Molecular Characterization of a Rare Case of Bilateral Vitreoretinal T Cell Lymphoma through Vitreous Liquid Biopsy
Abstract
1. Introduction
2. Methods
2.1. Case Selection
2.2. DNA Analysis
2.3. Next-Generation Sequencing
2.4. Next-Generation Sequencing Analysis
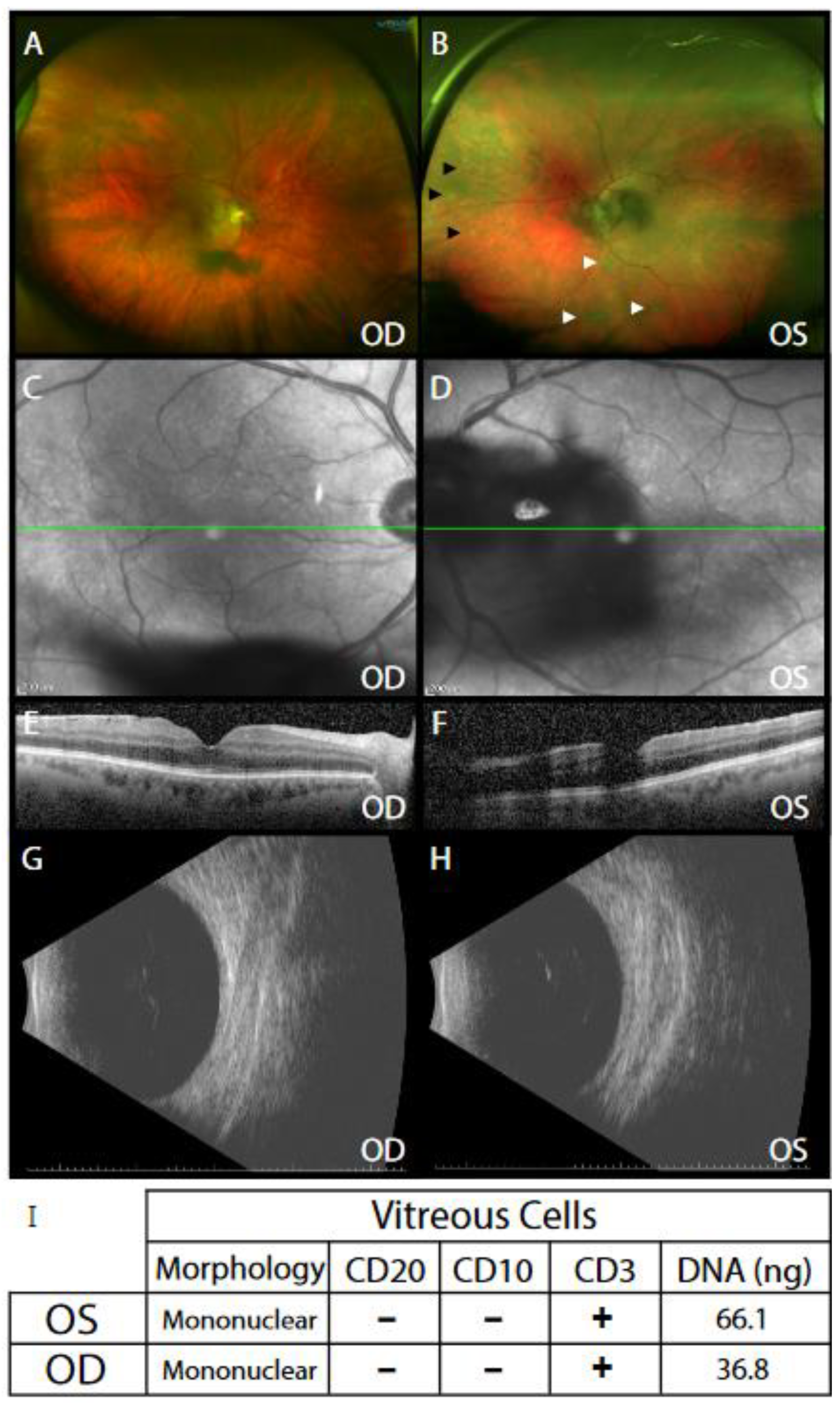
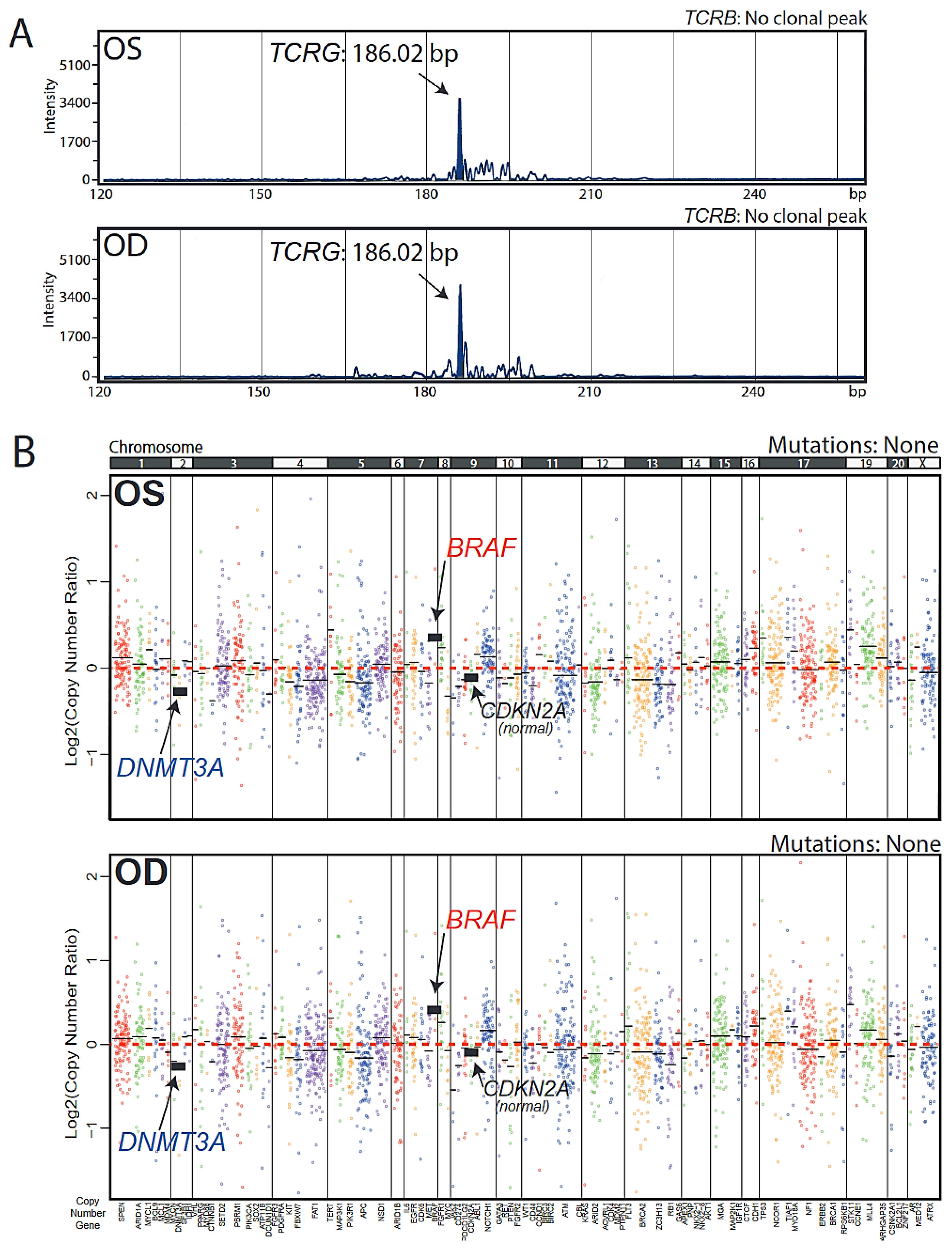
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, C.; Rubenstein, J.L.; Coupland, S.; Davis, J.L.; Harbour, J.W.; Johnston, P.B.; Cassoux, N.; Touitou, V.; Smith, J.R.; Batchelor, T.T.; et al. Primary Vitreoretinal Lymphoma: A Report from an International Primary Central Nervous System Lymphoma Collaborative Group Symposium. Oncologist 2011, 16, 1589–1599. [Google Scholar] [CrossRef] [PubMed]
- Pulido, J.S.; Johnston, P.B.; Nowakowski, G.S.; Castellino, A.; Raja, H. Correction to: The diagnosis and treatment of primary vitreoretinal lymphoma: A review. Int. J. Retin. Vitr. 2018, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Cani, A.K.; Hovelson, D.H.; Demirci, H.; Johnson, M.W.; Tomlins, S.A.; Rao, R.C. Next generation sequencing of vitreoretinal lymphomas from small-volume intraocular liquid biopsies: New routes to targeted therapies. Oncotarget 2016, 8, 7989–7998. [Google Scholar] [CrossRef] [PubMed]
- Hirschhorn, J.W.; Snider, J.S.; Lindsey, K.G.; Schandl, C.A. Molecular profiling of vitreous fluid by massively parallel sequencing: A case series. J. Am. Soc. Cytopathol. 2020, 9, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Fend, F.; Ferreri, A.J.M.; Coupland, S. How we diagnose and treat vitreoretinal lymphoma. Br. J. Haematol. 2016, 173, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.L. Intraocular lymphoma: A clinical perspective. Eye 2013, 27, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Hovelson, D.H.; McDaniel, A.S.; Cani, A.K.; Johnson, B.; Rhodes, K.; Williams, P.D.; Bandla, S.; Bien, G.; Choppa, P.; Hyland, F.; et al. Development and Validation of a Scalable Next-Generation Sequencing System for Assessing Relevant Somatic Variants in Solid Tumors. Neoplasia 2015, 17, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Conley, B.A.; Doroshow, J.H. Molecular Analysis for Therapy Choice: NCI MATCH. Semin. Oncol. 2014, 41, 297–299. [Google Scholar] [CrossRef] [PubMed]
- Cani, A.K.; Soliman, M.; Hovelson, D.H.; Liu, C.-J.; McDaniel, A.S.; Haller, M.J.; Bratley, J.V.; Rahrig, S.; Li, Q.; Briceño, C.A.; et al. Comprehensive genomic profiling of orbital and ocular adnexal lymphomas identifies frequent alterations in MYD88 and chromatin modifiers: New routes to targeted therapies. Mod. Pathol. 2016, 29, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Grasso, C.S.; Cani, A.K.; Hovelson, D.H.; Quist, M.J.; Douville, N.J.; Yadati, V.; Amin, A.M.; Nelson, P.S.; Betz, B.L.; Liu, C.-J.; et al. Integrative molecular profiling of routine clinical prostate cancer specimens. Ann. Oncol. 2015, 26, 1110–1118. [Google Scholar] [CrossRef]
- Warrick, J.I.; Hovelson, D.H.; Amin, A.; Liu, C.-J.; Cani, A.K.; McDaniel, A.S.; Yadati, V.; Quist, M.J.; Weizer, A.Z.; Brenner, J.C.; et al. Tumor evolution and progression in multifocal and paired non-invasive/invasive urothelial carcinoma. Virchows Archiv 2015, 466, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Brüggemann, M.; White, H.; Gaulard, P.; Garcia-Sanz, R.; Gameiro, P.; Oeschger, S.; Jasani, B.; Ott, M.; Delsol, G.; Orfao, A.; et al. Powerful strategy for polymerase chain reaction-based clonality assessment in T-cell malignancies Report of the BIOMED-2 Concerted Action BHM4 CT98-3936. Leukemia 2006, 21, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018, 24, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Almeida, A.C.; Abate, F.; Khiabanian, H.; Martinez-Escala, E.; Guitart, J.; Tensen, C.P.; Vermeer, M.H.; Rabadan, R.; Ferrando, A.; Palomero, T. The mutational landscape of cutaneous T cell lymphoma and Sezary syndrome. Nat Genet. 2015, 47, 1465–1470. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef]
- Watatani, Y.; Sato, Y.; Miyoshi, H.; Sakamoto, K.; Nishida, K.; Gion, Y.; Nagata, Y.; Shiraishi, Y.; Chiba, K.; Tanaka, H.; et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia 2019, 33, 2867–2883. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cani, A.K.; Toral, M.A.; Balikov, D.A.; Betz, B.L.; Hu, K.; Liu, C.-J.; Prifti, M.V.; Chinnaiyan, A.M.; Tomlins, S.A.; Mahajan, V.B.; et al. Molecular Characterization of a Rare Case of Bilateral Vitreoretinal T Cell Lymphoma through Vitreous Liquid Biopsy. Int. J. Mol. Sci. 2021, 22, 6099. https://doi.org/10.3390/ijms22116099
Cani AK, Toral MA, Balikov DA, Betz BL, Hu K, Liu C-J, Prifti MV, Chinnaiyan AM, Tomlins SA, Mahajan VB, et al. Molecular Characterization of a Rare Case of Bilateral Vitreoretinal T Cell Lymphoma through Vitreous Liquid Biopsy. International Journal of Molecular Sciences. 2021; 22(11):6099. https://doi.org/10.3390/ijms22116099
Chicago/Turabian StyleCani, Andi K., Marcus A. Toral, Daniel A. Balikov, Bryan L. Betz, Kevin Hu, Chia-Jen Liu, Matthew V. Prifti, Arul M. Chinnaiyan, Scott A. Tomlins, Vinit B. Mahajan, and et al. 2021. "Molecular Characterization of a Rare Case of Bilateral Vitreoretinal T Cell Lymphoma through Vitreous Liquid Biopsy" International Journal of Molecular Sciences 22, no. 11: 6099. https://doi.org/10.3390/ijms22116099
APA StyleCani, A. K., Toral, M. A., Balikov, D. A., Betz, B. L., Hu, K., Liu, C.-J., Prifti, M. V., Chinnaiyan, A. M., Tomlins, S. A., Mahajan, V. B., & Rao, R. C. (2021). Molecular Characterization of a Rare Case of Bilateral Vitreoretinal T Cell Lymphoma through Vitreous Liquid Biopsy. International Journal of Molecular Sciences, 22(11), 6099. https://doi.org/10.3390/ijms22116099






