MRTF: Basic Biology and Role in Kidney Disease
Abstract
1. Introduction
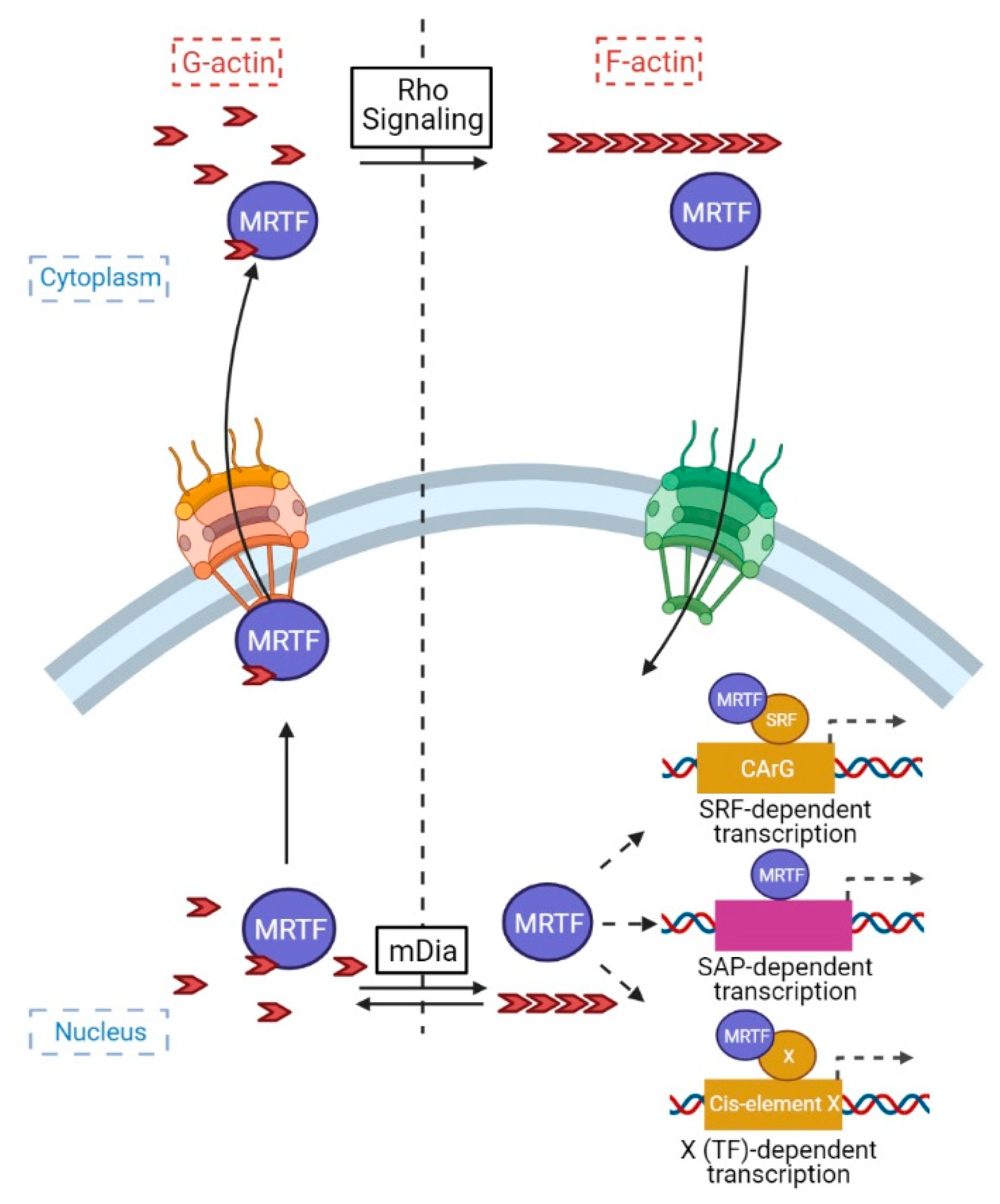
2. MRTFs: Their Discovery and Modus Operandi
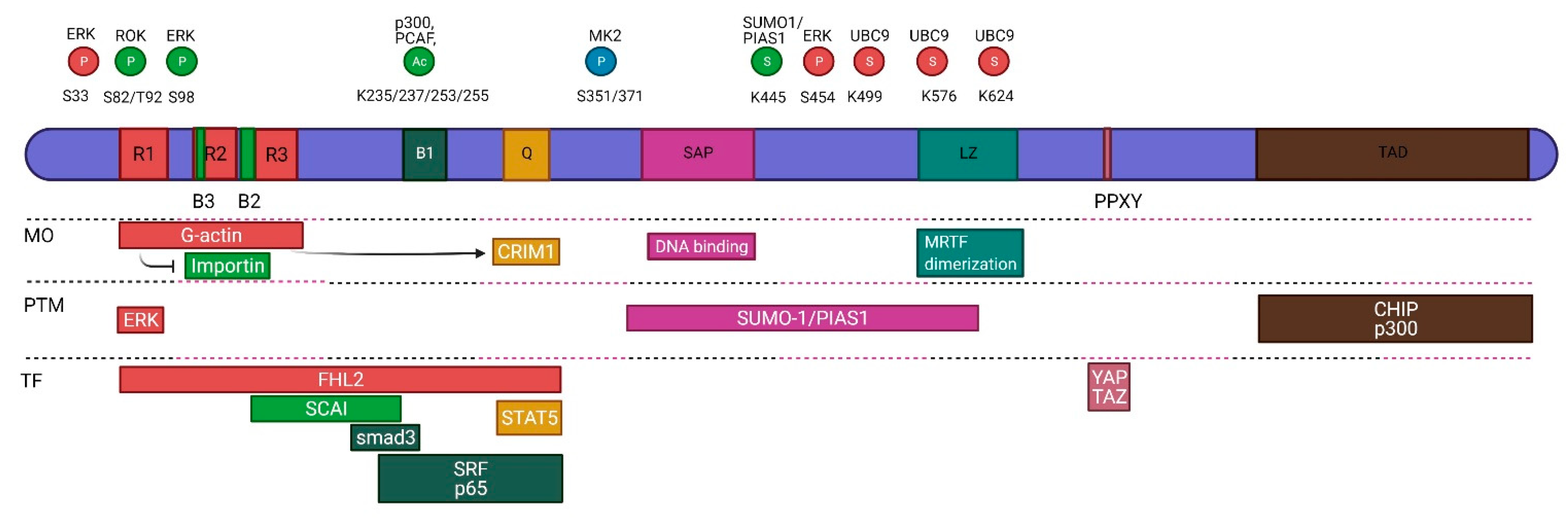
3. Structure
4. Regulation
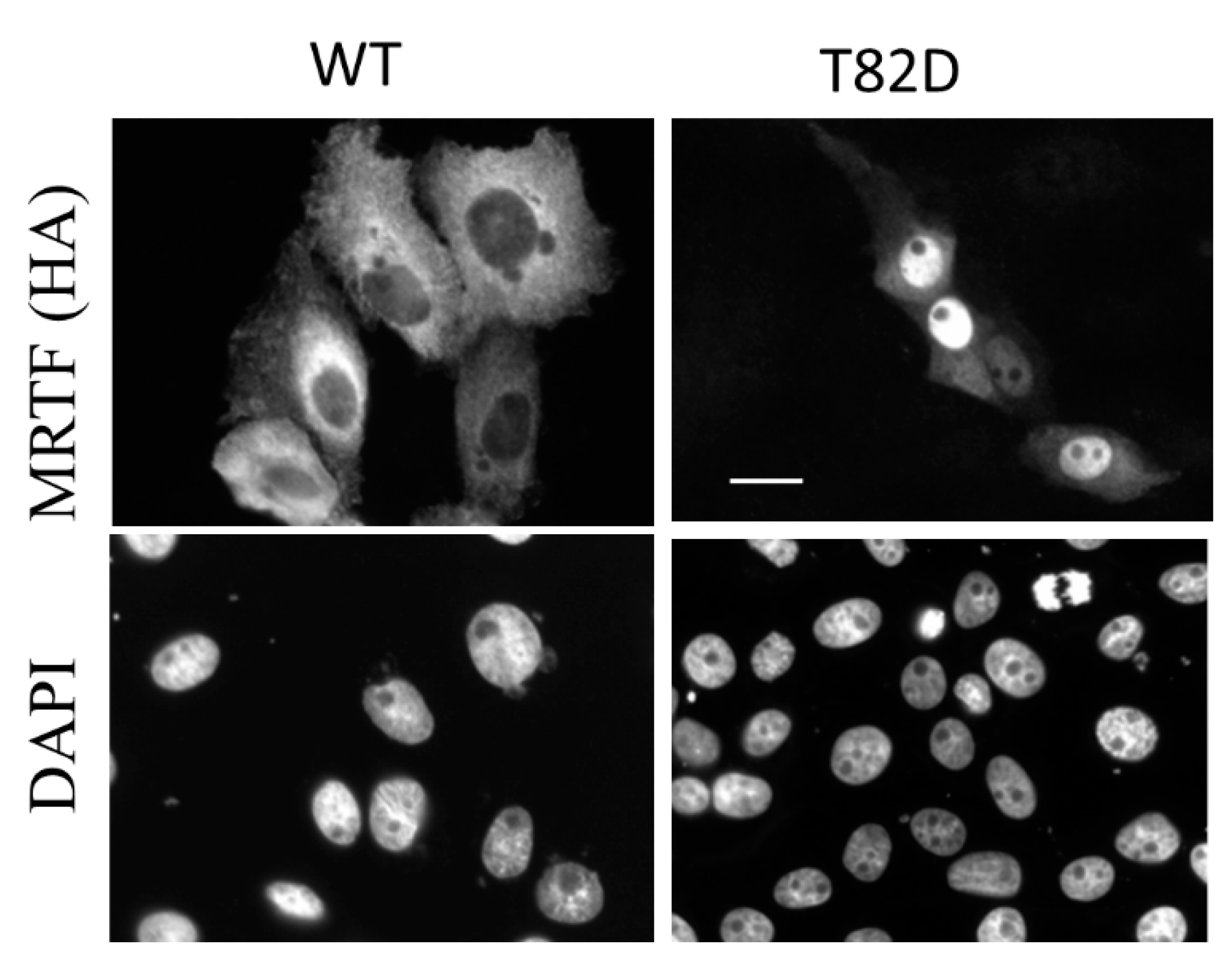
4.1. Posttranslational Modifications
4.2. Regulatory Protein Interactions
4.3. Regulation of MRTF Transcription
5. Targets, Functions, Actions
- (1)
- transcriptional co-activator for SRF or other TFs
- (2)
- regulator of epigenetic processes and chromosome organization
- (3)
- “moonlighting” protein, when it exerts its effect independent of its role in gene expression.
5.1. MRTF as a Transcriptional Coactivator
5.2. MRTF as an Epigenetic Modifier
5.3. ”Moonlighting” Functions of MRTF
5.4. MRTF Knockouts
6. MRTF in Kidney Diseases
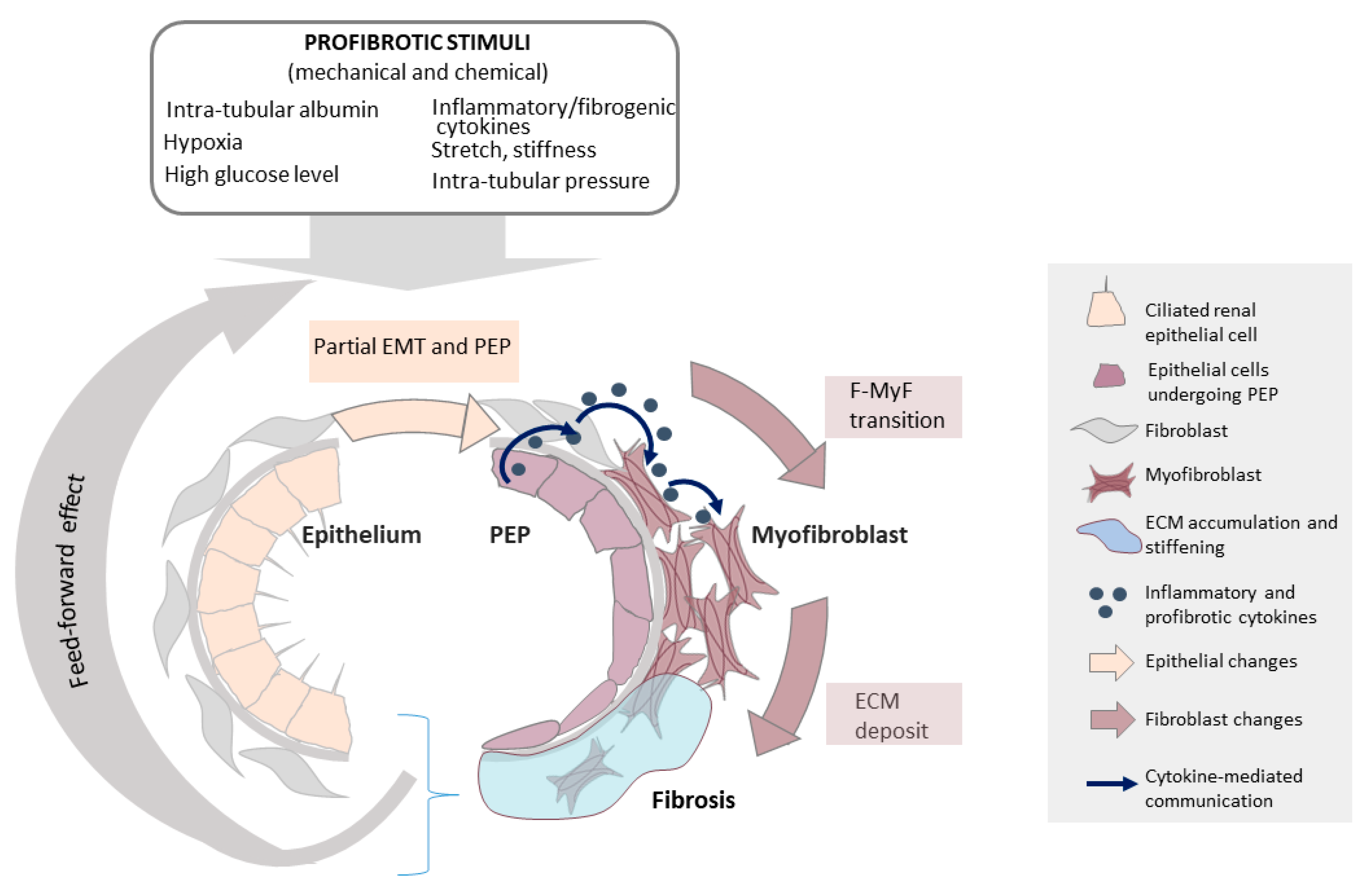
6.1. The Epithelium as a Key Initiator of Fibrosis
6.2. EMT/EMyT and PEP
6.3. MRTF in Animal Models of Kidney Disease
6.3.1. Diabetic Nephropathy (DN)
6.3.2. Obstructive Nephropathy (ON)
6.3.3. Acute Kidney Injury (AKI)
6.3.4. Polycystic Kidney Disease (PKD)—Solidifying a Hypothesis
6.3.5. Other Renal Diseases
7. Perspectives: MRTF in Therapy
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hall, A. Rho family GTPases. Biochem. Soc. Trans. 2012, 40, 1378–1382. [Google Scholar] [CrossRef]
- Perico, L.; Conti, S.; Benigni, A.; Remuzzi, G. Podocyte-actin dynamics in health and disease. Nat. Rev. Nephrol. 2016, 12, 692–710. [Google Scholar] [CrossRef] [PubMed]
- Schell, C.; Huber, T.B. The Evolving Complexity of the Podocyte Cytoskeleton. J. Am. Soc. Nephrol. 2017, 28, 3166–3174. [Google Scholar] [CrossRef]
- Blaine, J.; Dylewski, J. Regulation of the Actin Cytoskeleton in Podocytes. Cells 2020, 9, 1700. [Google Scholar] [CrossRef]
- Sever, S. Role of actin cytoskeleton in podocytes. Pediatr. Nephrol. 2020. [Google Scholar] [CrossRef]
- Moriyama, T.; Nagatoya, K. The Rho-ROCK system as a new therapeutic target for preventing interstitial fibrosis. Drug News Perspect. 2004, 17, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Guan, K.L. Polycystic kidney disease: A Hippo connection. Genes Dev. 2018, 32, 737–739. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.A.; Welsh, G.I. Podocyte RhoGTPases: New therapeutic targets for nephrotic syndrome? F1000Research 2019, 8. [Google Scholar] [CrossRef]
- Matsuda, J.; Asano-Matsuda, K.; Kitzler, T.M.; Takano, T. Rho GTPase regulatory proteins in podocytes. Kidney Int. 2021, 99, 336–345. [Google Scholar] [CrossRef]
- Kustermans, G.; Piette, J.; Legrand-Poels, S. Actin-targeting natural compounds as tools to study the role of actin cytoskeleton in signal transduction. Biochem. Pharmacol. 2008, 76, 1310–1322. [Google Scholar] [CrossRef]
- Olson, E.N.; Nordheim, A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 2010, 11, 353–365. [Google Scholar] [CrossRef]
- Sidorenko, E.; Vartiainen, M.K. Nucleoskeletal regulation of transcription: Actin on MRTF. Exp. Biol. Med. 2019, 244, 1372–1381. [Google Scholar] [CrossRef]
- Hoon, J.L.; Tan, M.H.; Koh, C.G. The Regulation of Cellular Responses to Mechanical Cues by Rho GTPases. Cells 2016, 5, 17. [Google Scholar] [CrossRef]
- Bros, M.; Haas, K.; Moll, L.; Grabbe, S. RhoA as a Key Regulator of Innate and Adaptive Immunity. Cells 2019, 8, 733. [Google Scholar] [CrossRef]
- Burridge, K.; Monaghan-Benson, E.; Graham, D.M. Mechanotransduction: From the cell surface to the nucleus via RhoA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180229. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Sebe, A.; Peterfi, Z.; Masszi, A.; Thirone, A.C.; Rotstein, O.D.; Nakano, H.; McCulloch, C.A.; Szaszi, K.; Mucsi, I.; et al. Cell contact-dependent regulation of epithelial-myofibroblast transition via the rho-rho kinase-phospho-myosin pathway. Mol. Biol. Cell 2007, 18, 1083–1097. [Google Scholar] [CrossRef] [PubMed]
- Small, E.M. The actin-MRTF-SRF gene regulatory axis and myofibroblast differentiation. J. Cardiovasc. Transl. Res. 2012, 5, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Gasparics, A.; Sebe, A. MRTFs- master regulators of EMT. Dev. Dyn. 2018, 247, 396–404. [Google Scholar] [CrossRef]
- Morita, T.; Mayanagi, T.; Sobue, K. Dual roles of myocardin-related transcription factors in epithelial mesenchymal transition via slug induction and actin remodeling. J. Cell Biol. 2007, 179, 1027–1042. [Google Scholar] [CrossRef]
- Masszi, A.; Speight, P.; Charbonney, E.; Lodyga, M.; Nakano, H.; Szaszi, K.; Kapus, A. Fate-determining mechanisms in epithelial-myofibroblast transition: Major inhibitory role for Smad3. J. Cell Biol. 2010, 188, 383–399. [Google Scholar] [CrossRef]
- Yu, O.M.; Miyamoto, S.; Brown, J.H. Myocardin-Related Transcription Factor A and Yes-Associated Protein Exert Dual Control in G Protein-Coupled Receptor- and RhoA-Mediated Transcriptional Regulation and Cell Proliferation. Mol. Cell. Biol. 2016, 36, 39–49. [Google Scholar] [CrossRef]
- Speight, P.; Kofler, M.; Szaszi, K.; Kapus, A. Context-dependent switch in chemo/mechanotransduction via multilevel crosstalk among cytoskeleton-regulated MRTF and TAZ and TGFbeta-regulated Smad3. Nat. Commun. 2016, 7, 11642. [Google Scholar] [CrossRef]
- Foster, C.T.; Gualdrini, F.; Treisman, R. Mutual dependence of the MRTF-SRF and YAP-TEAD pathways in cancer-associated fibroblasts is indirect and mediated by cytoskeletal dynamics. Genes Dev. 2017, 31, 2361–2375. [Google Scholar] [CrossRef]
- Aragona, M.; Panciera, T.; Manfrin, A.; Giulitti, S.; Michielin, F.; Elvassore, N.; Dupont, S.; Piccolo, S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 2013, 154, 1047–1059. [Google Scholar] [CrossRef]
- Graham, R.; Gilman, M. Distinct protein targets for signals acting at the c-fos serum response element. Science 1991, 251, 189–192. [Google Scholar] [CrossRef]
- Norman, C.; Runswick, M.; Pollock, R.; Treisman, R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988, 55, 989–1003. [Google Scholar] [CrossRef]
- Sartorelli, V.; Webster, K.A.; Kedes, L. Muscle-specific expression of the cardiac alpha-actin gene requires MyoD1, CArG-box binding factor, and Sp1. Genes Dev. 1990, 4, 1811–1822. [Google Scholar] [CrossRef]
- Sepulveda, J.L.; Belaguli, N.; Nigam, V.; Chen, C.Y.; Nemer, M.; Schwartz, R.J. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: Role for regulating early cardiac gene expression. Mol. Cell. Biol. 1998, 18, 3405–3415. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.; Treisman, R.; Garrett, N.; Mohun, T. Muscle-specific (CArG) and serum-responsive (SRE) promoter elements are functionally interchangeable in Xenopus embryos and mouse fibroblasts. Development 1989, 106, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Miano, J.M. Serum response factor: Toggling between disparate programs of gene expression. J. Mol. Cell Cardiol. 2003, 35, 577–593. [Google Scholar] [CrossRef]
- Hill, C.S.; Wynne, J.; Treisman, R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 1995, 81, 1159–1170. [Google Scholar] [CrossRef]
- Mack, C.P.; Somlyo, A.V.; Hautmann, M.; Somlyo, A.P.; Owens, G.K. Smooth muscle differentiation marker gene expression is regulated by RhoA-mediated actin polymerization. J. Biol. Chem. 2001, 276, 341–347. [Google Scholar] [CrossRef]
- Onuh, J.O.; Qiu, H. Serum response factor-cofactor interactions and their implications in disease. FEBS J. 2020, 288, 3120–3134. [Google Scholar] [CrossRef]
- Price, M.A.; Hill, C.; Treisman, R. Integration of growth factor signals at the c-fos serum response element. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1996, 351, 551–559. [Google Scholar]
- Treisman, R.; Alberts, A.S.; Sahai, E. Regulation of SRF activity by Rho family GTPases. Cold Spring Harb. Symp. Quant. Biol. 1998, 63, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Sotiropoulos, A.; Gineitis, D.; Copeland, J.; Treisman, R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 1999, 98, 159–169. [Google Scholar] [CrossRef]
- Wang, D.Z.; Li, S.; Hockemeyer, D.; Sutherland, L.; Wang, Z.; Schratt, G.; Richardson, J.A.; Nordheim, A.; Olson, E.N. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. USA 2002, 99, 14855–14860. [Google Scholar] [CrossRef]
- Miralles, F.; Posern, G.; Zaromytidou, A.I.; Treisman, R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 2003, 113, 329–342. [Google Scholar] [CrossRef]
- Selvaraj, A.; Prywes, R. Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J. Biol. Chem. 2003, 278, 41977–41987. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chang, P.S.; Wang, Z.; Sutherland, L.; Richardson, J.A.; Small, E.; Krieg, P.A.; Olson, E.N. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 2001, 105, 851–862. [Google Scholar] [CrossRef]
- Mercher, T.; Coniat, M.B.; Monni, R.; Mauchauffe, M.; Nguyen Khac, F.; Gressin, L.; Mugneret, F.; Leblanc, T.; Dastugue, N.; Berger, R.; et al. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc. Natl. Acad. Sci. USA 2001, 98, 5776–5779. [Google Scholar] [CrossRef]
- Ma, Z.; Morris, S.W.; Valentine, V.; Li, M.; Herbrick, J.A.; Cui, X.; Bouman, D.; Li, Y.; Mehta, P.K.; Nizetic, D.; et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 2001, 28, 220–221. [Google Scholar] [CrossRef]
- Cen, B.; Selvaraj, A.; Burgess, R.C.; Hitzler, J.K.; Ma, Z.; Morris, S.W.; Prywes, R. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 2003, 23, 6597–6608. [Google Scholar] [CrossRef]
- Descot, A.; Rex-Haffner, M.; Courtois, G.; Bluteau, D.; Menssen, A.; Mercher, T.; Bernard, O.A.; Treisman, R.; Posern, G. OTT-MAL is a deregulated activator of serum response factor-dependent gene expression. Mol. Cell. Biol. 2008, 28, 6171–6181. [Google Scholar] [CrossRef][Green Version]
- Sasazuki, T.; Sawada, T.; Sakon, S.; Kitamura, T.; Kishi, T.; Okazaki, T.; Katano, M.; Tanaka, M.; Watanabe, M.; Yagita, H.; et al. Identification of a novel transcriptional activator, BSAC, by a functional cloning to inhibit tumor necrosis factor-induced cell death. J. Biol. Chem. 2002, 277, 28853–28860. [Google Scholar] [CrossRef]
- Panayiotou, R.; Miralles, F.; Pawlowski, R.; Diring, J.; Flynn, H.R.; Skehel, M.; Treisman, R. Phosphorylation acts positively and negatively to regulate MRTF-A subcellular localisation and activity. eLife 2016, 5. [Google Scholar] [CrossRef]
- Muehlich, S.; Wang, R.; Lee, S.M.; Lewis, T.C.; Dai, C.; Prywes, R. Serum-induced phosphorylation of the serum response factor coactivator MKL1 by the extracellular signal-regulated kinase 1/2 pathway inhibits its nuclear localization. Mol. Cell. Biol. 2008, 28, 6302–6313. [Google Scholar] [CrossRef]
- Ronkina, N.; Lafera, J.; Kotlyarov, A.; Gaestel, M. Stress-dependent phosphorylation of myocardin-related transcription factor A (MRTF-A) by the p38(MAPK)/MK2 axis. Sci. Rep. 2016, 6, 31219. [Google Scholar] [CrossRef]
- Charbonney, E.; Speight, P.; Masszi, A.; Nakano, H.; Kapus, A. beta-catenin and Smad3 regulate the activity and stability of myocardin-related transcription factor during epithelial-myofibroblast transition. Mol. Biol. Cell 2011, 22, 4472–4485. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Fan, Y.; Zhang, H.; Zhang, Y.; She, M.; Gu, D.; Patterson, C.; Li, H. CHIP represses myocardin-induced smooth muscle cell differentiation via ubiquitin-mediated proteasomal degradation. Mol. Cell. Biol. 2009, 29, 2398–2408. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Wang, C.; Tang, R.; Chen, H.; Zhang, Z.; Tatsuguchi, M.; Wang, D.Z. Acetylation of myocardin is required for the activation of cardiac and smooth muscle genes. J. Biol. Chem. 2012, 287, 38495–38504. [Google Scholar] [CrossRef]
- He, H.; Wang, D.; Yao, H.; Wei, Z.; Lai, Y.; Hu, J.; Liu, X.; Wang, Y.; Zhou, H.; Wang, N.; et al. Transcriptional factors p300 and MRTF-A synergistically enhance the expression of migration-related genes in MCF-7 breast cancer cells. Biochem. Biophys. Res. Commun. 2015, 467, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, Z.; Fang, M.; Xu, Y. Acetylation of MKL1 by PCAF regulates pro-inflammatory transcription. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yuan, Q.; Cao, X.L.; Zhang, Y.; Min, Z.L.; Xu, S.Q.; Yu, Z.J.; Cheng, J.; Zhang, C.; Hu, X.M. Opposite effects of HDAC5 and p300 on MRTF-A-related neuronal apoptosis during ischemia/reperfusion injury in rats. Cell Death Dis. 2017, 8, e2624. [Google Scholar] [CrossRef]
- Li, Z.; Qin, H.; Li, J.; Yu, L.; Yang, Y.; Xu, Y. HADC5 deacetylates MKL1 to dampen TNF-alpha induced pro-inflammatory gene transcription in macrophages. Oncotarget 2017, 8, 94235–94246. [Google Scholar] [CrossRef]
- Zhang, M.; Urabe, G.; Little, C.; Wang, B.; Kent, A.M.; Huang, Y.; Kent, K.C.; Guo, L.W. HDAC6 Regulates the MRTF-A/SRF Axis and Vascular Smooth Muscle Cell Plasticity. JACC Basic Transl. Sci. 2018, 3, 782–795. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Z.; Guo, J.; Xu, Y. Deacetylation of MRTF-A by SIRT1 defies senescence induced down-regulation of collagen type I in fibroblast cells. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165723. [Google Scholar] [CrossRef]
- Nakagawa, K.; Kuzumaki, N. Transcriptional activity of megakaryoblastic leukemia 1 (MKL1) is repressed by SUMO modification. Genes Cells 2005, 10, 835–850. [Google Scholar] [CrossRef]
- Wang, J.; Li, A.; Wang, Z.; Feng, X.; Olson, E.N.; Schwartz, R.J. Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol. Cell. Biol. 2007, 27, 622–632. [Google Scholar] [CrossRef]
- Hinson, J.S.; Medlin, M.D.; Taylor, J.M.; Mack, C.P. Regulation of myocardin factor protein stability by the LIM-only protein FHL2. Am. J. Physiol. Heart Circ. Physiol. 2008, 295, H1067–H1075. [Google Scholar] [CrossRef]
- Philippar, U.; Schratt, G.; Dieterich, C.; Muller, J.M.; Galgoczy, P.; Engel, F.B.; Keating, M.T.; Gertler, F.; Schule, R.; Vingron, M.; et al. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol. Cell 2004, 16, 867–880. [Google Scholar] [CrossRef]
- Luchsinger, L.L.; Patenaude, C.A.; Smith, B.D.; Layne, M.D. Myocardin-related transcription factor-A complexes activate type I collagen expression in lung fibroblasts. J. Biol. Chem. 2011, 286, 44116–44125. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zhang, Q.; Fan, H.; Zhao, H.; Yang, Y. Myocardin-Related Transcription Factor A Mediates LPS-Induced iNOS Transactivation. Inflammation 2020, 43, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Prakash, J.; Nguyen, P.; Davis-Dusenbery, B.N.; Hill, N.S.; Layne, M.D.; Hata, A.; Lagna, G. Bone morphogenetic protein signaling in vascular disease: Anti-inflammatory action through myocardin-related transcription factor A. J. Biol. Chem. 2012, 287, 28067–28077. [Google Scholar] [CrossRef]
- Chen, Q.; Huang, J.; Gong, W.; Chen, Z.; Huang, J.; Liu, P.; Huang, H. MRTF-A mediated FN and ICAM-1 expression in AGEs-induced rat glomerular mesangial cells via activating STAT5. Mol. Cell Endocrinol. 2018, 460, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Xing, W.J.; Liao, X.H.; Wang, N.; Zhao, D.W.; Zheng, L.; Zheng, D.L.; Dong, J.; Zhang, T.C. MRTF-A and STAT3 promote MDA-MB-231 cell migration via hypermethylating BRSM1. IUBMB Life 2015, 67, 202–217. [Google Scholar] [CrossRef] [PubMed]
- Mouilleron, S.; Guettler, S.; Langer, C.A.; Treisman, R.; McDonald, N.Q. Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL. EMBO J. 2008, 27, 3198–3208. [Google Scholar] [CrossRef]
- Vartiainen, M.K.; Guettler, S.; Larijani, B.; Treisman, R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 2007, 316, 1749–1752. [Google Scholar] [CrossRef]
- Staus, D.P.; Weise-Cross, L.; Mangum, K.D.; Medlin, M.D.; Mangiante, L.; Taylor, J.M.; Mack, C.P. Nuclear RhoA signaling regulates MRTF-dependent SMC-specific transcription. Am. J. Physiol. Heart Circ. Physiol. 2014, 307, H379–H390. [Google Scholar] [CrossRef]
- Garcia-Mata, R.; Burridge, K. Catching a GEF by its tail. Trends Cell. Biol. 2007, 17, 36–43. [Google Scholar] [CrossRef]
- Joo, E.; Olson, M.F. Regulation and functions of the RhoA regulatory guanine nucleotide exchange factor GEF-H1. Small GTPases 2020, 1–14. [Google Scholar] [CrossRef]
- Denk-Lobnig, M.; Martin, A.C. Modular regulation of Rho family GTPases in development. Small GTPases 2019, 10, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Hinkel, R.; Trenkwalder, T.; Petersen, B.; Husada, W.; Gesenhues, F.; Lee, S.; Hannappel, E.; Bock-Marquette, I.; Theisen, D.; Leitner, L.; et al. MRTF-A controls vessel growth and maturation by increasing the expression of CCN1 and CCN2. Nat. Commun. 2014, 5, 3970. [Google Scholar] [CrossRef]
- Zhao, X.H.; Laschinger, C.; Arora, P.; Szaszi, K.; Kapus, A.; McCulloch, C.A. Force activates smooth muscle alpha-actin promoter activity through the Rho signaling pathway. J. Cell Sci. 2007, 120, 1801–1809. [Google Scholar] [CrossRef] [PubMed]
- Thirone, A.C.; Speight, P.; Zulys, M.; Rotstein, O.D.; Szaszi, K.; Pedersen, S.F.; Kapus, A. Hyperosmotic stress induces Rho/Rho kinase/LIM kinase-mediated cofilin phosphorylation in tubular cells: Key role in the osmotically triggered F-actin response. Am. J. Physiol. Cell. Physiol. 2009, 296, C463–C475. [Google Scholar] [CrossRef] [PubMed]
- Ly, D.L.; Waheed, F.; Lodyga, M.; Speight, P.; Masszi, A.; Nakano, H.; Hersom, M.; Pedersen, S.F.; Szaszi, K.; Kapus, A. Hyperosmotic stress regulates the distribution and stability of myocardin-related transcription factor, a key modulator of the cytoskeleton. Am. J. Physiol. Cell. Physiol. 2013, 304, C115–C127. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.W.; Chaudary, F.; Lee, W.; Copeland, J.W.; McCulloch, C.A. Force-induced myofibroblast differentiation through collagen receptors is dependent on mammalian diaphanous (mDia). J. Biol. Chem. 2010, 285, 9273–9281. [Google Scholar] [CrossRef] [PubMed]
- Lundquist, M.R.; Storaska, A.J.; Liu, T.C.; Larsen, S.D.; Evans, T.; Neubig, R.R.; Jaffrey, S.R. Redox modification of nuclear actin by MICAL-2 regulates SRF signaling. Cell 2014, 156, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Mouilleron, S.; Langer, C.A.; Guettler, S.; McDonald, N.Q.; Treisman, R. Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci. Signal. 2011, 4, ra40. [Google Scholar] [CrossRef]
- Hirano, H.; Matsuura, Y. Sensing actin dynamics: Structural basis for G-actin-sensitive nuclear import of MAL. Biochem. Biophys. Res. Commun. 2011, 414, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, R.; Rajakyla, E.K.; Vartiainen, M.K.; Treisman, R. An actin-regulated importin alpha/beta-dependent extended bipartite NLS directs nuclear import of MRTF-A. EMBO J. 2010, 29, 3448–3458. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Hayashi, K.; Iwasaki, K.; Fujioka, T.; Egusa, H.; Yatani, H.; Sobue, K. Nuclear import mechanism for myocardin family members and their correlation with vascular smooth muscle cell phenotype. J. Biol. Chem. 2010, 285, 37314–37323. [Google Scholar] [CrossRef]
- Hayashi, K.; Morita, T. Differences in the nuclear export mechanism between myocardin and myocardin-related transcription factor A. J. Biol. Chem. 2013, 288, 5743–5755. [Google Scholar] [CrossRef]
- Posern, G.; Sotiropoulos, A.; Treisman, R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell 2002, 13, 4167–4178. [Google Scholar] [CrossRef] [PubMed]
- Zaromytidou, A.I.; Miralles, F.; Treisman, R. MAL and ternary complex factor use different mechanisms to contact a common surface on the serum response factor DNA-binding domain. Mol. Cell. Biol. 2006, 26, 4134–4148. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. SAP—A putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000, 25, 112–114. [Google Scholar] [CrossRef]
- Kipp, M.; Gohring, F.; Ostendorp, T.; van Drunen, C.M.; van Driel, R.; Przybylski, M.; Fackelmayer, F.O. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 2000, 20, 7480–7489. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Yang, Y.; Yuan, Z.; Gao, Y.; Zhou, J.; Chen, Q.; Xu, Y. Myocardin-related transcription factor A mediates OxLDL-induced endothelial injury. Circ. Res. 2011, 108, 797–807. [Google Scholar] [CrossRef]
- Yang, Y.; Cheng, X.; Tian, W.; Zhou, B.; Wu, X.; Xu, H.; Fang, F.; Fang, M.; Xu, Y. MRTF-A steers an epigenetic complex to activate endothelin-induced pro-inflammatory transcription in vascular smooth muscle cells. Nucl. Acids Res. 2014, 42, 10460–10472. [Google Scholar] [CrossRef]
- Shao, J.; Xu, H.; Wu, X.; Xu, Y. Epigenetic activation of CTGF transcription by high glucose in renal tubular epithelial cells is mediated by myocardin-related transcription factor A. Cell Tissue Res. 2020, 379, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, G.; Yu, L.; Lin, L.; Liu, L.; Fang, M.; Xu, Y. An Interplay Between MRTF-A and the Histone Acetyltransferase TIP60 Mediates Hypoxia-Reoxygenation Induced iNOS Transcription in Macrophages. Front. Cell Dev. Biol 2020, 8, 484. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Morita, T. Importance of dimer formation of myocardin family members in the regulation of their nuclear export. Cell Struct. Funct. 2013, 38, 123–134. [Google Scholar] [CrossRef][Green Version]
- Weissbach, J.; Schikora, F.; Weber, A.; Kessels, M.; Posern, G. Myocardin-Related Transcription Factor A Activation by Competition with WH2 Domain Proteins for Actin Binding. Mol. Cell. Biol. 2016, 36, 1526–1539. [Google Scholar] [CrossRef] [PubMed]
- Kluge, F.; Weissbach, J.; Weber, A.; Stradal, T.; Posern, G. Regulation of MRTF-A by JMY via a nucleation-independent mechanism. Cell Commun. Signal. 2018, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Miranda, M.Z.; Bialik, J.F.; Speight, P.; Dan, Q.; Yeung, T.; Szaszi, K.; Pedersen, S.F.; Kapus, A. TGF-beta1 regulates the expression and transcriptional activity of TAZ protein via a Smad3-independent, myocardin-related transcription factor-mediated mechanism. J. Biol. Chem. 2017, 292, 14902–14920. [Google Scholar] [CrossRef]
- Lockman, K.; Hinson, J.S.; Medlin, M.D.; Morris, D.; Taylor, J.M.; Mack, C.P. Sphingosine 1-phosphate stimulates smooth muscle cell differentiation and proliferation by activating separate serum response factor co-factors. J. Biol. Chem. 2004, 279, 42422–42430. [Google Scholar] [CrossRef]
- Sebe, A.; Masszi, A.; Zulys, M.; Yeung, T.; Speight, P.; Rotstein, O.D.; Nakano, H.; Mucsi, I.; Szaszi, K.; Kapus, A. Rac, PAK and p38 regulate cell contact-dependent nuclear translocation of myocardin-related transcription factor. FEBS Lett. 2008, 582, 291–298. [Google Scholar] [CrossRef]
- Kojonazarov, B.; Novoyatleva, T.; Boehm, M.; Happe, C.; Sibinska, Z.; Tian, X.; Sajjad, A.; Luitel, H.; Kriechling, P.; Posern, G.; et al. p38 MAPK Inhibition Improves Heart Function in Pressure-Loaded Right Ventricular Hypertrophy. Am. J. Respir. Cell. Mol. Biol. 2017, 57, 603–614. [Google Scholar] [CrossRef]
- Ma, F.Y.; Sachchithananthan, M.; Flanc, R.S.; Nikolic-Paterson, D.J. Mitogen activated protein kinases in renal fibrosis. Front. Biosci. 2009, 1, 171–187. [Google Scholar] [CrossRef]
- Ruwanpura, S.M.; Thomas, B.J.; Bardin, P.G. Pirfenidone: Molecular Mechanisms and Potential Clinical Applications in Lung Disease. Am. J. Respir. Cell. Mol. Biol. 2020, 62, 413–422. [Google Scholar] [CrossRef]
- Haller, V.; Nahidino, P.; Forster, M.; Laufer, S.A. An updated patent review of p38 MAP kinase inhibitors (2014–2019). Exp. Opin. Ther. Pat. 2020, 30, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Martin-Garrido, A.; Brown, D.I.; Lyle, A.N.; Dikalova, A.; Seidel-Rogol, B.; Lassegue, B.; San Martin, A.; Griendling, K.K. NADPH oxidase 4 mediates TGF-beta-induced smooth muscle alpha-actin via p38MAPK and serum response factor. Free Radic. Biol. Med. 2011, 50, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Badorff, C.; Seeger, F.H.; Zeiher, A.M.; Dimmeler, S. Glycogen synthase kinase 3beta inhibits myocardin-dependent transcription and hypertrophy induction through site-specific phosphorylation. Circ. Res. 2005, 97, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Elberg, G.; Chen, L.; Elberg, D.; Chan, M.D.; Logan, C.J.; Turman, M.A. MKL1 mediates TGF-beta1-induced alpha-smooth muscle actin expression in human renal epithelial cells. Am. J. Physiol. Renal Physiol. 2008, 294, F1116–F1128. [Google Scholar] [CrossRef]
- Jeon, E.S.; Park, W.S.; Lee, M.J.; Kim, Y.M.; Han, J.; Kim, J.H. A Rho kinase/myocardin-related transcription factor-A-dependent mechanism underlies the sphingosylphosphorylcholine-induced differentiation of mesenchymal stem cells into contractile smooth muscle cells. Circ. Res. 2008, 103, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; San Martin, A.; Valdivia, A.; Martin-Garrido, A.; Griendling, K.K. Redox-Sensitive Regulation of Myocardin-Related Transcription Factor (MRTF-A) Phosphorylation via Palladin in Vascular Smooth Muscle Cell Differentiation Marker Gene Expression. PLoS ONE 2016, 11, e0153199. [Google Scholar] [CrossRef]
- Scott, R.W.; Olson, M.F. LIM kinases: Function, regulation and association with human disease. J. Mol. Med. 2007, 85, 555–568. [Google Scholar] [CrossRef]
- Geneste, O.; Copeland, J.W.; Treisman, R. LIM kinase and Diaphanous cooperate to regulate serum response factor and actin dynamics. J. Cell. Biol. 2002, 157, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Julian, L.; Olson, M.F. Rho-associated coiled-coil containing kinases (ROCK): Structure, regulation, and functions. Small GTPases 2014, 5, e29846. [Google Scholar] [CrossRef]
- Janota, C.S.; Calero-Cuenca, F.J.; Gomes, E.R. The role of the cell nucleus in mechanotransduction. Curr. Opin. Cell. Biol. 2020, 63, 204–211. [Google Scholar] [CrossRef]
- Ho, C.Y.; Jaalouk, D.E.; Vartiainen, M.K.; Lammerding, J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature 2013, 497, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Plessner, M.; Melak, M.; Chinchilla, P.; Baarlink, C.; Grosse, R. Nuclear F-actin formation and reorganization upon cell spreading. J. Biol. Chem. 2015, 290, 11209–11216. [Google Scholar] [CrossRef] [PubMed]
- Kassianidou, E.; Kalita, J.; Lim, R.Y.H. The role of nucleocytoplasmic transport in mechanotransduction. Exp. Cell Res. 2019, 377, 86–93. [Google Scholar] [CrossRef]
- Elosegui-Artola, A.; Andreu, I.; Beedle, A.E.M.; Lezamiz, A.; Uroz, M.; Kosmalska, A.J.; Oria, R.; Kechagia, J.Z.; Rico-Lastres, P.; Le Roux, A.L.; et al. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 2017, 171, 1397–1410 e14. [Google Scholar] [CrossRef]
- Hoffman, L.M.; Smith, M.A.; Jensen, C.C.; Yoshigi, M.; Blankman, E.; Ullman, K.S.; Beckerle, M.C. Mechanical stress triggers nuclear remodeling and the formation of transmembrane actin nuclear lines with associated nuclear pore complexes. Mol. Biol. Cell 2020, 31, 1774–1787. [Google Scholar] [CrossRef]
- Alam, S.G.; Zhang, Q.; Prasad, N.; Li, Y.; Chamala, S.; Kuchibhotla, R.; Kc, B.; Aggarwal, V.; Shrestha, S.; Jones, A.L.; et al. The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Sci. Rep. 2016, 6, 38063. [Google Scholar] [CrossRef]
- Pecci, A.; Ma, X.; Savoia, A.; Adelstein, R.S. MYH9: Structure, functions and role of non-muscle myosin IIA in human disease. Gene 2018, 664, 152–167. [Google Scholar] [CrossRef]
- Wei, Y.; Renard, C.A.; Labalette, C.; Wu, Y.; Levy, L.; Neuveut, C.; Prieur, X.; Flajolet, M.; Prigent, S.; Buendia, M.A. Identification of the LIM protein FHL2 as a coactivator of beta-catenin. J. Biol. Chem. 2003, 278, 5188–5194. [Google Scholar] [CrossRef]
- Li, S.Y.; Huang, P.H.; Tarng, D.C.; Lin, T.P.; Yang, W.C.; Chang, Y.H.; Yang, A.H.; Lin, C.C.; Yang, M.H.; Chen, J.W.; et al. Four-and-a-Half LIM Domains Protein 2 Is a Coactivator of Wnt Signaling in Diabetic Kidney Disease. J. Am. Soc. Nephrol. 2015, 26, 3072–3084. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.G.; Ma, H.J.; Guo, Z.Y.; Zhang, W.; Yang, X. FHL2 participates in renal interstitial fibrosis by altering the phenotype of renal tubular epithelial cells via regulating the beta-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2734–2741. [Google Scholar] [PubMed]
- Li, S.Y.; Chu, P.H.; Huang, P.H.; Hsieh, T.H.; Susztak, K.; Tarng, D.C. FHL2 mediates podocyte Rac1 activation and foot process effacement in hypertensive nephropathy. Sci. Rep. 2019, 9, 6693. [Google Scholar] [CrossRef]
- Duan, Y.; Qiu, Y.; Huang, X.; Dai, C.; Yang, J.; He, W. Deletion of FHL2 in fibroblasts attenuates fibroblasts activation and kidney fibrosis via restraining TGF-beta1-induced Wnt/beta-catenin signaling. J. Mol. Med. 2020, 98, 291–307. [Google Scholar] [CrossRef]
- Xu, H.; Wu, X.; Qin, H.; Tian, W.; Chen, J.; Sun, L.; Fang, M.; Xu, Y. Myocardin-Related Transcription Factor A Epigenetically Regulates Renal Fibrosis in Diabetic Nephropathy. J. Am. Soc. Nephrol. 2015, 26, 1648–1660. [Google Scholar] [CrossRef] [PubMed]
- Bialik, J.F.; Ding, M.; Speight, P.; Dan, Q.; Miranda, M.Z.; Di Ciano-Oliveira, C.; Kofler, M.M.; Rotstein, O.D.; Pedersen, S.F.; Szaszi, K.; et al. Profibrotic epithelial phenotype: A central role for MRTF and TAZ. Sci. Rep. 2019, 9, 4323. [Google Scholar] [CrossRef]
- Brandt, D.T.; Xu, J.; Steinbeisser, H.; Grosse, R. Regulation of myocardin-related transcriptional coactivators through cofactor interactions in differentiation and cancer. Cell Cycle 2009, 8, 2523–2527. [Google Scholar] [CrossRef] [PubMed]
- Fintha, A.; Gasparics, A.; Fang, L.; Erdei, Z.; Hamar, P.; Mozes, M.M.; Kokeny, G.; Rosivall, L.; Sebe, A. Characterization and role of SCAI during renal fibrosis and epithelial-to-mesenchymal transition. Am. J. Pathol. 2013, 182, 388–400. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Du, F.; He, Y.; Wei, Z.; Meng, C.; Xu, Y.; Zhou, H.; Wang, N.; Luo, X.G.; Ma, W.; et al. The Wnt-beta-catenin signaling regulated MRTF-A transcription to activate migration-related genes in human breast cancer cells. Oncotarget 2018, 9, 15239–15251. [Google Scholar] [CrossRef]
- Francisco, J.; Zhang, Y.; Jeong, J.I.; Mizushima, W.; Ikeda, S.; Ivessa, A.; Oka, S.; Zhai, P.; Tallquist, M.D.; Del Re, D.P. Blockade of Fibroblast YAP Attenuates Cardiac Fibrosis and Dysfunction Through MRTF-A Inhibition. JACC Basic Transl. Sci. 2020, 5, 931–945. [Google Scholar] [CrossRef]
- Gao, X.; Xu, D.; Li, S.; Wei, Z.; Li, S.; Cai, W.; Mao, N.; Jin, F.; Li, Y.; Yi, X.; et al. Pulmonary Silicosis Alters MicroRNA Expression in Rat Lung and miR-411-3p Exerts Anti-fibrotic Effects by Inhibiting MRTF-A/SRF Signaling. Mol. Ther. Nucl. Acids 2020, 20, 851–865. [Google Scholar] [CrossRef] [PubMed]
- Holstein, I.; Singh, A.K.; Pohl, F.; Misiak, D.; Braun, J.; Leitner, L.; Huttelmaier, S.; Posern, G. Post-transcriptional regulation of MRTF-A by miRNAs during myogenic differentiation of myoblasts. Nucleic Acids Res. 2020, 48, 8927–8942. [Google Scholar] [CrossRef] [PubMed]
- Cen, B.; Selvaraj, A.; Prywes, R. Myocardin/MKL family of SRF coactivators: Key regulators of immediate early and muscle specific gene expression. J. Cell Biochem. 2004, 93, 74–82. [Google Scholar] [CrossRef]
- Posern, G.; Treisman, R. Actin’ together: Serum response factor, its cofactors and the link to signal transduction. Trends Cell. Biol. 2006, 16, 588–596. [Google Scholar] [CrossRef]
- Parmacek, M.S. Myocardin-related transcription factors: Critical coactivators regulating cardiovascular development and adaptation. Circ. Res. 2007, 100, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Scharenberg, M.A.; Chiquet-Ehrismann, R.; Asparuhova, M.B. Megakaryoblastic leukemia protein-1 (MKL1): Increasing evidence for an involvement in cancer progression and metastasis. Int, J. Biochem. Cell Biol. 2010, 42, 1911–1914. [Google Scholar] [CrossRef] [PubMed]
- Quaggin, S.E.; Kapus, A. Scar wars: Mapping the fate of epithelial-mesenchymal-myofibroblast transition. Kidney Int. 2011, 80, 41–50. [Google Scholar] [CrossRef]
- Kuwahara, K.; Nakao, K. New molecular mechanisms for cardiovascular disease:transcriptional pathways and novel therapeutic targets in heart failure. J. Pharmacol. Sci. 2011, 116, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Janmey, P.A.; Wells, R.G.; Assoian, R.K.; McCulloch, C.A. From tissue mechanics to transcription factors. Differentiation 2013, 86, 112–120. [Google Scholar] [CrossRef]
- Sward, K.; Stenkula, K.G.; Rippe, C.; Alajbegovic, A.; Gomez, M.F.; Albinsson, S. Emerging roles of the myocardin family of proteins in lipid and glucose metabolism. J. Physiol. 2016, 594, 4741–4752. [Google Scholar] [CrossRef]
- Gau, D.; Roy, P. SRF’ing and SAP’ing—The role of MRTF proteins in cell migration. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Esnault, C.; Stewart, A.; Gualdrini, F.; East, P.; Horswell, S.; Matthews, N.; Treisman, R. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 2014, 28, 943–958. [Google Scholar] [CrossRef]
- Rozycki, M.; Lodyga, M.; Lam, J.; Miranda, M.Z.; Fatyol, K.; Speight, P.; Kapus, A. The fate of the primary cilium during myofibroblast transition. Mol. Biol. Cell 2014, 25, 643–657. [Google Scholar] [CrossRef]
- Masszi, A.; Di Ciano, C.; Sirokmany, G.; Arthur, W.T.; Rotstein, O.D.; Wang, J.; McCulloch, C.A.; Rosivall, L.; Mucsi, I.; Kapus, A. Central role for Rho in TGF-beta1-induced alpha-smooth muscle actin expression during epithelial-mesenchymal transition. Am. J. Physiol. Renal Physiol. 2003, 284, F911–F924. [Google Scholar] [CrossRef] [PubMed]
- Muehlich, S.; Rehm, M.; Ebenau, A.; Goppelt-Struebe, M. Synergistic induction of CTGF by cytochalasin D and TGFbeta-1 in primary human renal epithelial cells: Role of transcriptional regulators MKL1, YAP/TAZ and Smad2/3. Cell Signal. 2017, 29, 31–40. [Google Scholar] [CrossRef]
- Krawczyk, K.M.; Hansson, J.; Nilsson, H.; Krawczyk, K.K.; Sward, K.; Johansson, M.E. Injury induced expression of caveolar proteins in human kidney tubules—Role of megakaryoblastic leukemia 1. BMC Nephrol. 2017, 18, 320. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, A.; Zent, R. Integrins in kidney disease. J. Am. Soc. Nephrol. 2013, 24, 1034–1039. [Google Scholar] [CrossRef] [PubMed]
- Asparuhova, M.B.; Ferralli, J.; Chiquet, M.; Chiquet-Ehrismann, R. The transcriptional regulator megakaryoblastic leukemia-1 mediates serum response factor-independent activation of tenascin-C transcription by mechanical stress. FASEB J. 2011, 25, 3477–3488. [Google Scholar] [CrossRef] [PubMed]
- Parreno, J.; Raju, S.; Niaki, M.N.; Andrejevic, K.; Jiang, A.; Delve, E.; Kandel, R. Expression of type I collagen and tenascin C is regulated by actin polymerization through MRTF in dedifferentiated chondrocytes. FEBS Lett. 2014, 588, 3677–3684. [Google Scholar] [CrossRef] [PubMed]
- Iwanciw, D.; Rehm, M.; Porst, M.; Goppelt-Struebe, M. Induction of connective tissue growth factor by angiotensin II: Integration of signaling pathways. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 1782–1787. [Google Scholar] [CrossRef]
- Chaqour, B.; Goppelt-Struebe, M. Mechanical regulation of the Cyr61/CCN1 and CTGF/CCN2 proteins. FEBS J. 2006, 273, 3639–3649. [Google Scholar] [CrossRef]
- Mao, L.; Liu, L.; Zhang, T.; Wu, X.; Zhang, T.; Xu, Y. MKL1 mediates TGF-beta-induced CTGF transcription to promote renal fibrosis. J. Cell. Physiol. 2020, 235, 4790–4803. [Google Scholar] [CrossRef]
- Guan, T.H.; Chen, G.; Gao, B.; Janssen, M.R.; Uttarwar, L.; Ingram, A.J.; Krepinsky, J.C. Caveolin-1 deficiency protects against mesangial matrix expansion in a mouse model of type 1 diabetic nephropathy. Diabetologia 2013, 56, 2068–2077. [Google Scholar] [CrossRef]
- Yang, S.; Liu, L.; Xu, P.; Yang, Z. MKL1 inhibits cell cycle progression through p21 in podocytes. BMC Mol. Biol. 2015, 16, 1. [Google Scholar] [CrossRef][Green Version]
- Rozycki, M.; Bialik, J.F.; Speight, P.; Dan, Q.; Knudsen, T.E.; Szeto, S.G.; Yuen, D.A.; Szaszi, K.; Pedersen, S.F.; Kapus, A. Myocardin-related Transcription Factor Regulates Nox4 Protein Expression: Linking cytoskeletal organization to redox state. J. Biol. Chem. 2016, 291, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wu, X.; Xu, H.; Yu, L.; Zhang, X.; Li, L.; Jin, J.; Zhang, T.; Xu, Y. Myocardin-related transcription factor A (MRTF-A) contributes to acute kidney injury by regulating macrophage ROS production. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3109–3121. [Google Scholar] [CrossRef]
- Yoshino, J.; Monkawa, T.; Tsuji, M.; Inukai, M.; Itoh, H.; Hayashi, M. Snail1 is involved in the renal epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2007, 362, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Grande, M.T.; Sanchez-Laorden, B.; Lopez-Blau, C.; De Frutos, C.A.; Boutet, A.; Arevalo, M.; Rowe, R.G.; Weiss, S.J.; Lopez-Novoa, J.M.; Nieto, M.A. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat. Med. 2015, 21, 989–997. [Google Scholar] [CrossRef]
- Tang, W.B.; Zheng, L.; Yan, R.; Yang, J.; Ning, J.; Peng, L.; Zhou, Q.; Chen, L. miR302a-3p May Modulate Renal Epithelial-Mesenchymal Transition in Diabetic Kidney Disease by Targeting ZEB1. Nephron 2018, 138, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Davis-Dusenbery, B.N.; Chan, M.C.; Reno, K.E.; Weisman, A.S.; Layne, M.D.; Lagna, G.; Hata, A. down-regulation of Kruppel-like factor-4 (KLF4) by microRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-beta and bone morphogenetic protein 4. J. Biol. Chem. 2011, 286, 28097–28110. [Google Scholar] [CrossRef]
- Li, C.X.; Talele, N.P.; Boo, S.; Koehler, A.; Knee-Walden, E.; Balestrini, J.L.; Speight, P.; Kapus, A.; Hinz, B. MicroRNA-21 preserves the fibrotic mechanical memory of mesenchymal stem cells. Nat. Mater. 2017, 16, 379–389. [Google Scholar] [CrossRef]
- Hayashi, K.; Murai, T.; Oikawa, H.; Masuda, T.; Kimura, K.; Muehlich, S.; Prywes, R.; Morita, T. A novel inhibitory mechanism of MRTF-A/B on the ICAM-1 gene expression in vascular endothelial cells. Sci. Rep. 2015, 5, 10627. [Google Scholar] [CrossRef]
- Yu, L.; Fang, F.; Dai, X.; Xu, H.; Qi, X.; Fang, M.; Xu, Y. MKL1 defines the H3K4Me3 landscape for NF-kappaB dependent inflammatory response. Sci. Rep. 2017, 7, 191. [Google Scholar] [CrossRef]
- Weng, X.; Yu, L.; Liang, P.; Li, L.; Dai, X.; Zhou, B.; Wu, X.; Xu, H.; Fang, M.; Chen, Q.; et al. A crosstalk between chromatin remodeling and histone H3K4 methyltransferase complexes in endothelial cells regulates angiotensin II-induced cardiac hypertrophy. J. Mol. Cell Cardiol. 2015, 82, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Lockman, K.; Taylor, J.M.; Mack, C.P. The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ. Res. 2007, 101, e115–e123. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Yang, G.; Zhang, X.; Wang, P.; Weng, X.; Yang, Y.; Li, Z.; Fang, M.; Xu, Y.; Sun, A.; et al. Megakaryocytic Leukemia 1 Bridges Epigenetic Activation of NADPH Oxidase in Macrophages to Cardiac Ischemia-Reperfusion Injury. Circulation 2018, 138, 2820–2836. [Google Scholar] [CrossRef] [PubMed]
- Menendez, M.T.; Ong, E.C.; Shepherd, B.T.; Muthukumar, V.; Silasi-Mansat, R.; Lupu, F.; Griffin, C.T. BRG1 (Brahma-Related Gene 1) Promotes Endothelial Mrtf Transcription to Establish Embryonic Capillary Integrity. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1674–1682. [Google Scholar] [CrossRef] [PubMed]
- Malicki, J.J.; Johnson, C.A. The Cilium: Cellular Antenna and Central Processing Unit. Trends Cell. Biol. 2017, 27, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Dynlacht, B.D. The regulation of cilium assembly and disassembly in development and disease. Development 2018, 145. [Google Scholar] [CrossRef]
- Praetorius, H.A.; Spring, K.R. The renal cell primary cilium functions as a flow sensor. Curr. Opin. Nephrol. Hypertens. 2003, 12, 517–520. [Google Scholar] [CrossRef]
- Praetorius, H.A. The primary cilium as sensor of fluid flow: New building blocks to the model. A review in the theme: Cell signaling: Proteins, pathways and mechanisms. Am. J. Physiol. Cell. Physiol. 2015, 308, C198–C208. [Google Scholar] [CrossRef]
- Ke, Y.N.; Yang, W.X. Primary cilium: An elaborate structure that blocks cell division? Gene 2014, 547, 175–185. [Google Scholar] [CrossRef]
- Oh, E.C.; Vasanth, S.; Katsanis, N. Metabolic regulation and energy homeostasis through the primary Cilium. Cell Metab. 2015, 21, 21–31. [Google Scholar] [CrossRef]
- Wheway, G.; Nazlamova, L.; Hancock, J.T. Signaling through the Primary Cilium. Front. Cell Dev. Biol. 2018, 6, 8. [Google Scholar] [CrossRef]
- Braun, D.A.; Hildebrandt, F. Ciliopathies. Cold Spring Harb. Perspect. Biol. 2017, 9, a028191. [Google Scholar] [CrossRef]
- Menezes, L.F.; Germino, G.G. The pathobiology of polycystic kidney disease from a metabolic viewpoint. Nat. Rev. Nephrol. 2019, 15, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Douguet, D.; Patel, A.; Honore, E. Structure and function of polycystins: Insights into polycystic kidney disease. Nat. Rev. Nephrol. 2019, 15, 412–422. [Google Scholar] [CrossRef]
- McConnachie, D.J.; Stow, J.L.; Mallett, A.J. Ciliopathies and the Kidney: A Review. Am. J. Kidney Dis. 2021, 77, 410–419. [Google Scholar] [CrossRef]
- Kobayashi, T.; Dynlacht, B.D. Regulating the transition from centriole to basal body. J. Cell. Biol. 2011, 193, 435–444. [Google Scholar] [CrossRef]
- Vertii, A.; Hung, H.F.; Hehnly, H.; Doxsey, S. Human basal body basics. Cilia 2016, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, I.; Dynlacht, B.D. Cilium assembly and disassembly. Nat. Cell Biol. 2016, 18, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Breslow, D.K.; Holland, A.J. Mechanism and Regulation of Centriole and Cilium Biogenesis. Annu. Rev. Biochem. 2019, 88, 691–724. [Google Scholar] [CrossRef]
- Li, S.; Chang, S.; Qi, X.; Richardson, J.A.; Olson, E.N. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol. Cell. Biol. 2006, 26, 5797–5808. [Google Scholar] [CrossRef]
- Sun, Y.; Boyd, K.; Xu, W.; Ma, J.; Jackson, C.W.; Fu, A.; Shillingford, J.M.; Robinson, G.W.; Hennighausen, L.; Hitzler, J.K.; et al. Acute myeloid leukemia-associated Mkl1 (Mrtf-a) is a key regulator of mammary gland function. Mol. Cell. Biol. 2006, 26, 5809–5826. [Google Scholar] [CrossRef]
- Oh, J.; Richardson, J.A.; Olson, E.N. Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc. Natl. Acad. Sci. USA 2005, 102, 15122–15127. [Google Scholar] [CrossRef]
- Guo, B.; Lyu, Q.; Slivano, O.J.; Dirkx, R.; Christie, C.K.; Czyzyk, J.; Hezel, A.F.; Gharavi, A.G.; Small, E.M.; Miano, J.M. Serum Response Factor Is Essential for Maintenance of Podocyte Structure and Function. J. Am. Soc. Nephrol. 2018, 29, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Pakshir, P.; Hinz, B. The big five in fibrosis: Macrophages, myofibroblasts, matrix, mechanics, and miscommunication. Matrix Biol. 2018, 68–69, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From mechanisms to medicines. Nature 2020, 587, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, B.D. Mechanisms of Renal Fibrosis. Annu. Rev. Physiol. 2018, 80, 309–326. [Google Scholar] [CrossRef]
- Gewin, L.S. Renal fibrosis: Primacy of the proximal tubule. Matrix Biol. 2018, 68–69, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Grgic, I.; Duffield, J.S.; Humphreys, B.D. The origin of interstitial myofibroblasts in chronic kidney disease. Pediatr. Nephrol. 2012, 27, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, N.; Duffield, J.S. Myofibroblasts in Fibrotic Kidneys. Curr. Pathobiol. Rep. 2013, 1. [Google Scholar] [CrossRef][Green Version]
- Lovisa, S.; Zeisberg, M.; Kalluri, R. Partial Epithelial-to-Mesenchymal Transition and Other New Mechanisms of Kidney Fibrosis. Trends Endocrinol. Metab. 2016, 27, 681–695. [Google Scholar] [CrossRef]
- Pakshir, P.; Noskovicova, N.; Lodyga, M.; Son, D.O.; Schuster, R.; Goodwin, A.; Karvonen, H.; Hinz, B. The myofibroblast at a glance. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef]
- Grgic, I.; Campanholle, G.; Bijol, V.; Wang, C.; Sabbisetti, V.S.; Ichimura, T.; Humphreys, B.D.; Bonventre, J.V. Targeted proximal tubule injury triggers interstitial fibrosis and glomerulosclerosis. Kidney Int. 2012, 82, 172–183. [Google Scholar] [CrossRef]
- Pattaro, C.; Kottgen, A.; Teumer, A.; Garnaas, M.; Boger, C.A.; Fuchsberger, C.; Olden, M.; Chen, M.H.; Tin, A.; Taliun, D.; et al. Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet. 2012, 8, e1002584. [Google Scholar] [CrossRef]
- Okada, Y.; Sim, X.; Go, M.J.; Wu, J.Y.; Gu, D.; Takeuchi, F.; Takahashi, A.; Maeda, S.; Tsunoda, T.; Chen, P.; et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat. Genet. 2012, 44, 904–909. [Google Scholar] [CrossRef]
- Pattaro, C.; Teumer, A.; Gorski, M.; Chu, A.Y.; Li, M.; Mijatovic, V.; Garnaas, M.; Tin, A.; Sorice, R.; Li, Y.; et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat. Commun. 2016, 7, 10023. [Google Scholar] [CrossRef]
- Wuttke, M.; Kottgen, A. Insights into kidney diseases from genome-wide association studies. Nat. Rev. Nephrol. 2016, 12, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Hishida, A.; Nakatochi, M.; Akiyama, M.; Kamatani, Y.; Nishiyama, T.; Ito, H.; Oze, I.; Nishida, Y.; Hara, M.; Takashima, N.; et al. Japan Multi-Institutional Collaborative Cohort Study, G. Genome-Wide Association Study of Renal Function Traits: Results from the Japan Multi-Institutional Collaborative Cohort Study. Am. J. Nephrol. 2018, 47, 304–316. [Google Scholar] [CrossRef]
- Qiu, C.; Huang, S.; Park, J.; Park, Y.; Ko, Y.A.; Seasock, M.J.; Bryer, J.S.; Xu, X.X.; Song, W.C.; Palmer, M.; et al. Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat. Med. 2018, 24, 1721–1731. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.M.; Ng, Y.Y.; Hill, P.A.; Nikolic-Paterson, D.J.; Mu, W.; Atkins, R.C.; Lan, H.Y. Transforming growth factor-beta regulates tubular epithelial-myofibroblast transdifferentiation in vitro. Kidney Int. 1999, 56, 1455–1467. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Y. Dissection of key events in tubular epithelial to myofibroblast transition and its implications in renal interstitial fibrosis. Am. J. Pathol. 2001, 159, 1465–1475. [Google Scholar] [CrossRef]
- Masszi, A.; Fan, L.; Rosivall, L.; McCulloch, C.A.; Rotstein, O.D.; Mucsi, I.; Kapus, A. Integrity of cell-cell contacts is a critical regulator of TGF-beta 1-induced epithelial-to-myofibroblast transition: Role for beta-catenin. Am. J. Pathol. 2004, 165, 1955–1967. [Google Scholar] [CrossRef]
- Speight, P.; Nakano, H.; Kelley, T.J.; Hinz, B.; Kapus, A. Differential topical susceptibility to TGFbeta in intact and injured regions of the epithelium: Key role in myofibroblast transition. Mol. Biol. Cell 2013, 24, 3326–3336. [Google Scholar] [CrossRef]
- Dan, Q.; Shi, Y.; Rabani, R.; Venugopal, S.; Xiao, J.; Anwer, S.; Ding, M.; Speight, P.; Pan, W.; Alexander, R.T.; et al. Claudin-2 suppresses GEF-H1, RHOA, and MRTF, thereby impacting proliferation and profibrotic phenotype of tubular cells. J. Biol. Chem. 2019, 294, 15446–15465. [Google Scholar] [CrossRef] [PubMed]
- Gomez, E.W.; Chen, Q.K.; Gjorevski, N.; Nelson, C.M. Tissue geometry patterns epithelial-mesenchymal transition via intercellular mechanotransduction. J. Cell Biochem. 2010, 110, 44–51. [Google Scholar] [CrossRef]
- O’Connor, J.W.; Gomez, E.W. Cell adhesion and shape regulate TGF-beta1-induced epithelial-myofibroblast transition via MRTF-A signaling. PLoS ONE 2013, 8, e83188. [Google Scholar] [CrossRef]
- O’Connor, J.W.; Riley, P.N.; Nalluri, S.M.; Ashar, P.K.; Gomez, E.W. Matrix Rigidity Mediates TGFbeta1-Induced Epithelial-Myofibroblast Transition by Controlling Cytoskeletal Organization and MRTF-A Localization. J. Cell. Physiol. 2015, 230, 1829–1839. [Google Scholar] [CrossRef]
- Hinson, J.S.; Medlin, M.D.; Lockman, K.; Taylor, J.M.; Mack, C.P. Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H1170–H1180. [Google Scholar] [CrossRef]
- Poncelet, A.C.; Schnaper, H.W.; Tan, R.; Liu, Y.; Runyan, C.E. Cell phenotype-specific down-regulation of Smad3 involves decreased gene activation as well as protein degradation. J. Biol. Chem. 2007, 282, 15534–15540. [Google Scholar] [CrossRef]
- Iwano, M.; Plieth, D.; Danoff, T.M.; Xue, C.; Okada, H.; Neilson, E.G. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J. Clin. Investig. 2002, 110, 341–350. [Google Scholar] [CrossRef]
- Kalluri, R.; Neilson, E.G. Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Investig. 2003, 112, 1776–1784. [Google Scholar] [CrossRef]
- Duffield, J.S.; Humphreys, B.D. Origin of new cells in the adult kidney: Results from genetic labeling techniques. Kidney Int. 2011, 79, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Lovisa, S.; LeBleu, V.S.; Tampe, B.; Sugimoto, H.; Vadnagara, K.; Carstens, J.L.; Wu, C.C.; Hagos, Y.; Burckhardt, B.C.; Pentcheva-Hoang, T.; et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat. Med. 2015, 21, 998–1009. [Google Scholar] [CrossRef]
- Sheng, L.; Zhuang, S. New Insights Into the Role and Mechanism of Partial Epithelial-Mesenchymal Transition in Kidney Fibrosis. Front. Physiol. 2020, 11, 569322. [Google Scholar] [CrossRef]
- Fukuda, K.; Yoshitomi, K.; Yanagida, T.; Tokumoto, M.; Hirakata, H. Quantification of TGF-beta1 mRNA along rat nephron in obstructive nephropathy. Am. J. Physiol. Renal Physiol. 2001, 281, F513–F521. [Google Scholar] [CrossRef]
- Yang, L.; Besschetnova, T.Y.; Brooks, C.R.; Shah, J.V.; Bonventre, J.V. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat. Med. 2010, 16, 535–543. [Google Scholar] [CrossRef]
- Chen, Y.T.; Chang, F.C.; Wu, C.F.; Chou, Y.H.; Hsu, H.L.; Chiang, W.C.; Shen, J.; Chen, Y.M.; Wu, K.D.; Tsai, T.J.; et al. Platelet-derived growth factor receptor signaling activates pericyte-myofibroblast transition in obstructive and post-ischemic kidney fibrosis. Kidney Int. 2011, 80, 1170–1181. [Google Scholar] [CrossRef]
- Fabian, S.L.; Penchev, R.R.; St-Jacques, B.; Rao, A.N.; Sipila, P.; West, K.A.; McMahon, A.P.; Humphreys, B.D. Hedgehog-Gli pathway activation during kidney fibrosis. Am. J. Pathol. 2012, 180, 1441–1453. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.F.; Chiang, W.C.; Lai, C.F.; Chang, F.C.; Chen, Y.T.; Chou, Y.H.; Wu, T.H.; Linn, G.R.; Ling, H.; Wu, K.D.; et al. Transforming growth factor beta-1 stimulates profibrotic epithelial signaling to activate pericyte-myofibroblast transition in obstructive kidney fibrosis. Am. J. Pathol. 2013, 182, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Szeto, S.G.; Narimatsu, M.; Lu, M.; He, X.; Sidiqi, A.M.; Tolosa, M.F.; Chan, L.; De Freitas, K.; Bialik, J.F.; Majumder, S.; et al. YAP/TAZ Are Mechanoregulators of TGF-beta-Smad Signaling and Renal Fibrogenesis. J. Am. Soc. Nephrol. 2016, 27, 3117–3128. [Google Scholar] [CrossRef]
- Patel, S.; Tang, J.; Overstreet, J.M.; Anorga, S.; Lian, F.; Arnouk, A.; Goldschmeding, R.; Higgins, P.J.; Samarakoon, R. Rac-GTPase promotes fibrotic TGF-beta1 signaling and chronic kidney disease via EGFR, p53, and Hippo/YAP/TAZ pathways. FASEB J. 2019, 33, 9797–9810. [Google Scholar] [CrossRef]
- Sandbo, N.; Kregel, S.; Taurin, S.; Bhorade, S.; Dulin, N.O. Critical role of serum response factor in pulmonary myofibroblast differentiation induced by TGF-beta. Am. J. Respir. Cell. Mol. Biol. 2009, 41, 332–338. [Google Scholar] [CrossRef]
- Ni, J.; Dong, Z.; Han, W.; Kondrikov, D.; Su, Y. The role of RhoA and cytoskeleton in myofibroblast transformation in hyperoxic lung fibrosis. Free Radic. Biol. Med. 2013, 61, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Small, E.M.; Thatcher, J.E.; Sutherland, L.B.; Kinoshita, H.; Gerard, R.D.; Richardson, J.A.; Dimaio, J.M.; Sadek, H.; Kuwahara, K.; Olson, E.N. Myocardin-related transcription factor-a controls myofibroblast activation and fibrosis in response to myocardial infarction. Circ. Res. 2010, 107, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Sakai, N.; Chun, J.; Duffield, J.S.; Wada, T.; Luster, A.D.; Tager, A.M. LPA1-induced cytoskeleton reorganization drives fibrosis through CTGF-dependent fibroblast proliferation. FASEB J. 2013, 27, 1830–1846. [Google Scholar] [CrossRef]
- Velasquez, L.S.; Sutherland, L.B.; Liu, Z.; Grinnell, F.; Kamm, K.E.; Schneider, J.W.; Olson, E.N.; Small, E.M. Activation of MRTF-A-dependent gene expression with a small molecule promotes myofibroblast differentiation and wound healing. Proc. Natl. Acad. Sci. USA 2013, 110, 16850–16855. [Google Scholar] [CrossRef] [PubMed]
- Haak, A.J.; Tsou, P.S.; Amin, M.A.; Ruth, J.H.; Campbell, P.; Fox, D.A.; Khanna, D.; Larsen, S.D.; Neubig, R.R. Targeting the myofibroblast genetic switch: Inhibitors of myocardin-related transcription factor/serum response factor-regulated gene transcription prevent fibrosis in a murine model of skin injury. J. Pharmacol. Exp. Ther. 2014, 349, 480–486. [Google Scholar] [CrossRef]
- Yu-Wai-Man, C.; Treisman, R.; Bailly, M.; Khaw, P.T. The role of the MRTF-A/SRF pathway in ocular fibrosis. Invest. Ophthalmol. Vis. Sci. 2014, 55, 4560–4567. [Google Scholar] [CrossRef]
- Tian, W.; Hao, C.; Fan, Z.; Weng, X.; Qin, H.; Wu, X.; Fang, M.; Chen, Q.; Shen, A.; Xu, Y. Myocardin related transcription factor A programs epigenetic activation of hepatic stellate cells. J. Hepatol. 2015, 62, 165–174. [Google Scholar] [CrossRef]
- Lauriol, J.; Keith, K.; Jaffre, F.; Couvillon, A.; Saci, A.; Goonasekera, S.A.; McCarthy, J.R.; Kessinger, C.W.; Wang, J.; Ke, Q.; et al. RhoA signaling in cardiomyocytes protects against stress-induced heart failure but facilitates cardiac fibrosis. Sci. Signal. 2014, 7, ra100. [Google Scholar] [CrossRef]
- Rahaman, S.O.; Grove, L.M.; Paruchuri, S.; Southern, B.D.; Abraham, S.; Niese, K.A.; Scheraga, R.G.; Ghosh, S.; Thodeti, C.K.; Zhang, D.X.; et al. TRPV4 mediates myofibroblast differentiation and pulmonary fibrosis in mice. J. Clin. Investig. 2014, 124, 5225–5238. [Google Scholar] [CrossRef] [PubMed]
- Sisson, T.H.; Ajayi, I.O.; Subbotina, N.; Dodi, A.E.; Rodansky, E.S.; Chibucos, L.N.; Kim, K.K.; Keshamouni, V.G.; White, E.S.; Zhou, Y.; et al. Inhibition of myocardin-related transcription factor/serum response factor signaling decreases lung fibrosis and promotes mesenchymal cell apoptosis. Am. J. Pathol. 2015, 185, 969–986. [Google Scholar] [CrossRef] [PubMed]
- Shiwen, X.; Stratton, R.; Nikitorowicz-Buniak, J.; Ahmed-Abdi, B.; Ponticos, M.; Denton, C.; Abraham, D.; Takahashi, A.; Suki, B.; Layne, M.D.; et al. A Role of Myocardin Related Transcription Factor-A (MRTF-A) in Scleroderma Related Fibrosis. PLoS ONE 2015, 10, e0126015. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Fan, Z.; Li, J.; Hao, C.; Li, M.; Xu, H.; Wu, X.; Zhou, B.; Zhang, L.; Fang, M.; et al. Myocardin-related transcription factor A (MRTF-A) plays an essential role in hepatic stellate cell activation by epigenetically modulating TGF-beta signaling. Int, J. Biochem. Cell Biol. 2016, 71, 35–43. [Google Scholar] [CrossRef]
- Yu-Wai-Man, C.; Tagalakis, A.D.; Manunta, M.D.; Hart, S.L.; Khaw, P.T. Receptor-targeted liposome-peptide-siRNA nanoparticles represent an efficient delivery system for MRTF silencing in conjunctival fibrosis. Sci. Rep. 2016, 6, 21881. [Google Scholar] [CrossRef]
- Yokota, S.; Chosa, N.; Kyakumoto, S.; Kimura, H.; Ibi, M.; Kamo, M.; Satoh, K.; Ishisaki, A. ROCK/actin/MRTF signaling promotes the fibrogenic phenotype of fibroblast-like synoviocytes derived from the temporomandibular joint. Int. J. Mol. Med. 2017, 39, 799–808. [Google Scholar] [CrossRef]
- Yu-Wai-Man, C.; Spencer-Dene, B.; Lee, R.M.H.; Hutchings, K.; Lisabeth, E.M.; Treisman, R.; Bailly, M.; Larsen, S.D.; Neubig, R.R.; Khaw, P.T. Local delivery of novel MRTF/SRF inhibitors prevents scar tissue formation in a preclinical model of fibrosis. Sci. Rep. 2017, 7, 518. [Google Scholar] [CrossRef]
- Giehl, K.; Keller, C.; Muehlich, S.; Goppelt-Struebe, M. Actin-mediated gene expression depends on RhoA and Rac1 signaling in proximal tubular epithelial cells. PLoS ONE 2015, 10, e0121589. [Google Scholar] [CrossRef]
- Wang, Y.; Jia, L.; Hu, Z.; Entman, M.L.; Mitch, W.E.; Wang, Y. AMP-activated protein kinase/myocardin-related transcription factor-A signaling regulates fibroblast activation and renal fibrosis. Kidney Int. 2018, 93, 81–94. [Google Scholar] [CrossRef]
- Liang, M.; Yu, M.; Xia, R.; Song, K.; Wang, J.; Luo, J.; Chen, G.; Cheng, J. Yap/Taz Deletion in Gli(+) Cell-Derived Myofibroblasts Attenuates Fibrosis. J. Am. Soc. Nephrol. 2017, 28, 3278–3290. [Google Scholar] [CrossRef]
- Anorga, S.; Overstreet, J.M.; Falke, L.L.; Tang, J.; Goldschmeding, R.G.; Higgins, P.J.; Samarakoon, R. Deregulation of Hippo-TAZ pathway during renal injury confers a fibrotic maladaptive phenotype. FASEB J. 2018, 32, 2644–2657. [Google Scholar] [CrossRef]
- Song, X.; Di Giovanni, V.; He, N.; Wang, K.; Ingram, A.; Rosenblum, N.D.; Pei, Y. Systems biology of autosomal dominant polycystic kidney disease (ADPKD): Computational identification of gene expression pathways and integrated regulatory networks. Hum. Mol. Genet. 2009, 18, 2328–2343. [Google Scholar] [CrossRef]
- Cai, J.; Song, X.; Wang, W.; Watnick, T.; Pei, Y.; Qian, F.; Pan, D. A RhoA-YAP-c-Myc signaling axis promotes the development of polycystic kidney disease. Genes Dev. 2018, 32, 781–793. [Google Scholar] [CrossRef]
- Streets, A.J.; Prosseda, P.P.; Ong, A.C. Polycystin-1 regulates ARHGAP35-dependent centrosomal RhoA activation and ROCK signaling. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Qian, Q.; Hunter, L.W.; Du, H.; Ren, Q.; Han, Y.; Sieck, G.C. Pkd2+/− vascular smooth muscles develop exaggerated vasocontraction in response to phenylephrine stimulation. J. Am. Soc. Nephrol. 2007, 18, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wang, X.; Wu, J.; Qian, Q. Phenylephrine induces elevated RhoA activation and smooth muscle alpha-actin expression in Pkd2+/- vascular smooth muscle cells. Hypertens. Res. 2010, 33, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Chadha, V.; Warady, B.A. Epidemiology of pediatric chronic kidney disease. Adv. Chronic Kidney Dis. 2005, 12, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Klahr, S.; Morrissey, J. Obstructive nephropathy and renal fibrosis. Am. J. Physiol. Renal Physiol. 2002, 283, F861–F875. [Google Scholar] [CrossRef] [PubMed]
- Holterman, C.E.; Read, N.C.; Kennedy, C.R. Nox and renal disease. Clin. Sci. 2015, 128, 465–481. [Google Scholar] [CrossRef]
- Mia, S.; Federico, G.; Feger, M.; Pakladok, T.; Meissner, A.; Voelkl, J.; Groene, H.J.; Alesutan, I.; Lang, F. Impact of AMP-Activated Protein Kinase alpha1 Deficiency on Tissue Injury following Unilateral Ureteral Obstruction. PLoS ONE 2015, 10, e0135235. [Google Scholar] [CrossRef]
- Scholz, H.; Boivin, F.J.; Schmidt-Ott, K.M.; Bachmann, S.; Eckardt, K.U.; Scholl, U.I.; Persson, P.B. Kidney physiology and susceptibility to acute kidney injury: Implications for renoprotection. Nat. Rev. Nephrol. 2021. [Google Scholar] [CrossRef]
- Liangos, O. Drugs and AKI. Minerva Urol. Nefrol. 2012, 64, 51–62. [Google Scholar]
- Morrell, E.D.; Kellum, J.A.; Pastor-Soler, N.M.; Hallows, K.R. Septic acute kidney injury: Molecular mechanisms and the importance of stratification and targeting therapy. Crit. Care 2014, 18, 501. [Google Scholar] [CrossRef]
- Xu, Z.; Tang, Y.; Huang, Q.; Fu, S.; Li, X.; Lin, B.; Xu, A.; Chen, J. Systematic review and subgroup analysis of the incidence of acute kidney injury (AKI) in patients with COVID-19. BMC Nephrol. 2021, 22, 52. [Google Scholar] [CrossRef]
- Coca, S.G.; Singanamala, S.; Parikh, C.R. Chronic kidney disease after acute kidney injury: A systematic review and meta-analysis. Kidney Int. 2012, 81, 442–448. [Google Scholar] [CrossRef] [PubMed]
- You, Y.H.; Okada, S.; Ly, S.; Jandeleit-Dahm, K.; Barit, D.; Namikoshi, T.; Sharma, K. Role of Nox2 in diabetic kidney disease. Am. J. Physiol. Renal Physiol. 2013, 304, F840–F848. [Google Scholar] [CrossRef]
- Yu, L.; Weng, X.; Liang, P.; Dai, X.; Wu, X.; Xu, H.; Fang, M.; Fang, F.; Xu, Y. MRTF-A mediates LPS-induced pro-inflammatory transcription by interacting with the COMPASS complex. J. Cell Sci. 2014, 127, 4645–4657. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Vogel, V. Spatial confinement downsizes the inflammatory response of macrophages. Nat. Mater. 2018, 17, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Chapin, H.C.; Caplan, M.J. The cell biology of polycystic kidney disease. J. Cell. Biol. 2010, 191, 701–710. [Google Scholar] [CrossRef]
- Suwabe, T.; Shukoor, S.; Chamberlain, A.M.; Killian, J.M.; King, B.F.; Edwards, M.; Senum, S.R.; Madsen, C.D.; Chebib, F.T.; Hogan, M.C.; et al. Epidemiology of Autosomal Dominant Polycystic Kidney Disease in Olmsted County. Clin. J. Am. Soc. Nephrol. 2020, 15, 69–79. [Google Scholar] [CrossRef]
- Harris, P.C.; Torres, V.E. Polycystic kidney disease. Annu. Rev. Med. 2009, 60, 321–337. [Google Scholar] [CrossRef] [PubMed]
- Malekshahabi, T.; Khoshdel Rad, N.; Serra, A.L.; Moghadasali, R. Autosomal dominant polycystic kidney disease: Disrupted pathways and potential therapeutic interventions. J. Cell. Physiol. 2019, 234, 12451–12470. [Google Scholar] [CrossRef]
- Song, C.J.; Zimmerman, K.A.; Henke, S.J.; Yoder, B.K. Inflammation and Fibrosis in Polycystic Kidney Disease. Results Probl. Cell Differ. 2017, 60, 323–344. [Google Scholar]
- Xue, C.; Mei, C.L. Polycystic Kidney Disease and Renal Fibrosis. Adv. Exp. Med. Biol 2019, 1165, 81–100. [Google Scholar]
- Formica, C.; Peters, D.J.M. Molecular pathways involved in injury-repair and ADPKD progression. Cell Signal. 2020, 72, 109648. [Google Scholar] [CrossRef] [PubMed]
- Qian, Q.; Hunter, L.W.; Li, M.; Marin-Padilla, M.; Prakash, Y.S.; Somlo, S.; Harris, P.C.; Torres, V.E.; Sieck, G.C. Pkd2 haploinsufficiency alters intracellular calcium regulation in vascular smooth muscle cells. Hum. Mol. Genet. 2003, 12, 1875–1880. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.M.; Nissaisorakarn, P.; Husain, I.; Jim, B. Proteinuric Kidney Diseases: A Podocyte’s Slit Diaphragm and Cytoskeleton Approach. Front. Med. 2018, 5, 221. [Google Scholar] [CrossRef]
- Lim, B.J.; Yang, J.W.; Do, W.S.; Fogo, A.B. Pathogenesis of Focal Segmental Glomerulosclerosis. J. Pathol. Transl. Med. 2016, 50, 405–410. [Google Scholar] [CrossRef]
- Evelyn, C.R.; Wade, S.M.; Wang, Q.; Wu, M.; Iniguez-Lluhi, J.A.; Merajver, S.D.; Neubig, R.R. CCG-1423: A small-molecule inhibitor of RhoA transcriptional signaling. Mol. Cancer Ther. 2007, 6, 2249–2260. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Watanabe, B.; Nakagawa, Y.; Minami, S.; Morita, T. RPEL proteins are the molecular targets for CCG-1423, an inhibitor of Rho signaling. PLoS ONE 2014, 9, e89016. [Google Scholar] [CrossRef]
- Lisabeth, E.M.; Kahl, D.; Gopallawa, I.; Haynes, S.E.; Misek, S.A.; Campbell, P.L.; Dexheimer, T.S.; Khanna, D.; Fox, D.A.; Jin, X.; et al. Identification of Pirin as a Molecular Target of the CCG-1423/CCG-203971 Series of Antifibrotic and Antimetastatic Compounds. ACS Pharmacol. Transl. Sci. 2019, 2, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Evelyn, C.R.; Bell, J.L.; Ryu, J.G.; Wade, S.M.; Kocab, A.; Harzdorf, N.L.; Showalter, H.D.; Neubig, R.R.; Larsen, S.D. Design, synthesis and prostate cancer cell-based studies of analogs of the Rho/MKL1 transcriptional pathway inhibitor, CCG-1423. Bioorg. Med. Chem. Lett. 2010, 20, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.L.; Haak, A.J.; Wade, S.M.; Kirchhoff, P.D.; Neubig, R.R.; Larsen, S.D. Optimization of novel nipecotic bis(amide) inhibitors of the Rho/MKL1/SRF transcriptional pathway as potential anti-metastasis agents. Bioorg. Med. Chem. Lett. 2013, 23, 3826–3832. [Google Scholar] [CrossRef]
- Hutchings, K.M.; Lisabeth, E.M.; Rajeswaran, W.; Wilson, M.W.; Sorenson, R.J.; Campbell, P.L.; Ruth, J.H.; Amin, A.; Tsou, P.S.; Leipprandt, J.R.; et al. Pharmacokinetic optimitzation of CCG-203971: Novel inhibitors of the Rho/MRTF/SRF transcriptional pathway as potential antifibrotic therapeutics for systemic scleroderma. Bioorg. Med. Chem. Lett. 2017, 27, 1744–1749. [Google Scholar] [CrossRef]
- Kahl, D.J.; Hutchings, K.M.; Lisabeth, E.M.; Haak, A.J.; Leipprandt, J.R.; Dexheimer, T.; Khanna, D.; Tsou, P.S.; Campbell, P.L.; Fox, D.A.; et al. 5-Aryl-1,3,4-oxadiazol-2-ylthioalkanoic Acids: A Highly Potent New Class of Inhibitors of Rho/Myocardin-Related Transcription Factor (MRTF)/Serum Response Factor (SRF)-Mediated Gene Transcription as Potential Antifibrotic Agents for Scleroderma. J. Med. Chem. 2019, 62, 4350–4369. [Google Scholar] [CrossRef]
- Haak, A.J.; Appleton, K.M.; Lisabeth, E.M.; Misek, S.A.; Ji, Y.; Wade, S.M.; Bell, J.L.; Rockwell, C.E.; Airik, M.; Krook, M.A.; et al. Pharmacological Inhibition of Myocardin-related Transcription Factor Pathway Blocks Lung Metastases of RhoC-Overexpressing Melanoma. Mol. Cancer Ther. 2017, 16, 193–204. [Google Scholar] [CrossRef] [PubMed]
| Enzyme | Site Modified | Domain Modified | Binding Site | Effect on MRTF Localization/Stability | Effect on MRTF Transcriptional Activity | Reference |
|---|---|---|---|---|---|---|
| Phosphorylation | ||||||
| ERK | S98 | between RPEL 1 and 2 | RPEL1 | Promotes MRTF nuclear import/prevents nuclear | + | [46] |
| ERK | S33 | N-term of RPEL1 | N/D | Promotes MRTF nuclear export | N/D | [46] |
| ERK | S454 | Between SAP and LZ | N/D | Promotes G-actin binding and MRTF export | N/D | [47] |
| P38 | N/D | N/D | N/D | + | ||
| MK2 | S351/371 | Between Q and SAP | N/D | N/D | No effect | [48] |
| ROK | S82/T92 | RPEL 1 | + | |||
| GSK3β | N/D | N/D | Binds MRTF via Smad3-dependent mechanism | Promotes MRTF ubiquitin-mediated degradation | − | [49] |
| Ubiquitination | ||||||
| Ubiquitinase | ||||||
| CHIP | N/D | N/D | TAD | Promotes MRTF ubiquitin-mediated degradation | − | [50] |
| Acetylation | ||||||
| Histone Acetyl Transferase (HAT) | ||||||
| P300 | K235/237/253/255 | N-term | Binds myocardin C-term | N/D | −/+ | [51] |
| Lysines are conserved between myocardin and MRTF | B1 | Binds MRTFA C-term (TAD) | N/D | + | [52] | |
| pCAF | Lysines are conserved between myocardin and MRTF | B1 | N/D | Promotes MRTF nuclear translocation | + | [53] |
| Histone Deacetyl Transferase | ||||||
| HDAC5 | N/D | N/D | Binds MRTF (domain undescribed) | Prevents MRTF-A nuclear translocation | − | [54,55] |
| HDAC6 | N/D | N/D | Binds MRTF (domain undescribed) | Regulates MRTF-A total protein (supresses) | − | [56] |
| SIRT1 | Lysines are conserved between myocardin and MRTF | B1 | Binds MRTF (domain undescribed) | N/D | + | [57] |
| SUMOylation | ||||||
| UBC9 | K499, 576, and 624 | C-term region (C-term LZ?) | N/D | No effect | − | [58] |
| SUMO-1/PIAS1 | K445 | C-term region (C-term LZ?) | 385–586 aa (C-term after SAP?) | No effect | + | [59] |
| Transcription Factor | Domain Bound | Effect on MRTF Localization/Stability | Effect on MRTF Transcriptional Activity | Reference |
|---|---|---|---|---|
| SRF | B1/Q | N/D | + | [37] |
| FHL2 | N-term (RPEL/B1-3/Q) | Increased myocardin protein levels | + | [60,61] |
| N-term (RPEL/B1-3/Q) | Increased MRTF-A protein levels | + | ||
| B1/Q | Decreased MRTF-B nuclear localization | − | ||
| YAP/TAZ | C-term (PPxY) | Decreased MRTF nuclear accumulation | −/+ | [22] |
| Smad3 | B1 | Promotes MRTF degradation | non-CArg = +; CArG = − | [19,20] |
| SP1 | N/D | N/D | + | [62] |
| NFκB/p65 | B1/Q | N/D | −/+ | [63,64] |
| Stat5 | Q | N/D | non-CArG/ICAM-1 = + | [65] |
| Stat3 | N/D | N/D | + | [66] |
| Epigenetic Modifier | Modification Type | Gene | Effect on Gene Activity | Reference |
|---|---|---|---|---|
| Methylation | ||||
| SET1 | H3K4 trimethyl transferase | Proinflammatory genes | + | [161] |
| Ash2/Wdr5 | H3K4 trimethyl transferase | Endothelin, COL1A1/COL1A2 | + | [123,162] |
| KDM3A | H3K9 demethylase | CTGF | + | [90] |
| Jmjd1a | H3K9 demethylase | SMC differentiation markers | − | [163] |
| Acetylation | ||||
| p300 | H3K18/H3K27 acetyltransferase | COL1A1/COL1A2 | + | [123] |
| TIP60 | H4K16 acetyltransferase | iNOS | + | [91] |
| MOF | H4K16 acetyltransferase | NOX1/4 | + | [164] |
| Disease | Animal/Cell Model | Experimental Conditions | Suggested Mechanim | Reference |
|---|---|---|---|---|
| Diabetic nephropathy | REC cell model and WT rat | SCAI overexpression, rat UUO | SCAI → blocks MRTF-A → locks fibrosis | [126] |
| Mrtf-a KO mice and fibroblasts | In vivo (STZ, high fat diet), in vitro (STZ, high glucose) | MRTF-A is necessary to recruit histone acetyl- transferase and methyl- transferase to collagen promoters and activate type I collagen transcription | [123] | |
| MRTF-A KO mice and | In vivo (STZ, high fat diet) In vitro (STZ, high glucose) | MRTF-A regulates histone acetylation and methylation on the CTGF promoter, partially through interacting with KDM3A | [90] | |
| Obstructive nephropathy | WT mice, REC cell model | UUO, in vitro functional studies | Epithelial MRTF-A links cytoskeletal and organization to redox state, through NOX4 | [153] |
| AMPK1α KO conditional (fibroblast) | UUO | AMPK1α → cofilin →F-actin → nuclear MRTF-A | [239] | |
| WT mice | UUO, MRTF-A inhibitor (CCG1423) | RhoA → MRTF-A → TAZ → PEP → fibrogenesis | [124,220,240,241] | |
| WT mice | UUO+ SCAI inhibition | SCAI → blocks MRTF-A → blocks fibrosis | [126] | |
| Acute kidney injury | Macrophage-specific MRTF-A KO mice | Ischemia-reperfusion lipopolysaccharide | MRTF-A → MYST1 → H4K16Ac at NOX → ROS | [154] |
| Polycystic kidney disease | PKD patients | Microarray comparison | MRTF-A/SRF transcription network is upregulated | [242] |
| PKD1 KO in tubules | Loss of PKD → LARG → RhoA → YAP/TAZ → c-Myc → cystogenesis | [243] | ||
| PKD1 patients PKD1 KO mice | ROK-inhibitor (hydroxyfasudyl) treatment | Loss of PKD → ArhGAP35 → RhoA/ROK | [244] | |
| Pkd2+/−vascular smooth muscle | phenylephrin stimulation | Loss of PKD → RhoA → F-actin → αSMA | [245,246] | |
| Pkd1 and Pkd2 KO mice | Expression and localization studies | Increased MRTF expression and nuclear localization | Kapus lab, unpublished data |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miranda, M.Z.; Lichner, Z.; Szászi, K.; Kapus, A. MRTF: Basic Biology and Role in Kidney Disease. Int. J. Mol. Sci. 2021, 22, 6040. https://doi.org/10.3390/ijms22116040
Miranda MZ, Lichner Z, Szászi K, Kapus A. MRTF: Basic Biology and Role in Kidney Disease. International Journal of Molecular Sciences. 2021; 22(11):6040. https://doi.org/10.3390/ijms22116040
Chicago/Turabian StyleMiranda, Maria Zena, Zsuzsanna Lichner, Katalin Szászi, and András Kapus. 2021. "MRTF: Basic Biology and Role in Kidney Disease" International Journal of Molecular Sciences 22, no. 11: 6040. https://doi.org/10.3390/ijms22116040
APA StyleMiranda, M. Z., Lichner, Z., Szászi, K., & Kapus, A. (2021). MRTF: Basic Biology and Role in Kidney Disease. International Journal of Molecular Sciences, 22(11), 6040. https://doi.org/10.3390/ijms22116040






