1. Introduction
Urothelial carcinoma of the urinary bladder (UCB) can be grossly subdivided into non-muscle invasive (NMIBC) and muscle-invasive tumors (MIBC). The overall rate of progression from NMIBC to MIBC lies between 20% and 30% [
1]. A scoring system, based on clinical and pathologic criteria, aims to predict progression by subdividing NMIBC into low-, intermediate-, and high-risk tumors [
2]. While low-risk tumors have a small likelihood of recurrence and progression, high-risk tumors, such as pT1 or carcinoma in situ (CIS), are known to become muscle-invasive in 50% of patients, if left untreated [
3]. The most accurate important prognostic index is the current WHO 2016 histologic grading system, where a high tumor grade strongly correlates with progression to a muscle-invasive stage [
4]. High throughput sequencing analyses provided a comprehensive molecular characterization of bladder cancer [
5]. Additionally, alterations in molecular pathways have proven to be characteristic for either high- or low-risk disease. For example, FGFR3 mutations are commonly found in papillary, non-invasive, low-grade carcinomas [
4]. A remaining task is to discover further molecules that are directly involved in tumor progression, with the hope of identifying potential therapeutic targets.
To identify such molecules, in vitro models are available to assess different aspects of malignancy in bladder cancer (BC) cell lines. Cell viability, migration, and invasion of cancer cells can be monitored using classic endpoint assays, such as the MTT and the Boyden chamber assays. However, a more recent electrophysiological method now enables real-time monitoring and objective quantification of the invasive potential of BC cells. Results obtained with the xCELLigence Real-Time Cell Analysis (RTCA) device have shown strong correlations with conventional endpoint methods mentioned above [
6,
7]. The higher sensitivity of RTCA methods allows for a better identification of new molecules involved in UCB progression and metastatic development [
8].
One such possible molecule is the interleukin-1-receptor antagonist IL1RA (encoded by the
IL1RN gene). It is a potent competitive antagonist to interleukin-1 (IL1) and mainly involved in regulating inflammation [
9]. IL1RA was previously identified as an IL1 inhibitor in the supernatants of human cells [
9]. This isoform now is referred to as secreted, sIL1RA. Besides the secreted isoform, additional IL1RA intracellular isoforms have been described in various cell types, including epithelial cells [
10,
11,
12,
13]. Although there is broad evidence showing that sIL1RA acts as a competitive inhibitor of IL1-alpha and -beta by binding to cell-surface receptors, there is scarce information on the role of the cytoplasmic IL1RA proteins [
11]. Genetic polymorphisms in the
IL1RN gene have been linked to an increased risk of developing gastric cancer [
14], prostate cancer [
15], and UCB [
16]. A downregulation of
IL1RN expression that coincides with the degree of malignancy has been observed in colorectal cancer [
17], chronic myeloid leukemia [
18], and in oral squamous cell carcinoma [
19]. Immunohistochemical staining of UCB tissue samples has yielded similar results, finding significantly reduced IL1RA expression in MIBC samples compared to NMIBC and control samples [
20].
Here we show that low expression of IL1RA correlates with a higher invasive capacity of bladder cancer cell lines and vice versa. Consequently, ectopic expression of IL1RA impairs migration and invasive capacities of aggressive UCB cells. Importantly, congruent results were observed in different experimental systems, such as RTCA analysis, Boyden chamber assays, and an ex vivo porcine organ culture model. Moreover, we show that recombinant IL1RA blocks IL1B-dependent endothelial barrier breakdown in the impedance-based assay. Taken together, the present data indicate that the IL1 family plays an important role in UCB metastatic progression.
3. Discussion
Considerable progress in cancer therapy has been documented over the last years; however, patients with metastatic UCB still suffer from a lack of curative therapies [
28]. For the colonization of distant sites, cancer cells must leave the primary tumor, then enter and exit the blood circulation. This process of intravasation and extravasation is termed trans-endothelial migration (TEM) and the vascular endothelial layer represents a critical barrier for tumor cells [
29]. In contrast to the usually leaky and dysfunctional tumor neo-vessels, the endothelial surface in distant organs is intact and it is highly probable that TEM is an active process promoted by cytokines, growth factors, and vascular permeability regulators. In response to these environmental stimuli, ECs adapt to the new conditions and undergo activation (endothelial cell activation, ECA), which in turn creates proinflammatory and procoagulatory intravascular conditions. ECA enables multiple heterotypic adhesive interactions of tumor cells with platelets, leucocytes, and the endothelium, facilitating subsequent tumor cell docking on the endothelium and extravasation [
30,
31,
32]. Therapeutic intervention in this intravascular multicellular crosstalk and the accompanying local hypercoagulation, for example, using low molecular heparins, has emerged as a promising option for attenuating the risk of venous thrombo-embolism and metastatic dissemination [
33,
34].
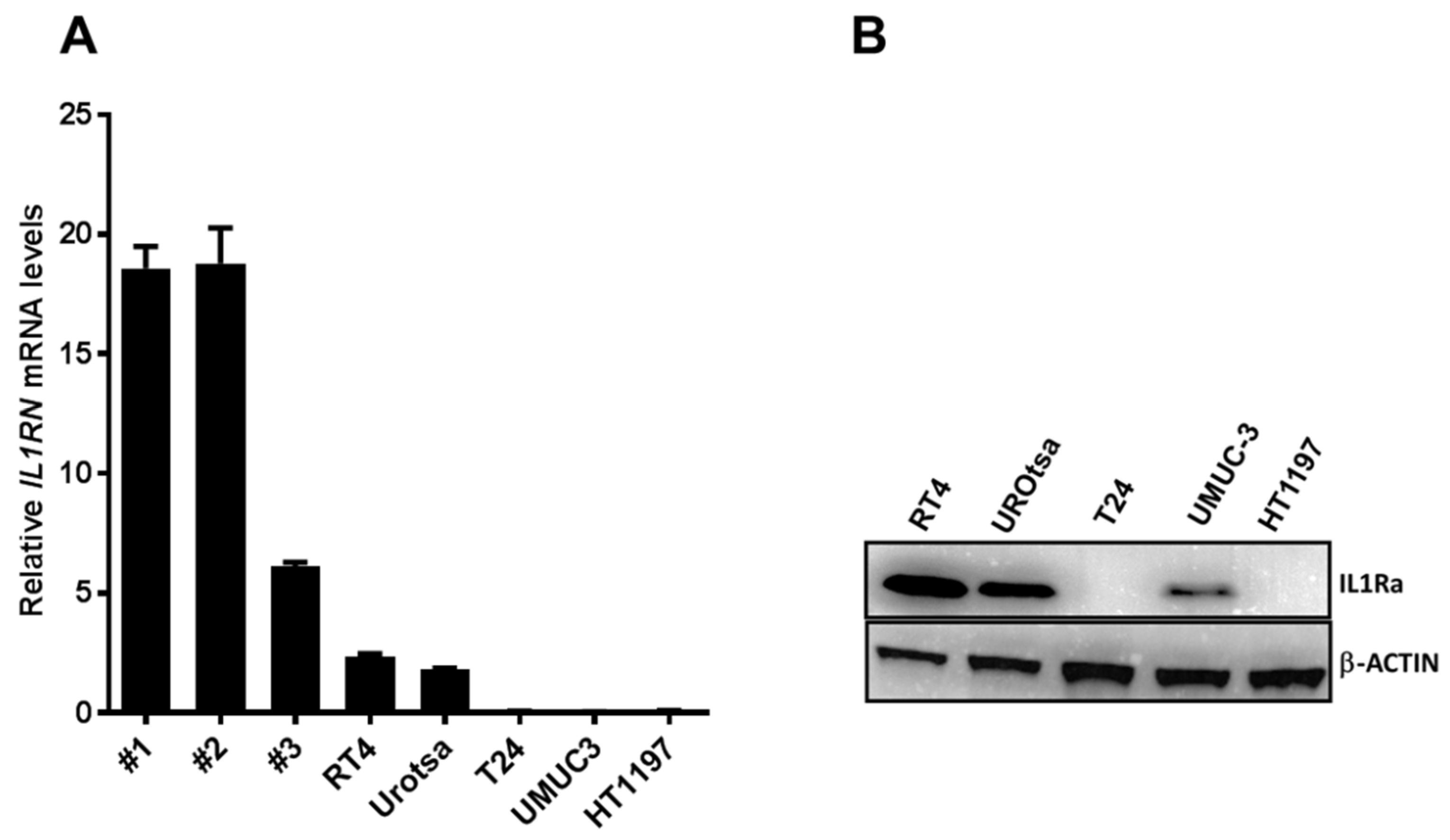
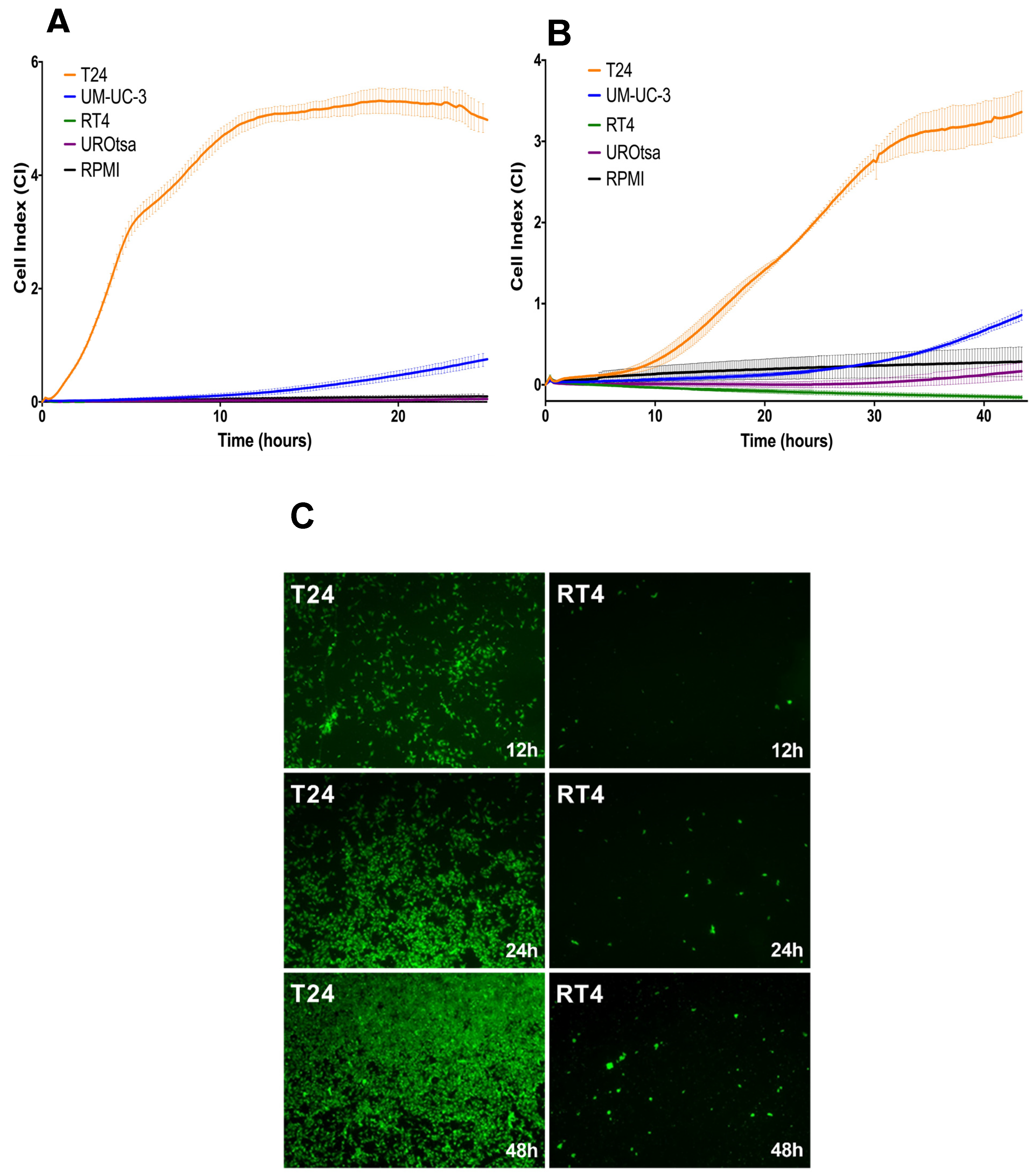
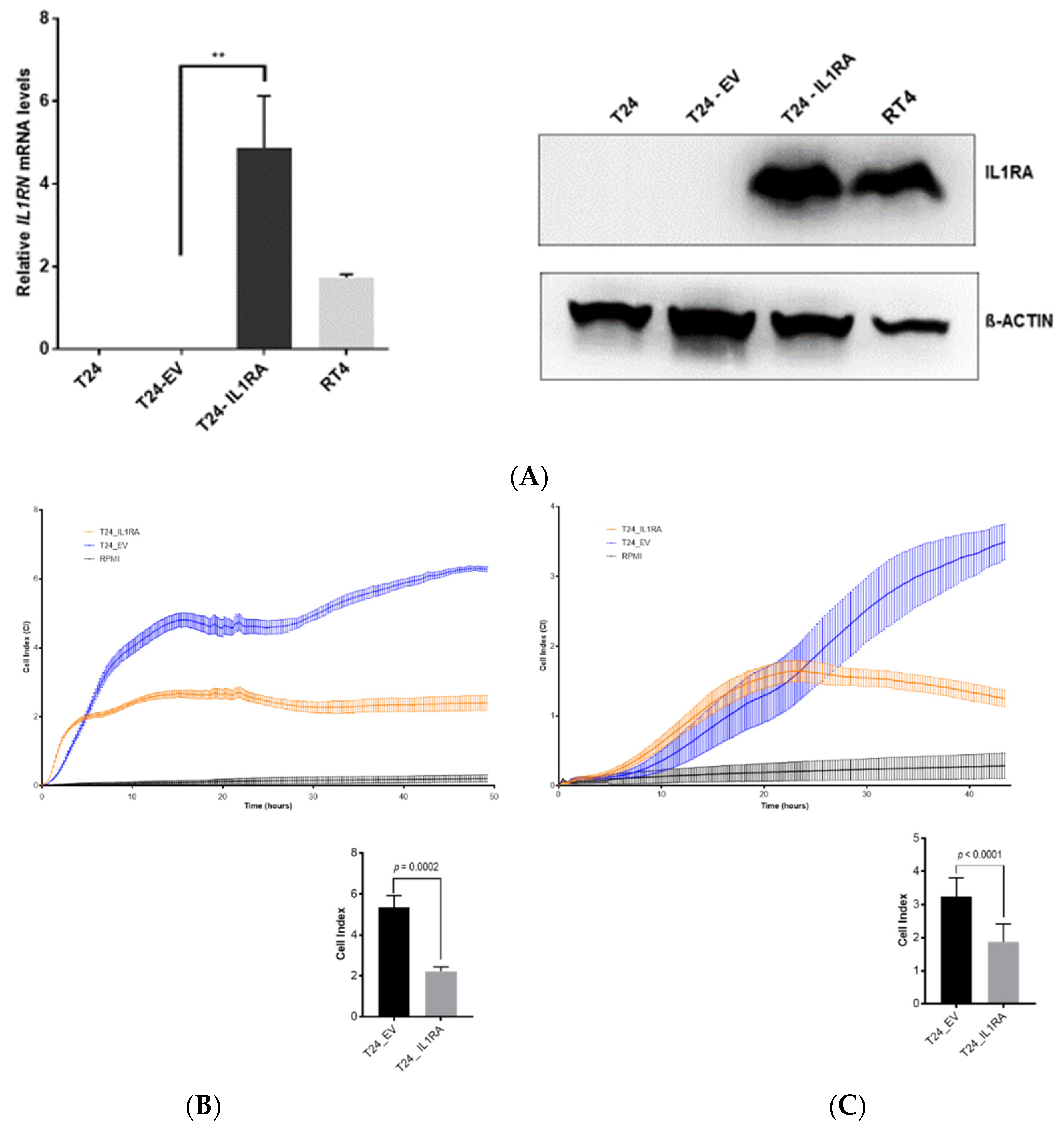
In the present study, we investigated the role of IL1RA in the context of ECA and as a potential modulator of migration and tissue invasion in UCB. We show that low
IL1RN mRNA levels correlate with increased migration and tissue invasion capacities of bladder cancer cell lines. Conversely,
IL1RN mRNA levels were higher in semi-benign UROtsa and non-invasive RT4 cell lines as well as in primary epithelial cells derived from apparently healthy ureter tissue, supporting the idea that IL1RA may impair invasiveness of bladder cancer cells. Notably, independent experimental approaches (Boyden chamber, impedance measurement, and ex vivo porcine invasion assay) provided congruent results, confirming the impact of ectopic IL1RA on cell invasiveness and migration. While the established Boyden chamber is a simple to perform end-point technique to evaluate the migration and invasion capacities, impedance sensing allows continuous real-time monitoring of the integrity and concomitant barrier function of cell layers, and thus represents a useful adjunct to classical end-point assays. Here, we combined the xCELLigence impedance sensing system with surface-coating of the well with Matrigel or a HUVEC monolayer. In both approaches, ectopic IL1RA in the highly invasive T24 cells lead to a markedly reduced invasive potential and attenuated cell migration compared to the parental cell line or cells transfected with an empty vector. Importantly, we also used the ex vivo porcine bladder organ culture model [
35], which allows the evaluation of tumor cell invasiveness under semi in vivo conditions [
21]. Again, IL1RA overexpression led to a significant loss of invasive potential of the tested cells, strongly supporting the conclusion that IL1RA impairs the invasive and migration capacities of bladder cancer cells. However, while overexpression of
IL1RN in T24 cells attenuated invasiveness, shRNA-mediated knockdown of
IL1RN in the non-invasive RT4 cells did not result in an increased invasiveness using RTCA (not shown). It remains to be elucidated whether IL1RA acts in concert with additional factors whose altered expression and/or functionality cooperates with IL1RA for efficient modulation of cell invasiveness.
The functions of IL1RA were mainly studied in the context of IL1 activity, where secreted IL1RA (sIL1RA) antagonizes the effects of IL1A or IL1B through inhibition of IL1 binding to surface receptors. In addition to sIL1RA, three intracellular IL1RA (icIL1RA) isoforms have been described [
10,
12]. The functions of the icIL1RA isoforms are still not clear. In line with our previous report [
20], the present data show that low IL1RA levels in bladder cancer cell lines correlate with their increased invasive potential. On the other hand, we find that the endogenous IL1RA (RT4 and UROtsa) as well as the ectopic IL1RA (T24-IL1RA) can be measured in the supernatant of cells, indicating that it is secreted. At this time-point, we do not have a conclusive explanation how exactly IL1RA impairs invasion and migration capacity of the cells. The vasculature is one of the first obstacles to overcome for tumor cells during invasion of distant tissues. In our investigation, we used a HUVEC monolayer to fully cover the wells of the impedance sensing system as a model for the vascular endothelium. Addition of IL1B led to a decline in HUVEC barrier function, which could be reversed by the addition of IL1RA in a dose-dependent manner (
Figure 4B). Thus, IL1 might be an important promoter of TEM, and external (secreted) IL1RA acts in an antagonistic manner impairing IL1B TEM activity.
Taken together, our data are in line with our previous report showing a reduced expression of IL1RA in human bladder cancer samples in comparison to healthy urothelium. Importantly, we reported that reduced IL1RA expression correlated with adverse pathologic characteristics [
20]. Furthermore, polymorphisms in the
IL1RN gene were associated with recurrence after BCG immunotherapy and susceptibility to bladder cancer [
36]. More aspects linked to the IL1 pathway have so far been described in UCB, such as IL1B-induced cisplatin-resistance by up-regulation of Aldo-keto reductase 1C1 [
24], IL1 dependent intra-tumoral androgen receptor (AR) signaling, T-cell attraction, and recruitment of tumor-associated fibroblasts [
26]. We think that molecular subtyping of bladder cancer will become an element of clinical routine in the upcoming years. For instance, it was shown that patients with luminal MIBC do not appear to derive much clinical benefit from neoadjuvant chemotherapy compared to basal type MIBC. On the contrary, patients with luminal tumors that are infiltrated with stromal cells seem to be sensitive to immune checkpoint inhibitors such as anti PD-L1. Since the IL1 axis is a strong regulator of inflammatory response, one might speculate that loss of IL1RA in high-grade bladder cancer might play a greater role in those tumors with marked immune cell infiltration. Finding a correlation between IL1/IL1RA expression and molecular subtypes of bladder cancer would be an interesting topic for further research.
In summary, IL1RA might have an anti-tumorigenic role in bladder cancer by attenuating tumor cell migration and invasion while preserving the integrity of vascular endothelial barriers. As impacts on migration and invasion capability were observed in the absence of a vascular endothelial layer, it is tempting to speculate that IL1RA may also contribute to cells’ invasive capacity by a yet unknown intracellular mechanism. It is also conceivable that IL1RA acts in an autocrine manner to modulate the cells’ invasion and migration capacity. While the exact molecular pathways and downstream targets remain to be uncovered, our study was able to show the possible relevance of IL1RA for tumor progression in UCB using a variety of experimental methods, including an ex vivo organ culture model and in vitro cell culture experiments. Further in vivo trials are necessary to evaluate the potential of IL1RA as a potential therapeutic target in bladder cancer patients.
4. Materials and Methods
4.1. Cell Lines and Primary Human Urothelium
The human UCB cell lines T24, UM-UC3, and RT4 were obtained from the European Collection of Authenticated Cell Cultures (ECACC). The simian virus 40 (SV40) large T antigen immortalized UROtsa cell line served as a model for benign urothelium (gift of Dr. Phillip Erben, PhD, Mannheim). All cell lines were kept in RPMI 1640 medium (ThermoFischer Scientific, Waltham, MA, USA), supplemented with 10% fetal calf serum (FCS) (Sigma-Aldrich, St. Louis, MO, USA) and 1% penicillin/streptomycin (Pan Biotech, Aidenbach, Germany). The medium for UROtsa cells was additionally supplemented with 1% GlutaMAX (ThermoFischer Scientific, Waltham, MA, USA) and 1% non-essential amino acids (MerckMillipor, Darmstadt, Germany). Human umbilical vein endothelial cells (HUVEC) were obtained from umbilical cord veins as described previously [
37] and grown in culture medium composed of two-thirds M199 supplemented with 10% heat-inactivated fetal calf serum, 1% antibiotics (penicillin and streptomycin), and one third EGM-2 (Lonza, Basel, Switzerland). HUVECs were maintained at 37 °C, 5% CO
2. Cells were cultured at 37 °C in a humid atmosphere with 5% CO
2. Primary human urothelial cells were prepared from benign ureter tissue of patients who underwent nephrectomy, with informed consent. Briefly, tissues were placed on ice directly after surgery and treated with stripper medium (500 mL HBSS without Ca
2+Mg
2+, Gibco 14170-088, 10 mM Hepes, Gibco 15630-056, and 0.1% EDTA) overnight. Epithelial cells were scratched and processed for RNA isolation immediately. The study was approved by the local Ethics Committee (ethical approval number: 239/18).
4.2. Cloning of IL1RN into the pBabe-Puro Plasmid Vector and Ectopic Expression in T24 Cells
A PCR product encoding the IL1RN gene was amplified from RT4 cDNA using the Phusion High-Fidelity PCR Kit (ThermoFischer Scientific, Waltham, MA, USA). The cDNA was generated by reverse transcription (RT) using 1 μg total RNA from RT4 cell cells with the GoScriptTM Reverse Transcription System (Promega, Fitchburg, WI, USA) in a 20 µL reaction mixture. The PCR reaction was conducted with 5 µL of the RT reaction and the cloning primers (forward primer: 5′-CGATGCGGATCCGAGGCCCTCCCCATGGCTTTAG-3′ and reverse primer 5′-CGATGCGTCGACGGCAGTACTACTCGTCCTCC-3′), which included BamHI and SalI restriction enzyme recognition sites, respectively. Cycling conditions: initial denaturation at 98 °C for 30 s, 25 cycles of 98 °C for 10 s, 55 °C for 30 s, followed by a final extension at 72 °C for 10 min. The PCR product was run on an agarose gel, followed by gel purification with the QIAquick® Gel Extraction Kit (QIAGEN, Hilden, Germany). Both the expression vector pBabe-Puro and the purified PCR product were digested with the restriction enzymes BamHI-HF® and SalI-HF® (New England Biolabs, Ipswich, MA, USA) and ligated using Quick Ligase (New England Biolabs, Ipswich, MA, USA). The ligation mix was transformed into Stbl 3 chemically competent Escherichia coli (ThermoFischer Scientific, Waltham, MA, USA) and the plasmid DNA was isolated from a single bacterial colony with the PureYield™ Plasmid Miniprep System (Promega, Fitchburg, MA, USA). The cloning of IL1RN was validated by Sanger sequencing using pBABE-5′ or pBABE-3′ sequencing primers (Eurofins, Ebersberg, Germany). Plasmid DNA was amplified with the NucleoBond® Xtra Midi/Maxi Kit (Macherey-Nagel, Düren, Germany). For virus production, HEK293 cells were cotransfected with the IL1RN expression vector, the pCMV-Gag-Pol and pCMV-VSV-G plasmids. The virus was harvested after 48 h, filtered twice through a 0.45 µm filter (Sarstedt, Nürnbrecht, Germany), and used to infect T24 cells. The infected target cells were selected with 1 μg/mL Puromycin (ThermoFischer Scientific, Waltham, MA, USA) for 7 days and permanent selection was continued with 0.5 μg/mL Puromycin.
4.3. Determination of IL1RA by ELISA
An amount of 1.5 × 107 cells were seeded in T25 flasks (Sarstedt, Nürnbrecht, Germany) to achieve full confluency and to minimize effects by cell numbers due to different proliferation capacities of the cells. An amount of 1 mL supernatant (SN) was transferred into Eppendorf tubes (E-tubes) at 24 h and 48 h post-plating and immediately placed on ice for 2 min. Samples were then centrifuged at 1400 rpm for 10 min to pellet potential contaminating cells and 800 µL of the clean SN were transferred into new E-tubes and stored in −80 °C until use by ELISA. The ELISA was determined according to the protocol provided by the supplier, using 100 µL of the SN. The standard curve was determined by the recombinant IL1RA provided with the kit. The absorbance of the samples was read at 450 nm.
4.4. Quantitative Real-Time PCR (RT-qPCR)
Cells were harvested and total RNA was isolated with the RNeasy
® Mini Kit (QIAGEN, Hilden, Germany). Reverse transcription was performed with the GoScript
TM Reverse Transcription System (Promega, Fitchburg, MA, USA) using 100 ng total RNA. Diluted (1:10) cDNA was incubated with IL1RN-specific primers (IL1RN-F1: 5′-TGTTCCCATTCTTGCATGGC and IL1RN-R1: 5′-GCAGCATGGAGGCTGGTCAG) and the iTaq™ Universal SYBR
® Green Supermix (Bio-Rad, Hercules, CA, USA). The Viia™ 7 Real-Time PCR-System (Applied Biosystems, Foster City, CA, USA) was used for quantitative real-time PCR (RT-qPCR) of
IL1RN and
GAPDH [
38]. The following conditions were used for quantitative real-time PCR: 1 × 50 °C for 2 min; 1 × 95 °C for 10 min; 40 × 95 °C for 15 s and 60 °C for 1 min, followed by 1 × 95 °C for 15 s, 1 × 60 °C for 1 min, 1 × 95 °C for 30 s, and 1 × 60 °C for 15 s. ΔCT-values were calculated using
GAPDH as reference values to reflect the relative mRNA level in the BC cell lines. RT-qPCR results were obtained from three individual experiments, each performed in triplicate.
4.5. Western Blot
Cells were washed in DPBS (ThermoFischer Scientific, Waltham, MA, USA) and detached with trypsin (Pan Biotech, Aidenbach, Germany). The cell pellet was resuspended in RIPA buffer (Sigma-Aldrich, St. Louis, MO, USA) containing phosphatase and protease inhibitors (Roche Applied Science, Penzberg, Germany). Cells were mechanically destroyed, and the cell lysate solution was centrifuged at 14,000 rpm for 40 min at 4 °C. The protein concentration was determined by Bicinchoninic acid (BCA) assay. The protein amount was adjusted to 20 μg and the sample volume brought up to 20 μL with RIPA buffer. Additionally, 4 μL of 4× Laemmli Buffer (Bio-Rad, Hercules, CA, USA) with 5% 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA) were added to each sample. The proteins were denatured at 96 °C for 10 min after being separated on a 10% acrylamide gel (Bio-Rad, Hercules, CA, USA) at 80 V for 2 h. The proteins were transferred to a PVDF membrane (Merck, Darmstadt, Germany) via electroblotting at 300 mA for 2 h. The membrane was blocked in TBS-Tween 20 (TBS-T) containing 5% skim milk for 45 min, followed by incubation with a primary monoclonal antibody overnight at 4 °C in TBS-T containing 5% skim milk (Sigma-Aldrich, St. Louis, MO, USA). The following primary antibodies were used: rabbit anti-IL1RA (1:500; Sigma-Aldrich, St. Louis, MO, USA, HPA001482) and mouse anti-β-actin (1:10,000; Sigma-Aldrich, St. Louis, MO, USA, A2228). Membranes were washed three times in TBS-T and subsequently incubated with an anti-rabbit/anti-mouse horseradish peroxidase (HRP)-linked secondary antibody (1:2000; Cell Signaling Technology, Danvers, MA, USA) for 2 h at room temperature. Membranes were then washed three times in TBS-T, incubated with AceGlowTM chemiluminescence substrate (VWR, Radnor, PA, USA), and antibody signals were visualized by chemiluminescence detection (Fusion Fx, Vilbert Lourmat, Collégien, France).
4.6. Classical Boyden Chamber Invasion Assay
Cultures were split to reach 80% confluency and then starved with serum-free RPMI 1640 for 24 h. Pore culture inserts (8 μm; Sarstedt, Nürnbrecht, Germany) were placed into a 24-well plate (Sarstedt, Nürnbrecht, Germany) and subsequently coated with 100 μL pre-thawed Matrigel (250 μg/mL) (Corning Incorporated, Corning, NY, USA). The plates were left to incubate for 30 min at 37 °C. The bottom wells were filled with 800 μL RPMI 1640 containing 10% FCS as chemoattractant. Cells were washed with DPBS, detached with trypsin, and counted with a Neubauer hemocytometer. The cell suspension concentration was adjusted to 1 × 105 cells/mL and 100 μL of cell suspension was added to each well and spread evenly by means of soft shaking. The plate was incubated at 37 °C under normal culture conditions. After 48 h, the inserts were removed, washed with DPBS, fixed with 500 μL 5% glutaraldehyde solution (Sigma-Aldrich, St. Louis, MO, USA) for 30 min, washed with DPBS, stained with 500 μL 0.2% crystal violet solution (Sigma-Aldrich, St. Louis, MO, USA) for 30 min, and rinsed with tap water. A cotton swab was used to remove non-migrated cells from the upper side of the insert membrane. The membranes were dried overnight and were then cut from the insert using a scalpel. Membranes were mounted face-down on a microscope slide with Entellan® (Merck, Darmstadt, Germany) and sealed with a cover-slip. The positively stained cells were evaluated for each membrane by taking pictures of five random fields at 10× magnification and counting the cells manually using “ImageJ”. The results of the Boyden chamber experiments were obtained from four biological replicates.
4.7. Modified Boyden Chamber Invasion Assay
To simulate interactions of tumor cells with the vascular endothelium, the Boyden chamber experiment was modified in a set of experiments. Wells with 8 µm pores were coated with 100 µL 0.1% gelatin; 2 × 105 HUVECs were seeded and wells incubated for 48 h until a confluent monolayer was obtained as confirmed by microscopy. Bladder cancer cells (RT4 or T24) were stained with CellTracker™ Green CMFDA Dye (Invitrogen, Carlsbad, CA, USA) according to the instructions of the manufacturer and kept in starvation medium for 24 h. Full growth medium supplemented with FCS as a chemoattractant was put in the lower part of the transwell while 1 × 105 stained and starved RT4 or T24 cells were seeded unto the HUVEC layer. After 18 h incubation, the membrane of the lower part of the well containing the migrated tumor cells was cut, washed, fixed, and prepared for further immunofluorescence imaging.
4.8. Real-Time Impedance Sensing for Migration, Invasion, and Endothelial Barrier Function
Cell invasion and migration was continuously monitored using the impedance-based measuring device xCELLigence RTCA DP instrument (ACEA Biosciences, San Diego, CA, USA). The CIM-Plate 16 (ACEA Biosciences, San Diego, CA, USA) contains 16 wells that are divided into upper and lower chambers by a microporous polyethylene terephthalate (PET) membrane with 8 µm pores, allowing cells to adhere to microelectrodes on the underside of the membrane [
39]. The number of adherent cells causes a change in impedance that is reflected by an increase in Cell Index (CI). The CI directly correlates with the cells’ migratory potential [
7,
39,
40]. When monitoring cell invasion, the upper chamber was additionally coated with 250 μg/mL pre-thawed Matrigel (Corning Incorporated, Corning, NY, USA) diluted in cold, serum-free RPMI 1640 and incubated for 4 h at 37 °C. Under these conditions, CI directly correlates with the cells’ invasive potential.
Initially, the plate was prepared by coating the electrodes with 1% gelatin in DPBS followed by an incubation period of 45 min under the tissue culture hood. The gelatin was removed, and the electrodes were washed once with sterile water. The lower chamber was filled with RPMI 1640 containing 10% FCS as a chemoattractant for migrating/invading cells. The upper chamber was filled with serum-free RPMI. The plate was left to equilibrate at 37 °C for 1 h in the RTCA DP-Analyzer.
Cells were passaged two days prior and starved one day prior to the experiment, reaching a final confluency of 70–80%. During plate equilibration, cells were washed with DPBS, detached with trypsin, and counted with a Neubauer hemocytometer. Cells were centrifuged for 3 min at 1200 rpm at 24 °C. The cell pellet was resuspended in serum-free RPMI to obtain a concentration of 4 × 105 cells/mL.
After plate equilibration, background measurements were recorded with the RTCA DP device. Subsequently, 100 μL of the cell suspension was carefully added to each well of the upper chamber to avoid bubble formation. The CIM-Plate 16 was placed in the RTCA DP-Analyzer and the CI was monitored over 48 h at 10–15 min interval measurements. Cell migration experiments were repeated twice and cell invasion experiments were repeated three times, both in quadruplicate.
The xCELLigence RTCA DP instrument (ACEA Biosciences, San Diego, CA, USA) was also used with the E-Plate 16 to assess the integrity of a HUVEC monolayer in response to treatment with IL1B and IL1RA. The electrodes were coated with 1% gelatin in DPBS and left to incubate at 37 °C for 1 h. The gelatin was removed, and the electrodes were washed once with sterile water. Each well was filled with HUVEC medium and incubated at 37 °C for 1 h for plate equilibration and background measurement. HUVECs with a maximum passage number of 5 were then seeded in each well at a seeding density of 30,000 cells/ well. The E-Plate 16 was left under the tissue culture hood to equilibrate for 30 min and then placed in the RTCA DP-Analyzer to begin with the impedance readings. The HUVECs were left to attach to the electrodes and form a monolayer within 20–24 h. This could be confirmed by a leveling out of the CI. IL1B (25 μg/mL; R&D systems, Wiesbaden, Germany) was diluted in RPMI to a final concentration of 1 ng/mL. IL1RA (100 μg/mL; R&D systems, Wiesbaden, Germany) was diluted in RPMI 1640 to final concentrations of 0.3 ng/mL, 3 ng/mL, 30 ng/mL, or 300 ng/mL. Half of the HUVEC medium was removed from each well and IL1B and IL1RA suspensions were added to each well. The E-Plate 16 was placed back into the RTCA device and the impedance readings were continued. The CI was monitored for another 20–24 h. For analysis, the CI was normalized to a time-point after IL1B and IL1RA addition. The experiment was repeated three times in quadruplicate.
4.9. Porcine Bladder Invasion Model
Porcine bladder was supplied by a local abattoir (Ulmer Fleisch GmbH, Ulm, Germany) and kept on ice during transport. The porcine bladder was cut open and washed three times in DPBS containing 10% penicillin/streptomycin. It was placed in 0.5% dispase II solution (Sigma-Aldrich, St. Louis, MO, USA) overnight at 4 °C to detach the urothelial cells from the basement membrane. The next day, the urothelium was removed with the blunt end of a scalpel and any loose connective tissue was removed with sterile scissors. The porcine bladder was cut into 0.5 cm2 pieces and each piece was placed onto a 70 μm cell strainer placed inside a 6-well plate containing 5.5 mL Waymouth’s medium (ThermoFischer Scientific, Waltham, MA, USA). The medium only covered the underside of the stromal tissue pieces, leading to an air-liquid interface. Silicon O-rings (diameter = 5 mm) were placed on the deepithelialized surface of each tissue piece. The porcine bladder was kept under normal culture conditions overnight. The next day, target cells were harvested and the seeding number adjusted to 1–1.5 × 106 cells per tissue piece. The cell suspension was pipetted into the area enclosed by the silicon O-ring, ensuring that most cells would stay in contact with the stroma. The medium of the 6-well plate was changed to Keratinocyte-SFM (1X) (ThermoFischer Scientific, Waltham, MA, USA). After two days, the medium was changed to RPMI 1640 media, supplemented with 10% fetal calf serum (FCS), 1% penicillin/streptomycin, 1% GlutaMAX, and 1% non-essential amino acids. The tissue pieces were kept under normal culture conditions. The wells were washed with DPBS containing 10% penicillin/streptomycin and the medium was changed on alternate days. After two weeks, the tissue was removed and fixed in 10% formalin solution. Samples were dehydrated and embedded in paraffin wax. Vertical tissue cross-sections (3 μm) were dried overnight and stained with hematoxylin and eosin (Waldeck, Münster, Germany) for histological evaluation. Pictures were taken with the Axio Imager M2 microscope and Axiocam 105 color (Zeiss, Oberkochen, Germany).
4.10. Statistical Analysis
Results were expressed as mean ± SD. Statistical significance was determined by comparing values using an unpaired two-tailed t test. All statistical analyses were carried out using GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA).