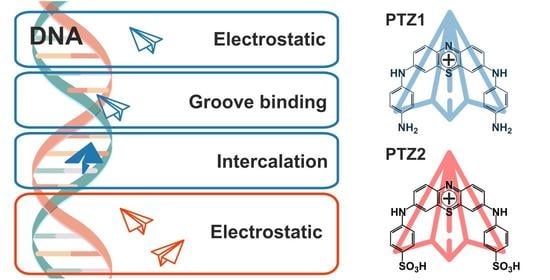
Arylamine Analogs of Methylene Blue: Substituent Effect on Aggregation Behavior and DNA Binding
Abstract
1. Introduction
2. Results and Discussion
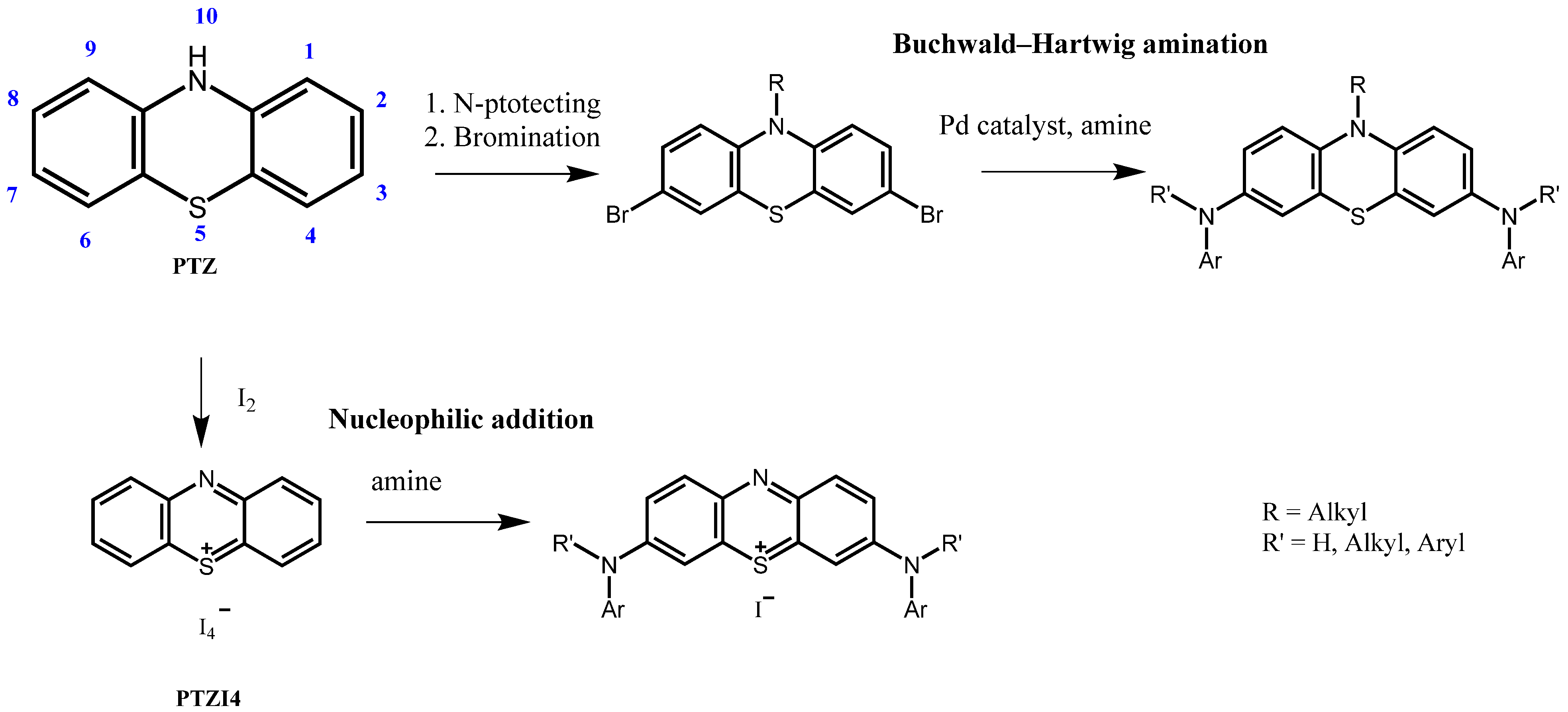
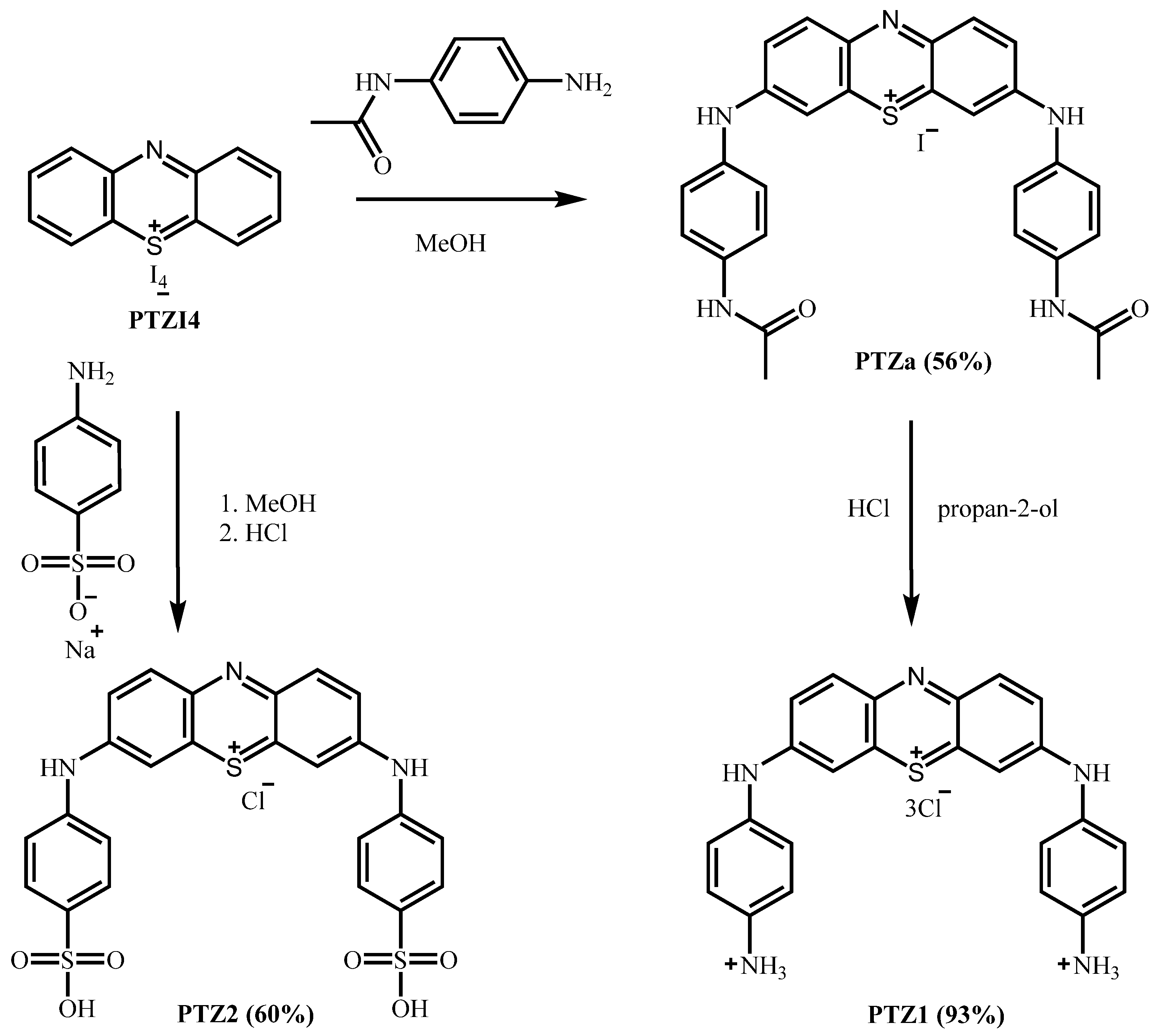
2.1. Synthesis of PTZ1, PTZ2
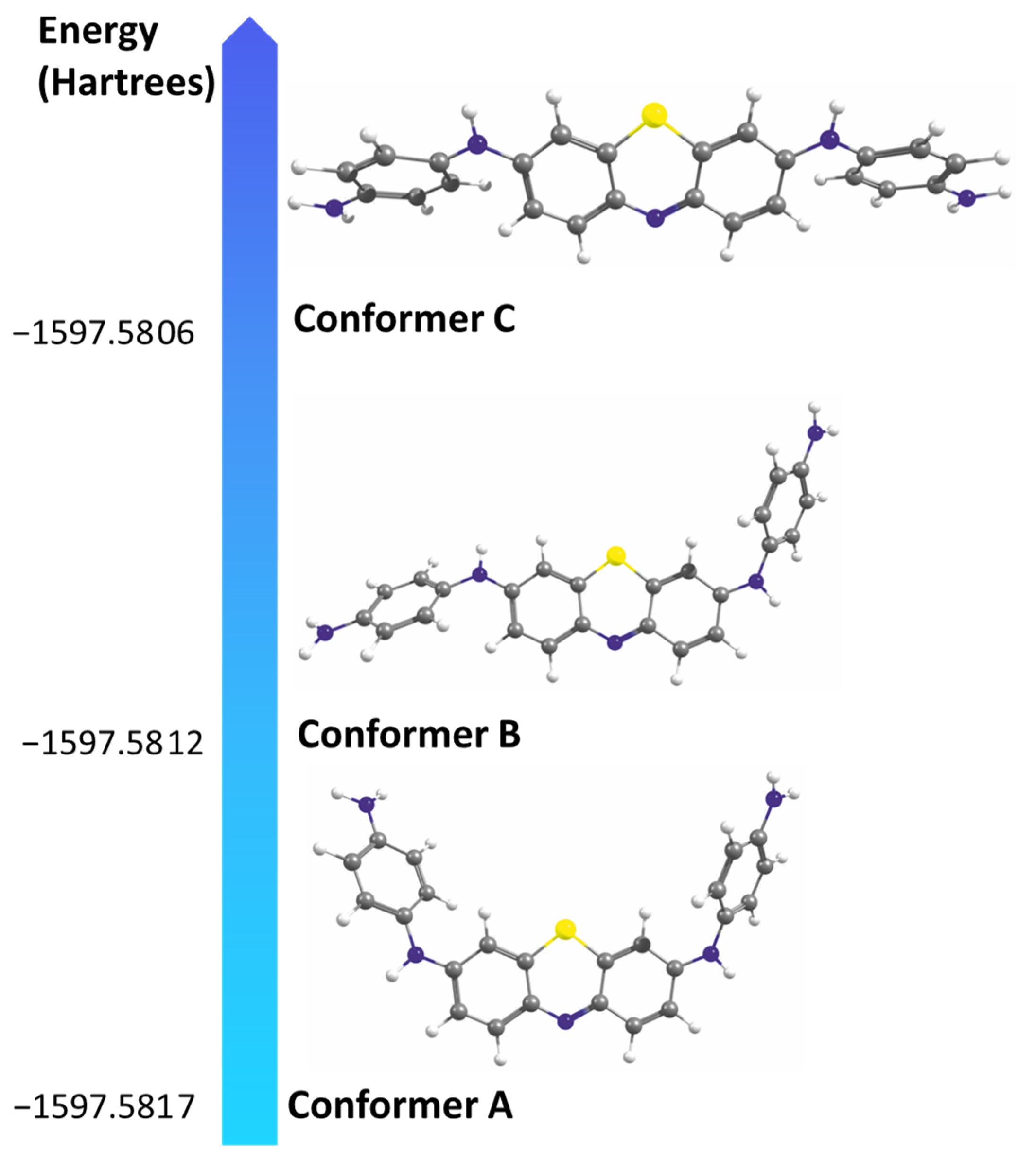
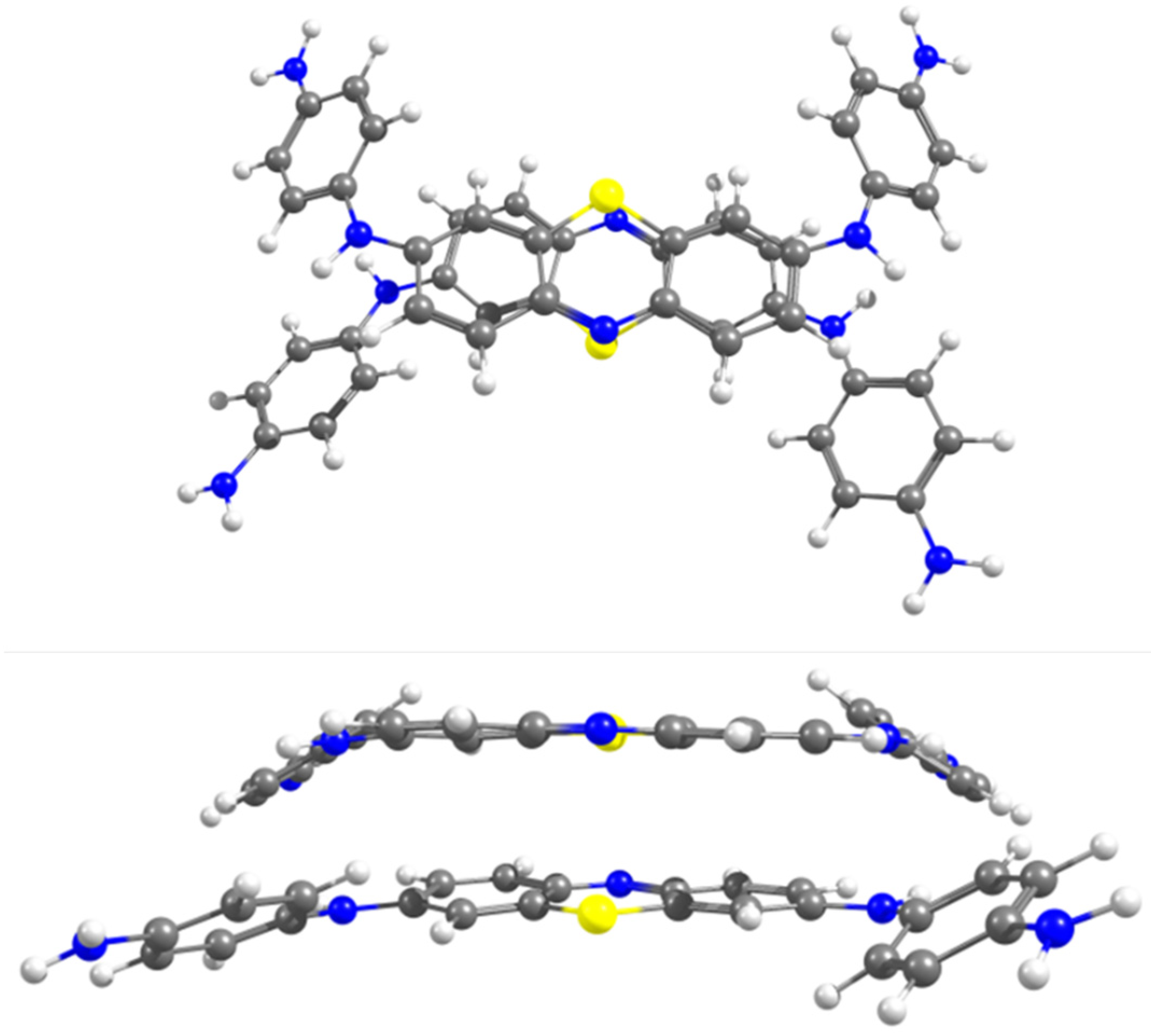
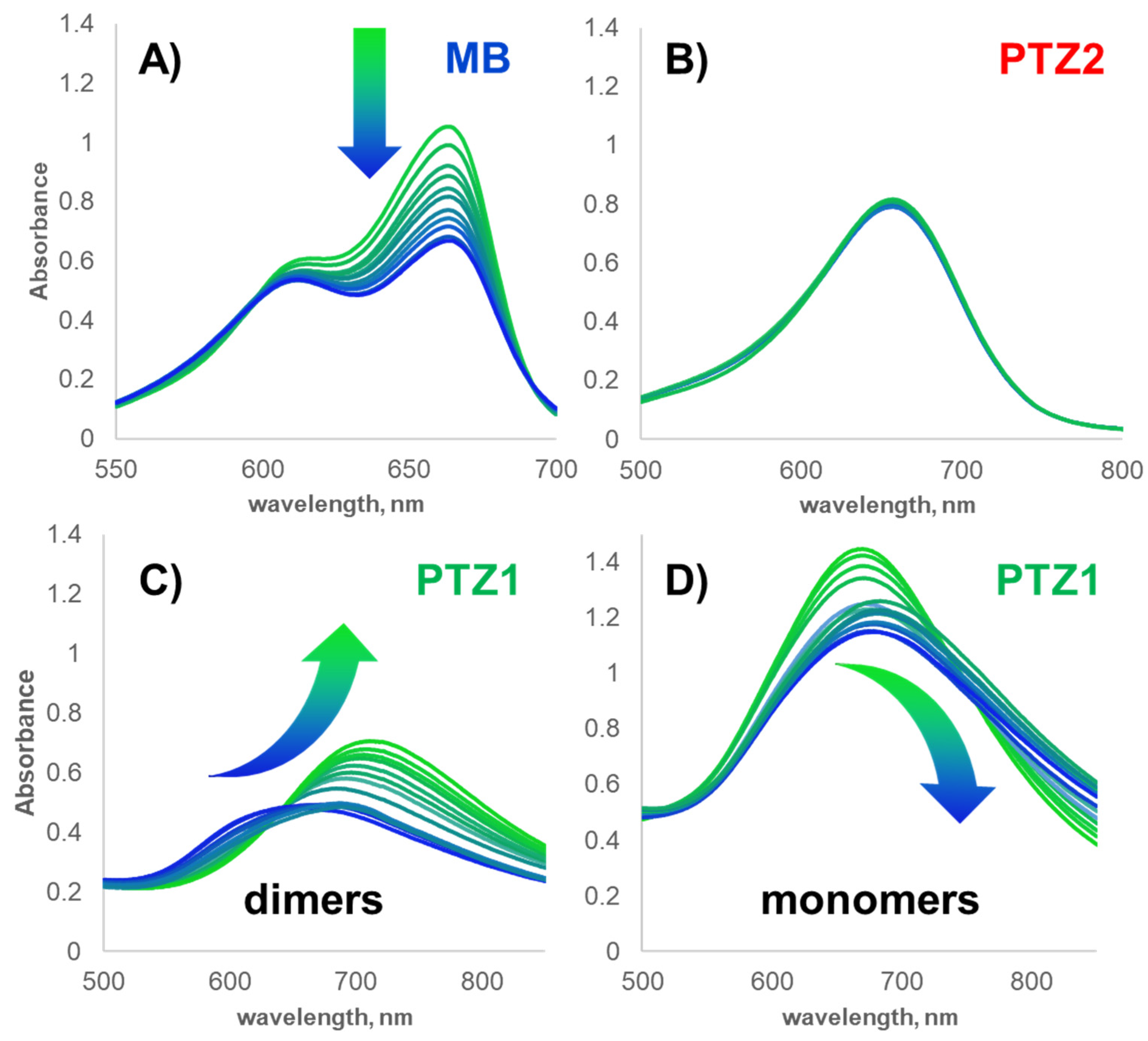
2.2. Aggregation Properties—Dimer Formation
2.3. Interaction with DNA
2.3.1. UV-Vis Spectroscopy
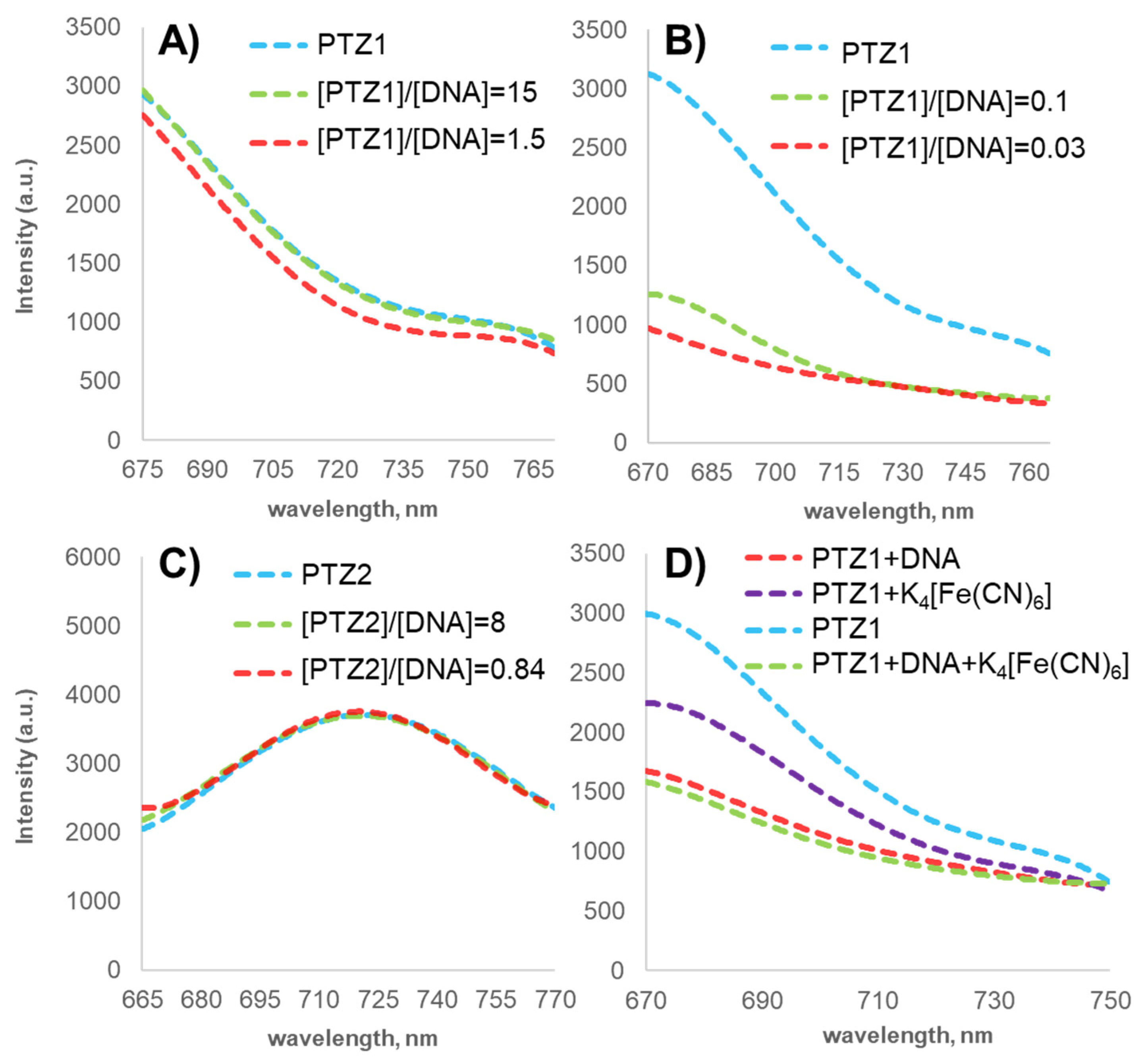
2.3.2. Fluorescence Spectroscopy
3. Materials and Methods
3.1. General Experimental Information
3.2. Synthesis of PTZ1 and PTZ2
3.2.1. 3,7-Bis((4-acetamidophenyl)amino)phenothiazin-5-ium iodide (PTZa)
3.2.2. 3,7-Bis((4-aminophenyl)amino)phenothiazin-5-ium chloride dihydrochloride (PTZ1)
3.2.3. 3,7-Bis((4-sulfophenyl)amino)phenothiazin-5-ium chloride (PTZ2)
3.3. DNA Binding Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gibson, D.S. Drug–DNA interactions and novel drug design. Pharm. J. 2002, 2, 275–276. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug–DNA interactions and their study by UV–Visible, fluorescence spectroscopies and cyclic voltammetry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef]
- Rehman, S.U.; Sarwar, T.; Husain, M.A.; Ishqi, H.M.; Tabish, M. Studying non-covalent drug–DNA interactions. Arch. Biochem. Biophys. 2015, 576, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Rescifina, A.; Zagni, C.; Varrica, M.G.; Pistarà, V.; Corsaro, A. Recent advances in small organic molecules as DNA intercalating agents: Synthesis, activity, and modeling. Eur. J. Med. Chem. 2014, 74, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, A.F.; Terra, L.F.; Wailemann, R.A.M.; Oliveira, T.C.; Gomes, V.D.M.; Mineiro, M.F.; Meotti, F.C.; Bruni-Cardoso, A.; Baptista, M.S.; Labriola, L. Methylene blue photodynamic therapy induces selective and massive cell death in human breast cancer cells. BMC Cancer 2017, 17. [Google Scholar] [CrossRef]
- Wu, P.-T.; Lin, C.-L.; Lin, C.-W.; Chang, N.-C.; Tsai, W.-B.; Yu, J. Methylene-Blue-Encapsulated Liposomes as Photodynamic Therapy Nano Agents for Breast Cancer Cells. Nanomaterials 2018, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Trevisan, E.; Menegazzi, R.; Zabucchi, G.; Troian, B.; Prato, S.; Vita, F.; Rapozzi, V.; Grandolfo, M.; Borelli, V. Effect of methylene blue photodynamic therapy on human neutrophil functional responses. J. Photochem. Photobiol. B Biol. 2019, 199, 111605. [Google Scholar] [CrossRef]
- Tuite, E.; Kelly, J.M. The interaction of methylene blue, azure B, and thionine with DNA: Formation of complexes with polynucleotides and mononucleotides as model systems. Biopolymers 1995, 35, 419–433. [Google Scholar] [CrossRef]
- Nafisi, S.; Saboury, A.A.; Keramat, N.; Neault, J.-F.; Tajmir-Riahi, H.-A. Stability and structural features of DNA intercalation with ethidium bromide, acridine orange and methylene blue. J. Mol. Struct. 2007, 827, 35–43. [Google Scholar] [CrossRef]
- Biebricher, A.S.; Heller, I.; Roijmans, R.F.H.; Hoekstra, T.P.; Peterman, E.J.G.; Wuite, G.J.L. The impact of DNA intercalators on DNA and DNA-processing enzymes elucidated through force-dependent binding kinetics. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Rohs, R.; Sklenar, H.; Lavery, A.R.; Röder, B. Methylene Blue Binding to DNA with Alternating GC Base Sequence: A Modeling Study. J. Am. Chem. Soc. 2000, 122, 2860–2866. [Google Scholar] [CrossRef]
- Ganeev, R.; Zvyagin, A.; Ovchinnikov, O.; Smirnov, M. Peculiarities of the nonlinear optical absorption of Methylene blue and Thionine in different solvents. Dye. Pigment. 2018, 149, 236–241. [Google Scholar] [CrossRef]
- Wainwright, M. The development of phenothiazinium photosensitisers. Photodiagn. Photodyn. Ther. 2005, 2, 263–272. [Google Scholar] [CrossRef]
- Rodrigues, G.B.; Dias-Baruffi, M.; Holman, N.; Wainwright, M.; Braga, G.U. In vitro photodynamic inactivation of Candida species and mouse fibroblasts with phenothiazinium photosensitisers and red light. Photodiagn. Photodyn. Ther. 2013, 10, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, M.; McLean, A. Rational design of phenothiazinium derivatives and photoantimicrobial drug discovery. Dye. Pigment. 2017, 136, 590–600. [Google Scholar] [CrossRef]
- Pereira, L.M.; Mota, C.M.; Baroni, L.; Da Costa, C.M.B.; Brochi, J.C.V.; Wainwright, M.; Mineo, T.W.P.; Braga, G.Ú.L.; Yatsuda, A.P. Inhibitory action of phenothiazinium dyes against Neospora caninum. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Santos, H.F.; Dos Santos, C.G.; Nascimento, O.R.; Reis, A.; Lanfredi, A.J.; De Oliveira, H.P.; Nantes-Cardoso, I.L. Charge separation of photosensitized phenothiazines for applications in catalysis and nanotechnology. Dye. Pigment. 2020, 177, 108314. [Google Scholar] [CrossRef]
- Pereţeanu, I.S.; Müller, T.J.J. Synthesis and electronic properties of 3,7-dianilino substituted N-hexyl phenothiazines. Org. Biomol. Chem. 2013, 11, 5127. [Google Scholar] [CrossRef]
- Wainwright, M.; Grice, N.J.; Pye, L.E. Phenothiazine photosensitizers: Part 2. 3,7-Bis(arylamino)phenothiazines. Dye. Pigment. 1999, 42, 45–51. [Google Scholar] [CrossRef]
- Lu, Y.-T.; Arai, C.; Ge, J.-F.; Ren, W.-S.; Kaiser, M.; Wittlin, S.; Brun, R.; Lu, J.-M.; Ihara, M. Synthesis and in vitro antiprotozoal activities of water-soluble, inexpensive phenothiazinium chlorides. Dye. Pigment. 2011, 89, 44–48. [Google Scholar] [CrossRef]
- Tiravia, M.; Sabuzi, F.; Cirulli, M.; Pezzola, S.; Di Carmine, G.; Cicero, D.O.; Floris, B.; Conte, V.; Galloni, P. 3,7-Bis(N -methyl-N -phenylamino)phenothiazinium Salt: Improved Synthesis and Aggregation Behavior in Solution. Eur. J. Org. Chem. 2019, 20, 3208–3216. [Google Scholar] [CrossRef]
- Khadieva, A.; Gorbachuk, V.; Shurpik, D.; Stoikov, I. Synthesis of Tris-pillar [5] arene and Its Association with Phenothiazine Dye: Colorimetric Recognition of Anions. Molecules 2019, 24, 1807. [Google Scholar] [CrossRef]
- Khadieva, A.I.; Gorbachuk, V.V.; Evtugyn, G.A.; Belyakova, S.V.; Latypov, R.R.; Drobyshev, S.V.; Stoikov, I.I. Phenyliminophenothiazine based self-organization of polyaniline nanowires and application as redox probe in electrochemical sensors. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Kuzin, Y.I.; Padnya, P.L.; Stoikov, I.I.; Gorbatchuk, V.V.; Stoikov, D.I.; Khadieva, A.I.; Evtugyn, G.A. Electrochemical behavior of the monomeric and polymeric forms of N-phenyl-3-(phenylimino)-3H-phenothiazin-7-amine. Electrochim. Acta 2020, 345, 136195. [Google Scholar] [CrossRef]
- Wainwright, M.; Meegan, K.; Loughran, C.; Giddens, R.M. Phenothiazinium photosensitisers, Part VI: Photobactericidal asymmetric derivatives. Dye. Pigment. 2009, 82, 387–391. [Google Scholar] [CrossRef]
- Khadieva, A.I.; Gorbachuk, V.V.; Stoikov, I.I. Synthesis and supramolecular self-assembly of phenothiazine functionalized by carboxyphenyl fragments. Russ. Chem. Bull. 2020, 69, 333–338. [Google Scholar] [CrossRef]
- Peterson, B.M.; Shen, L.; Lopez, G.J.; Gannett, C.N.; Ren, N.; Abruña, H.D.; Fors, B.P. Elucidation of the electrochemical behavior of phenothiazine-based polyaromatic amines. Tetrahedron 2019, 75, 4244–4249. [Google Scholar] [CrossRef]
- Oka, H.; Terane, M.; Kiyohara, Y.; Tanaka, H. Synthesis and magnetic behavior of stable organic open-shell polymers containing phenothiazine cation radicals as spin resources. Polyhedron 2007, 26, 1895–1900. [Google Scholar] [CrossRef]
- Andreani, F.; Bizzarri, P.C.; Casa, C.D.; Fiorini, M.; Salatelli, E. Ladder oligophenothiazines by direct thionation ofN-Arylanilino Derivatives. J. Heterocycl. Chem. 1991, 28, 295–299. [Google Scholar] [CrossRef]
- Kuzin, Y.I.; Khadieva, A.I.; Padnya, P.L.; Khannanov, A.A.; Kutyreva, M.P.; Stoikov, I.I.; Evtugyn, G.A. Electrochemistry of new derivatives of phenothiazine: Electrode kinetics and electropolymerization conditions. Electrochim. Acta 2021, 375, 137985. [Google Scholar] [CrossRef]
- Marin, L.; Bejan, A.; Shova, S. Phenothiazine based co-crystals with enhanced luminescence. Dye. Pigment. 2020, 175, 108164. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, C.; Li, X.; Liang, M.; Bian, W.; Zhang, Y.; Wang, K.; Xue, P. Fluorescence response of cruciform D–π–A–π–D phenothiazine derivatives to mechanical force. CrystEngComm 2019, 21, 4192–4199. [Google Scholar] [CrossRef]
- Wu, J.; Zeng, Z.; Chen, Q.; Zheng, J.; Xu, C. Thiophene and phenothiazine electrochromic copolymers with dual-state emission via tuning the distance of π-π stacking. Opt. Mater. 2019, 96, 109346. [Google Scholar] [CrossRef]
- Patil, K.; Pawar, R.; Talap, P. Self-aggregation of Methylene Blue in aqueous medium and aqueous solutions of Bu4NBr and urea. Phys. Chem. Chem. Phys. 2000, 2, 4313–4317. [Google Scholar] [CrossRef]
- Czímerová, A.; Bujdák, J.; Gáplovský, A. The aggregation of thionine and methylene blue dye in smectite dispersion. Colloids Surf. A Physicochem. Eng. Asp. 2004, 243, 89–96. [Google Scholar] [CrossRef]
- Ballard, R.E.; Park, C.H. Optical absorption bandshapes of Acridine Orange, Thionine, and Methylene Blue in monomeric and dimeric states. J. Chem. Soc. A 1970, 1340–1343. [Google Scholar] [CrossRef]
- Wainwright, M.; Amaral, L. Review: The phenothiazinium chromophore and the evolution of antimalarial drugs. Trop. Med. Int. Health 2005, 10, 501–511. [Google Scholar] [CrossRef]
- Vardevanyan, P.O.; Antonyan, A.P.; Parsadanyan, M.A.; Shahinyan, M.A.; Hambardzumyan, L.A. Mechanisms for Binding between Methylene Blue and DNA. J. Appl. Spectrosc. 2013, 80, 595–599. [Google Scholar] [CrossRef]
- Benesi, H.A.; Hildebrand, J.H. A Spectrophotometric Investigation of the Interaction of Iodine with Aromatic Hydrocarbons. J. Am. Chem. Soc. 1949, 71, 2703–2707. [Google Scholar] [CrossRef]
- Hajian, R.; Hossaini, P.; Mehrayin, Z.; Woi, P.M.; Shams, N. DNA-binding studies of valrubicin as a chemotherapy drug using spectroscopy and electrochemical techniques. J. Pharm. Anal. 2017, 7, 176–180. [Google Scholar] [CrossRef]
- Thordarson, P. Determining association constants from titration experiments in supramolecular chemistry. Chem. Soc. Rev. 2010, 40, 1305–1323. [Google Scholar] [CrossRef] [PubMed]
- Hibbert, D.B.; Thordarson, P. The death of the Job plot, transparency, open science and online tools, uncertainty estimation methods and other developments in supramolecular chemistry data analysis. Chem. Commun. 2016, 52, 12792–12805. [Google Scholar] [CrossRef]
- Bindfit v0.5 (Open Data Fit, 2016). Available online: http://app.supramolecular.org/bindfit/ (accessed on 3 May 2021).
- Wilson, B.; Fernandez, M.-J.; Lorente, A.; Grant, K.B. Synthesis and DNA interactions of a bis-phenothiazinium photosensitizer. Org. Biomol. Chem. 2008, 6, 4026. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Mati, S.S.; Kumar, G.S. Insights on the interaction of phenothiazinium dyes methylene blue and new methylene blue with synthetic duplex RNAs through spectroscopy and modeling. J. Photochem. Photobiol. B Biol. 2020, 204, 111804. [Google Scholar] [CrossRef]
- Juris, A.; Manfrin, M.F.; Maestri, M.; Serpone, N. Luminescence quenching of tris(2,2′-bipyridine) complexes of chromium(III), ruthenium(II), and osmium(II) by cyanide complexes. Inorg. Chem. 1978, 17, 2258–2261. [Google Scholar] [CrossRef]
- Creutz, C.; Sutin, N. Electron-transfer reactions of excited states. Reductive quenching of the tris(2,2′-bipyridine)ruthenium(II) luminescence. Inorg. Chem. 1976, 15, 496–499. [Google Scholar] [CrossRef]
- Zhang, L.Z.; Tang, G.-Q. The binding properties of photosensitizer methylene blue to herring sperm DNA: A spectroscopic study. J. Photochem. Photobiol. B Biol. 2004, 74, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Kumar, G.S. Spectroscopic studies on the binding interaction of phenothiazinium dyes toluidine blue O, azure A and azure B to DNA. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 107, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Kumar, G.S. Thionine Interaction to DNA: Comparative Spectroscopic Studies on Double Stranded Versus Single Stranded DNA. J. Fluoresc. 2011, 22, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; He, X.-W. Studies of interaction between Safranine T and double helix DNA by spectral methods. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 1998, 54, 883–892. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Masters, B.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Kluwer-Plenum: New York, NY, USA, 2008. [Google Scholar]
- Neese, F. The ORCA program system. WIRES Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Neese, F. Software update: The ORCA program system, version 4.0. WIRES Comput. Mol. Sci. 2017, 8. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khadieva, A.; Mostovaya, O.; Padnya, P.; Kalinin, V.; Grishaev, D.; Tumakov, D.; Stoikov, I. Arylamine Analogs of Methylene Blue: Substituent Effect on Aggregation Behavior and DNA Binding. Int. J. Mol. Sci. 2021, 22, 5847. https://doi.org/10.3390/ijms22115847
Khadieva A, Mostovaya O, Padnya P, Kalinin V, Grishaev D, Tumakov D, Stoikov I. Arylamine Analogs of Methylene Blue: Substituent Effect on Aggregation Behavior and DNA Binding. International Journal of Molecular Sciences. 2021; 22(11):5847. https://doi.org/10.3390/ijms22115847
Chicago/Turabian StyleKhadieva, Alena, Olga Mostovaya, Pavel Padnya, Valeriy Kalinin, Denis Grishaev, Dmitrii Tumakov, and Ivan Stoikov. 2021. "Arylamine Analogs of Methylene Blue: Substituent Effect on Aggregation Behavior and DNA Binding" International Journal of Molecular Sciences 22, no. 11: 5847. https://doi.org/10.3390/ijms22115847
APA StyleKhadieva, A., Mostovaya, O., Padnya, P., Kalinin, V., Grishaev, D., Tumakov, D., & Stoikov, I. (2021). Arylamine Analogs of Methylene Blue: Substituent Effect on Aggregation Behavior and DNA Binding. International Journal of Molecular Sciences, 22(11), 5847. https://doi.org/10.3390/ijms22115847