teng, E.M.; Bushong, E.A.; Aimone, J.B.; Zhao, C.; Consiglio, A.; van Praag, H.; Martone, M.E.; Ellisman, M.H.; Gage, F.H. Synapse formation on neurons born in the adult hippocampus. Nat. Neurosci. 2007, 10, 727–734. [Google Scholar] [CrossRef] [PubMed]
- van Deijk, A.L.F.; Camargo, N.; Timmerman, J.; Heistek, T.; Brouwers, J.F.; Mogavero, F.; Mansvelder, H.D.; Smit, A.B.; Verheijen, M.H. Astrocyte lipid metabolism is critical for synapse development and function in vivo. Glia 2017, 65, 670–682. [Google Scholar] [CrossRef]
- Kolomeets, N.S.; Orlovskaya, D.D.; Uranova, N.A. Decreased numerical density of CA3 hippocampal mossy fiber synapses in schizophrenia. Synapse 2007, 61, 615–621. [Google Scholar] [CrossRef]
- Coronel-Oliveros, C.M.; Pacheco-Calderón, R. Prenatal exposure to ketamine in rats: Implications on animal models of schizophrenia. Dev. Psychobiol. 2018, 60, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Amaral, D.G.; Witter, M.P. The three-dimensional organization of the hippocampal formation: A review of anatomical data. Neuroscience 1989, 31, 571–591. [Google Scholar] [CrossRef]
- Brivanlou, I.H.; Dantzker, J.L.; Stevens, C.F.; Callaway, E.M. Topographic specificity of functional connections from hippocampal CA3 to CA1. Proc. Natl. Acad. Sci. USA 2004, 101, 2560–2565. [Google Scholar] [CrossRef]
- Xie, Y.-C.; Yao, Z.-H.; Yao, X.-L.; Pan, J.-Z.; Zhang, S.-F.; Zhang, Y.; Hu, J.-C. Glucagon-like peptide-2 receptor is involved in spatial cognitive dysfunction in rats after chronic cerebral hypoperfusion. J. Alzheimer. Dis. 2018, 66, 1559–1576. [Google Scholar] [CrossRef]
- Kobayashi, M.; Taniura, H.; Yoshikawa, K. Ectopic expression of necdin induces differentiation of mouse neuroblastoma cells. J. Biol. Chem. 2002, 277, 42128–42135. [Google Scholar] [CrossRef]
- Takazaki, R.; Nishimura, I.; Yoshikawa, K. Necdin is required for terminal differentiation and survival of primary dorsal root ganglion neurons. Exp. Cell Res. 2002, 277, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Abel, K.M.; Drake, R.; Goldstein, J.M. Sex differences in schizophrenia. Int. Rev. Psychiatry 2010, 22, 417–428. [Google Scholar] [CrossRef]
- Nopoulos, P.; Flaum, M.; Andreasen, N.C. Sex differences in brain morphology in schizophrenia. Am. J. Psychiatry 1997, 154, 1648–1654. [Google Scholar] [CrossRef] [PubMed]
- Mendrek, A.; Mancini-Marïe, A. Sex/gender differences in the brain and cognition in schizophrenia. Neurosci. Biobehav. Rev. 2016, 67, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Zaidel, D.W.; Esiri, M.M.; Harrison, P.J. Size, shape, and orientation of neurons in the left and right hippocampus: Investigation of normal asymmetries and alterations in schizophrenia. Am. J. Psychiatry 1997, 154, 812–818. [Google Scholar] [CrossRef]
- Zheng, F.; Li, C.; Zhang, D.; Cui, D.; Wang, Z.; Qiu, J. Study on the sub-regions volume of hippocampus and amygdala in schizophrenia. Quant. Imaging Med. Surg. 2019, 9, 1025. [Google Scholar] [CrossRef]
- Geddes, A.E.; Huang, X.-F.; Newell, K.A. GluN2B protein deficits in the left, but not the right, hippocampus in schizophrenia. BMC Psychiatry. 2014, 14, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Delamillieure, P.; Constans, J.M.; Fernandez, J.; Brazo, P.; Benali, K.; Courthéoux, P.; Thibaut, F.; Petit, M.; Dollfus, S. Proton magnetic resonance spectroscopy (1H MRS) in schizophrenia: Investigation of the right and left hippocampus, thalamus, and prefrontal cortex. Schizophre. Bull. 2002, 28, 329–339. [Google Scholar] [CrossRef]
- Chiapponi, C.; Piras, F.; Fagioli, S.; Girardi, P.; Caltagirone, C.; Spalletta, G. Hippocampus age-related microstructural changes in schizophrenia: A case–control mean diffusivity study. Schizophr. Res. 2014, 157, 214–217. [Google Scholar] [CrossRef]
- Pujol, N.; Penadés, R.; Junqué, C.; Dinov, I.; Fu, C.; Catalán, R.; Ibarretxe-Bilbao, N.; Bargalló, N.; Bernardo, M.; Toga, A. Hippocampal abnormalities and age in chronic schizophrenia: Morphometric study across the adult lifespan. Br. J. Psychiatry. 2014, 205, 369–375. [Google Scholar] [CrossRef]
- Thoma, R.J.; Monnig, M.; Hanlon, F.M.; Miller, G.A.; Petropoulos, H.; Mayer, A.R.; Yeo, R.; Euler, M.; Lysne, P.; Moses, S.N. Hippocampus volume and episodic memory in schizophrenia. J. Int. Neuropsychol. Soc. 2009, 15, 182. [Google Scholar] [CrossRef]
- National Research Council. Guide for the Care and Use of Laboratory Animals, 8th ed.; The National Academies Press: Washington, DC, USA, 2011. [Google Scholar] [CrossRef]
- Kim, J.-S.; Lee, H.-J.; Kim, J.C.; Kang, S.S.; Bae, C.-S.; Shin, T.; Jin, J.-K.; Kim, S.H.; Wang, H.; Moon, C. Transient impairment of hippocampus-dependent learning and memory in relatively low-dose of acute radiation syndrome is associated with inhibition of hippocampal neurogenesis. J. Radiat. Res. 2008, 49, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Ang, M.J.; Kang, S.; Moon, C. Melatonin alters neuronal architecture and increases cysteine-rich protein 1 signaling in the male mouse hippocampus. J. Neurosci. Res. 2020, 98, 2333–2348. [Google Scholar] [CrossRef]
- Sholl, D.A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 1953, 87, 387–406. [Google Scholar] [PubMed]
- Morley, B.J.; Mervis, R. Dendritic spine alterations in the hippocampus and parietal cortex of alpha7 nicotinic acetylcholine receptor knockout mice. Neuroscience 2013, 233, 54–63. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Chakraborti, A.; Allen, A.; Allen, B.; Rosi, S.; Fike, J.R. Cranial irradiation alters dendritic spine density and morphology in the hippocampus. PLoS ONE 2012, 7, e40844. [Google Scholar] [CrossRef] [PubMed]
- Boston University Research Support. La Morte’s Power Calculator. Available online: https://www.bu.edu/researchsupport/compliance/animal-care/working-with-animals/research/sample-size-calculations-iacuc/ (accessed on 15 April 2021).

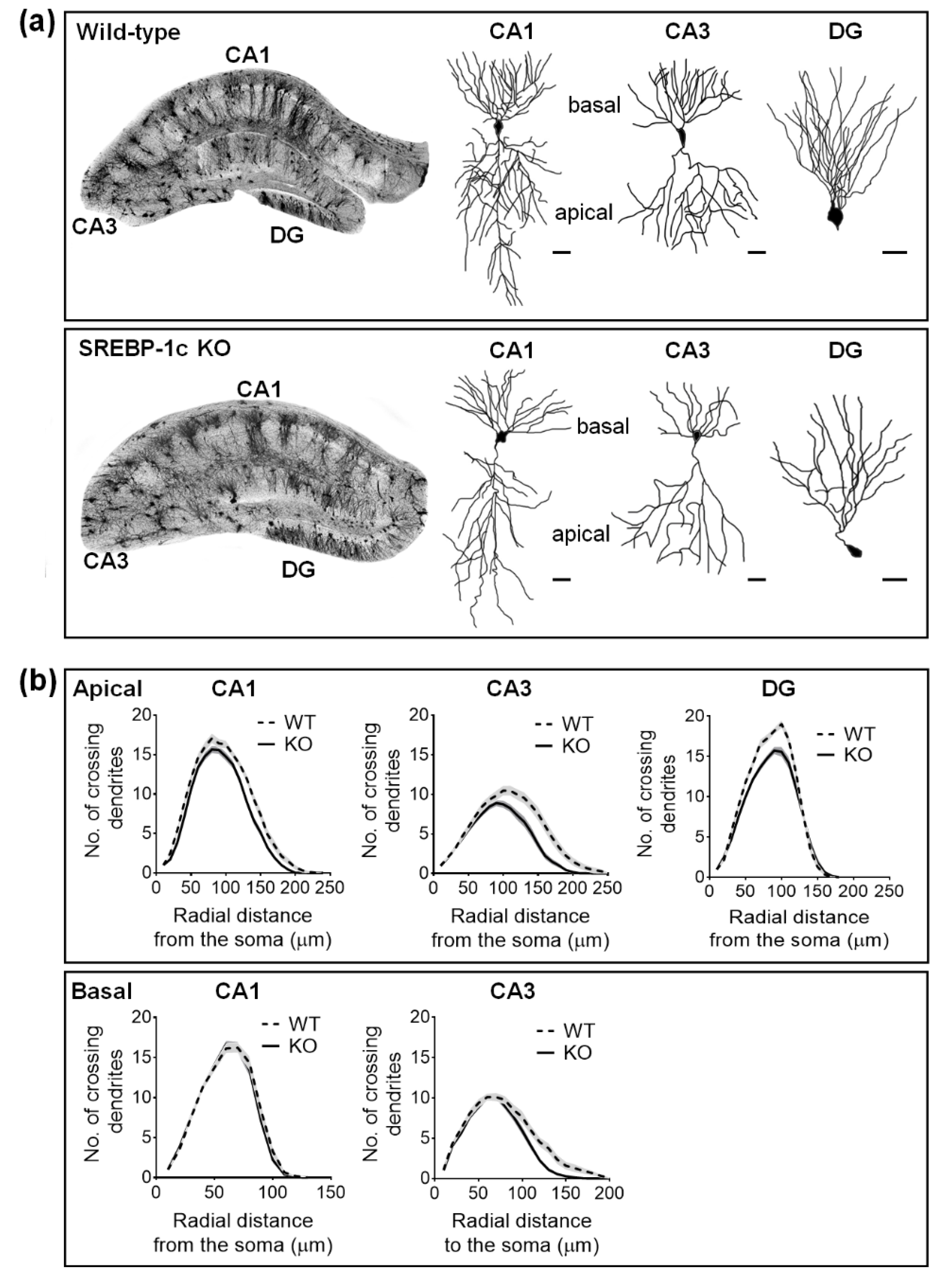


| Sholl Radii Distance (μm) 1 | Šidăk’s Post Hoc Test per Sholl Radii Distance | Two-Way ANOVA | |
|---|---|---|---|
| CA1 apical | 130, 140, 150 | p = 0.0001, p = 0.0007, p = 0.0121 | Finteraction (23, 1794) = 1.741, p = 0.0161 |
| basal | n.s. | n.s. | Finteraction (12, 936) = 0.8516, p = 0.5969 |
| CA3 apical | 110, 120, 130, 140, 150, 160, 170, 180 | p = 0.0371, p = 0.0046, p = 0.0004, p < 0.0001, p < 0.0001, p < 0.0001, p = 0.0004, p = 0.0257 | Finteraction (24,1872) = 3.985, p < 0.0001 |
| basal | 110, 120, 130, 140 | p = 0.0243, p = 0.0023, p = 0.0002, p = 0.0490 | Finteraction (19,1482) = 2.098, p = 0.0037 |
| DG | 70, 80, 90, 100, 110 | p = 0.0014, p = 0.0149, p = 0.0189, p < 0.0001, p = 0.0025 | Finteraction (17, 1326) = 4.031, p < 0.0001 |
| WT | KO | Student’s t-Test | |
|---|---|---|---|
| Total dendritic length (μm) | |||
| CA1 apical | 1811.3 ± 296.7 | 1516 ± 251.8 | t(78) = 4.794, p < 0.0001 |
| basal | 964.3 ± 229.5 | 940.8 ± 160.4 | t(78) = 0.531, p = 0.5971 |
| CA3 apical | 1281 ± 398 | 924 ± 308.2 | t(78) = 4.482, p < 0.0001 |
| basal | 967.5 ± 301.2 | 783 ± 156.1 | t(78) = 3.440, p = 0.0009 |
| DG | 1576 ± 245.1 | 1374 ± 278.5 | t(78) = 3.448, p = 0.0009 |
| Dendritic branch points | |||
| CA1 apical | 25.9 ± 6.7 | 22.0 ± 5.0 | t(78) = 2.956, p = 0.0041 |
| basal | 17.3 ± 6.3 | 16.4 ± 3.6 | t(78) = 0.719, p = 0.4741 |
| CA3 apical | 18.1 ± 5.1 | 10.6 ± 3.9 | t(78) = 7.201, p < 0.0001 |
| basal | 11.8 ± 4.5 | 8.5 ± 3.9 | t(78) = 3.469, p = 0.0009 |
| DG | 28.3 ± 5.7 | 19.7 ± 5.5 | t(78) = 3.282, p = 0.0015 |
| Dendritic thickness (μm) | |||
| CA1 apical | 0.845 ± 0.190 | 0.890 ± 0.185 | t(78) = 1.031, p = 0.3056 |
| basal | 0.864 ± 0.206 | 0.916 ± 0.189 | t(78) = 1.168, p = 0.2462 |
| CA3 apical | 0.976 ± 0.325 | 0.664 ± 0.202 | t(78) = 5.162, p < 0.0001 |
| basal | 0.775 ± 0.244 | 0.689 ± 0.190 | t(78) = 1.760, p = 0.0823 |
| DG | 1.117 ± 0.291 | 0.994 ± 0.228 | t(78) = 2.013, p = 0.0387 |
| WT | KO | Student’s t-Test | |
|---|---|---|---|
| CA1 apical | 24.8 ± 6.0 | 25.7 ± 5.1 | t(78) = 0.7302, p = 0.4675 |
| basal | 23.1 ± 5.7 | 23.2 ± 4.5 | t(78) = 0.0146, p = 0.9884 |
| CA3 apical | 24.9 ± 7.5 | 22.0 ± 4.4 | t(78) = 2.108, p = 0.0382 |
| basal | 23.6 ± 6.2 | 21.9 ± 5.5 | t(78) = 1.362, p = 0.1770 |
| DG | 27.7 ± 6.5 | 27.1 ± 7.1 | t(78) = 0.4203, p = 0.6754 |
| Mushroom Spines (%) | Stubby Spines (%) | Thin Spines (%) | Two-Way ANOVA | |
|---|---|---|---|---|
| CA1 apical | ||||
| WT | 2.2 ± 2.6 | 18.7 ± 11.2 | 79.1 ± 11.8 | Finteraction (2, 156) = 5.126, p = 0.0070 |
| KO | 3.4 ± 4.9 | 13.0 ± 7.8 | 83.6 ± 7.9 | |
| p = 0.9026 1 | p = 0.0082 1 | p = 0.0487 1 | ||
| CA1 basal | ||||
| WT | 4.0 ± 4.7 | 14.9 ± 9.3 | 81.0 ± 10 | Finteraction (2, 156) = 3.003, p = 0.0525 |
| KO | 3.3 ± 3.2 | 11.7 ± 7.9 | 84.9 ± 8.2 | |
| p = 0.9630 1 | p = 0.1753 1 | p = 0.0647 1 | ||
| CA3 apical | ||||
| WT | 6.4 ± 6.3 | 19.7 ± 14.8 | 73.8 ± 15.8 | Finteraction (2, 156) = 3.231, p = 0.0422 |
| KO | 3.9 ± 3.9 | 16.2 ± 8.4 | 79.9 ± 9.8 | |
| p = 0.6465 1 | p = 0.3666 1 | p = 0.0357 1 | ||
| CA3 basal | ||||
| WT | 6.3 ± 7.8 | 16.3 ± 9.4 | 77.4 ± 11.5 | Finteraction (2, 156) = 1.599, p = 0.2054 |
| KO | 2.7 ± 4.3 | 19.7 ± 12 | 77.6 ± 12.8 | |
| p = 0.3040 1 | p = 0.3488 1 | p = 0.9997 1 | ||
| DG | ||||
| WT | 4.2 ± 3.9 | 12.3 ± 6.7 | 83.4 ± 8.3 | Finteraction (2, 156) = 1.224, p = 0.2970 |
| KO | 5.1 ± 5.2 | 9.9 ± 6.8 | 84.9 ± 8.7 | |
| p = 0.9285 1 | p = 0.3283 1 | p = 0.6861 1 | ||
| Dendritic Length | Dendritic Branch Point | Dendritic Diameter | Spine Density | Two-Way ANOVA | |
|---|---|---|---|---|---|
| CA1 apical | −0.16 ± 0.022 | −0.25 ± 0.029 | 0.051 ± 0.035 | 0.037 ± 0.032 | Finteraction (12, 780) = 3.718, p < 0.0001 |
| basal | −0.02 ± 0.026 | −0.004 ± 0.043 | 0.06 ± 0.035 | 0.0007 ± 0.031 | |
| CA3 apical | −0.28 ± 0.038 | −0.33 ± 0.042 | −0.32 ± 0.033 | −0.12 ± 0.028 | |
| basal | −0.19 ± 0.026 | −0.24 ± 0.045 | −0.11 ± 0.039 | −0.08 ± 0.037 | |
| DG | −0.13 ± 0.028 | −0.28 ± 0.035 | −0.11 ± 0.032 | −0.02 ± 0.04 |
| CA1 Apical | CA1 Basal | CA3 Apical | CA3 Basal | DG | |
|---|---|---|---|---|---|
| CA1 apical | 0.0024 | <0.0001 | 0.0330 | 0.2481 | |
| CA1 basal | <0.0001 | <0.0001 | <0.0001 | ||
| CA3 apical | 0.0001 | <0.0001 | |||
| CA3 basal | 0.9978 | ||||
| DG |
| WT | KO | Šidăk’s Post Hoc Test | Two-Way ANOVA | |
|---|---|---|---|---|
| CA1 | 1.40 ± 0.25 1 | 1.31 ± 0.13 1 | p = 0.8182 | Genotype: F(1, 8) = 6.392, p = 0.0354 Subregion: F(2, 16) = 18.86, p < 0.0001 Interaction: F(2, 16) = 5.725, p = 0.0133 |
| CA3 | 1.59 ± 0.07 1 | 1.15 ± 0.25 1 | p = 0.001 | |
| DG | 1.05 ± 0.10 1 | 1 ± 0.08 1 | p = 0.9429 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ang, M.J.; Lee, S.; Wada, M.; Weerasinghe-Mudiyanselage, P.D.E.; Kim, S.-H.; Shin, T.; Jeon, T.-I.; Im, S.-S.; Moon, C. SREBP-1c Deficiency Affects Hippocampal Micromorphometry and Hippocampus-Dependent Memory Ability in Mice. Int. J. Mol. Sci. 2021, 22, 6103. https://doi.org/10.3390/ijms22116103
Ang MJ, Lee S, Wada M, Weerasinghe-Mudiyanselage PDE, Kim S-H, Shin T, Jeon T-I, Im S-S, Moon C. SREBP-1c Deficiency Affects Hippocampal Micromorphometry and Hippocampus-Dependent Memory Ability in Mice. International Journal of Molecular Sciences. 2021; 22(11):6103. https://doi.org/10.3390/ijms22116103
Chicago/Turabian StyleAng, Mary Jasmin, Sueun Lee, Mai Wada, Poornima D. E. Weerasinghe-Mudiyanselage, Sung-Ho Kim, Taekyun Shin, Tae-Il Jeon, Seung-Soon Im, and Changjong Moon. 2021. "SREBP-1c Deficiency Affects Hippocampal Micromorphometry and Hippocampus-Dependent Memory Ability in Mice" International Journal of Molecular Sciences 22, no. 11: 6103. https://doi.org/10.3390/ijms22116103
APA StyleAng, M. J., Lee, S., Wada, M., Weerasinghe-Mudiyanselage, P. D. E., Kim, S.-H., Shin, T., Jeon, T.-I., Im, S.-S., & Moon, C. (2021). SREBP-1c Deficiency Affects Hippocampal Micromorphometry and Hippocampus-Dependent Memory Ability in Mice. International Journal of Molecular Sciences, 22(11), 6103. https://doi.org/10.3390/ijms22116103








