Plasma Extracellular Vesicle miRNAs Can Identify Lung Cancer, Current Smoking Status, and Stable COPD
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics of the Four Case Groups
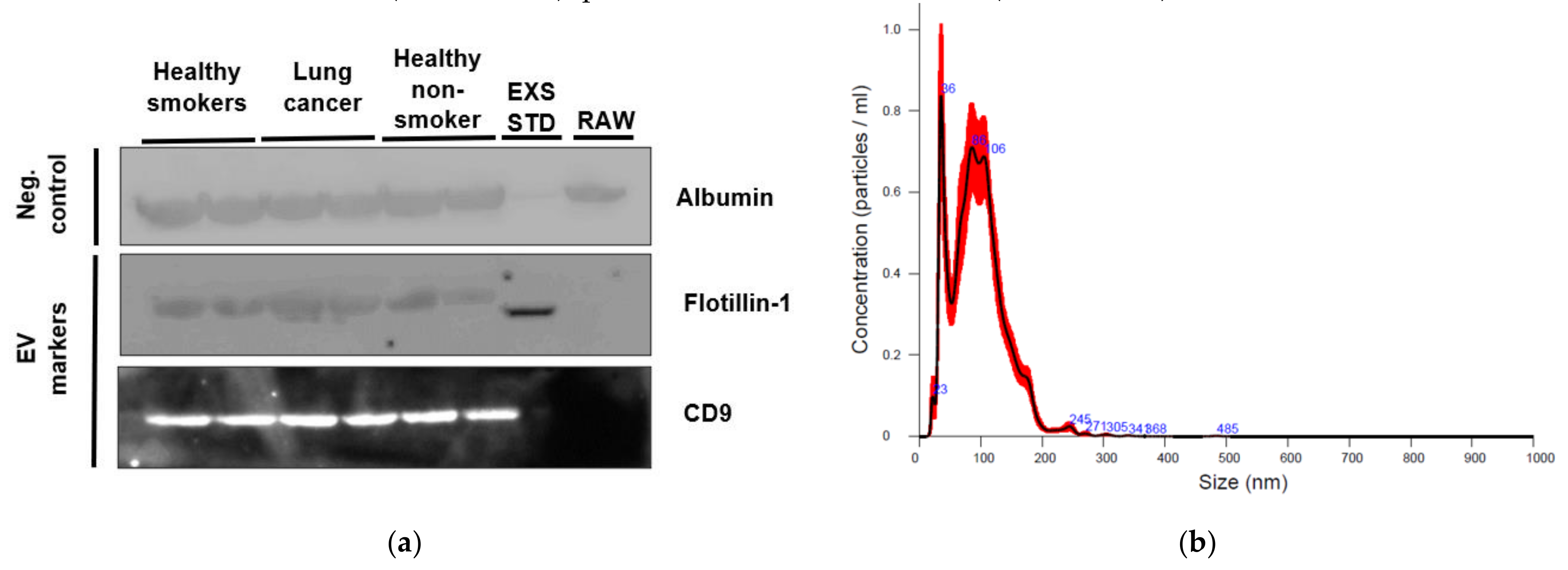
2.2. Characterization of EVs from Healthy Non-Smoking, Healthy Smoking, Lung Cancer, and Stable COPD Participants Plasma
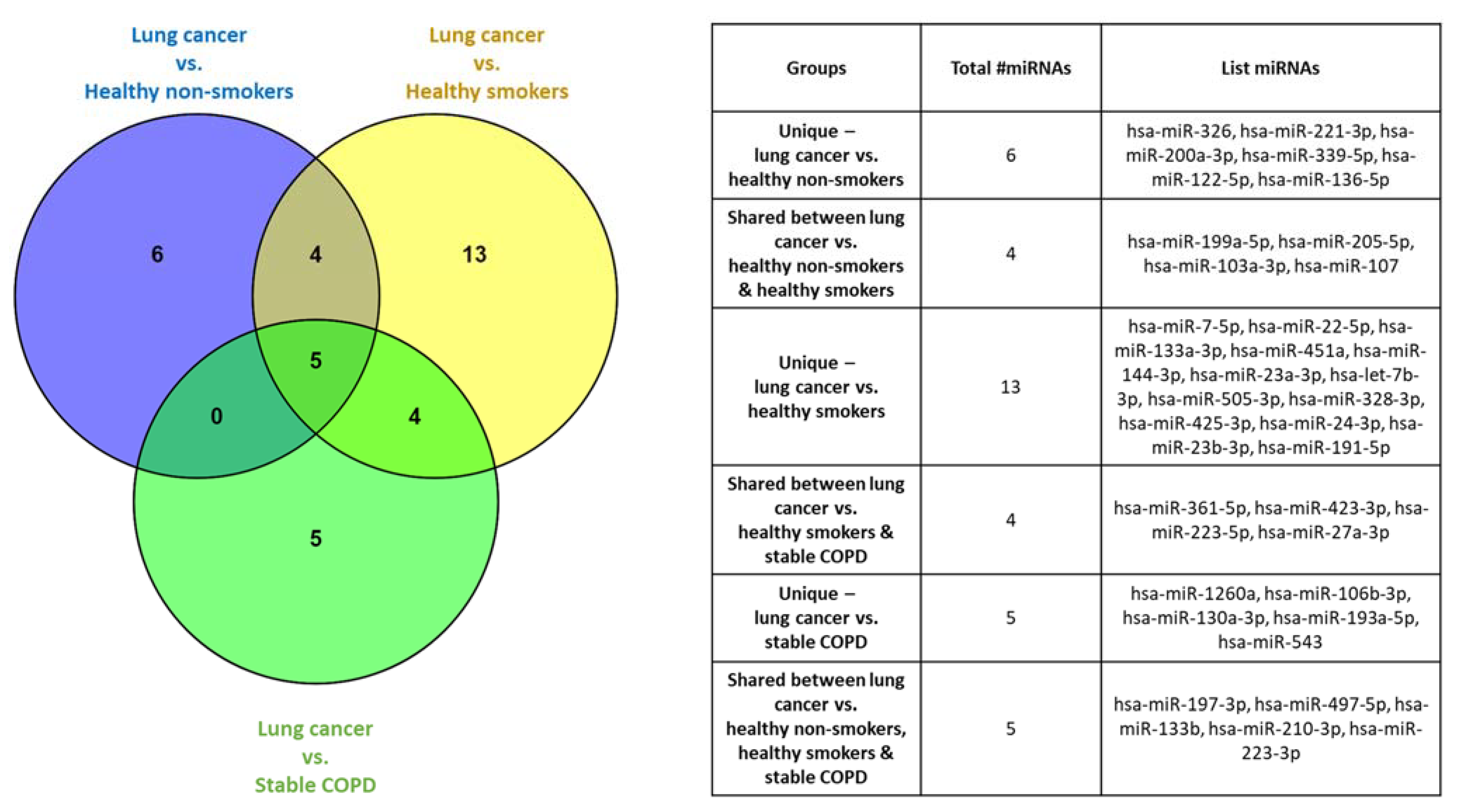
2.3. Plasma-Derived EV miRNA Profiling Identified Significantly Dysregulated miRNAs Specific for Select Cohorts
2.4. Correlations Identified between Significantly Dysregulated miRNAs and Clinical Characteristics
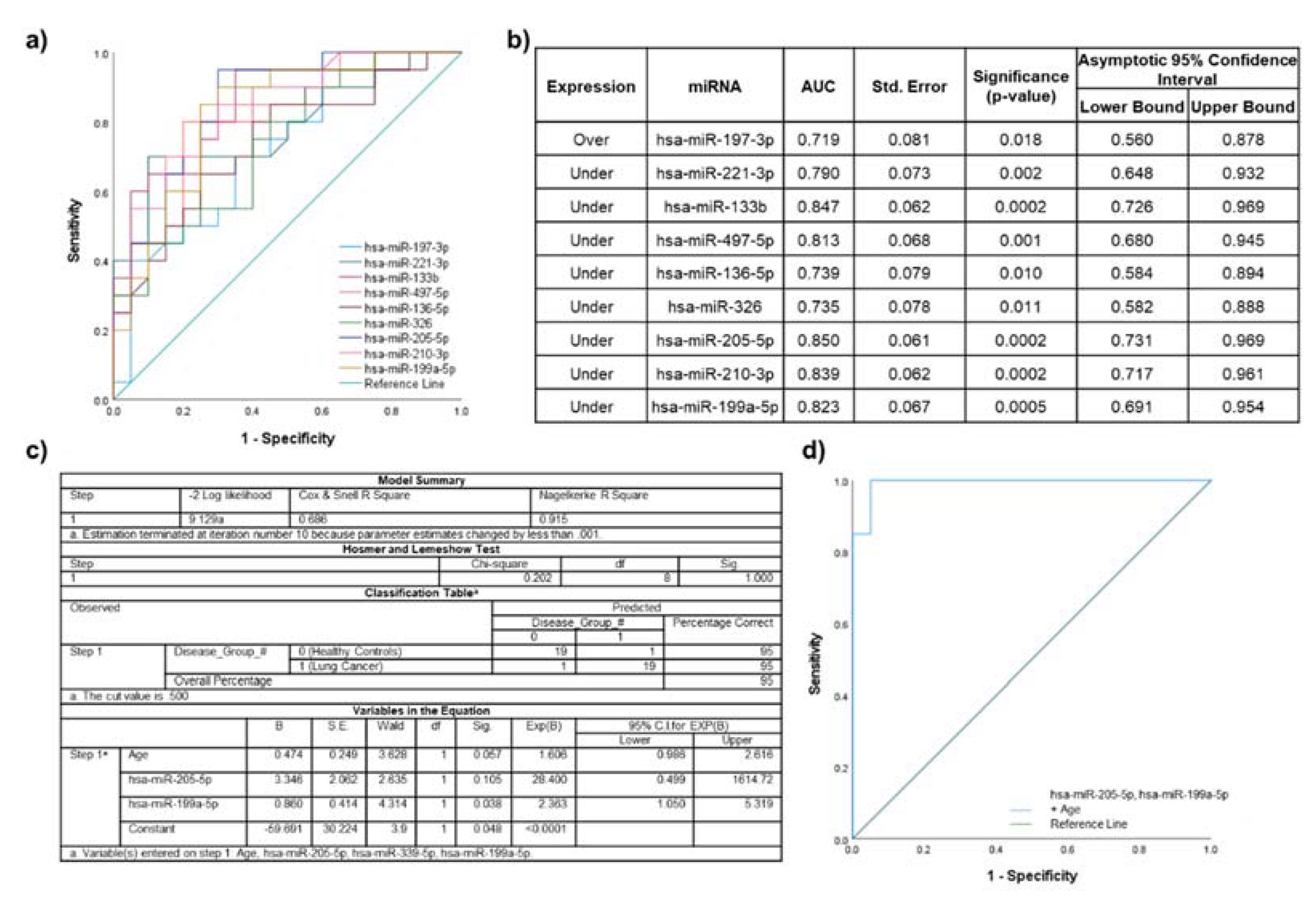
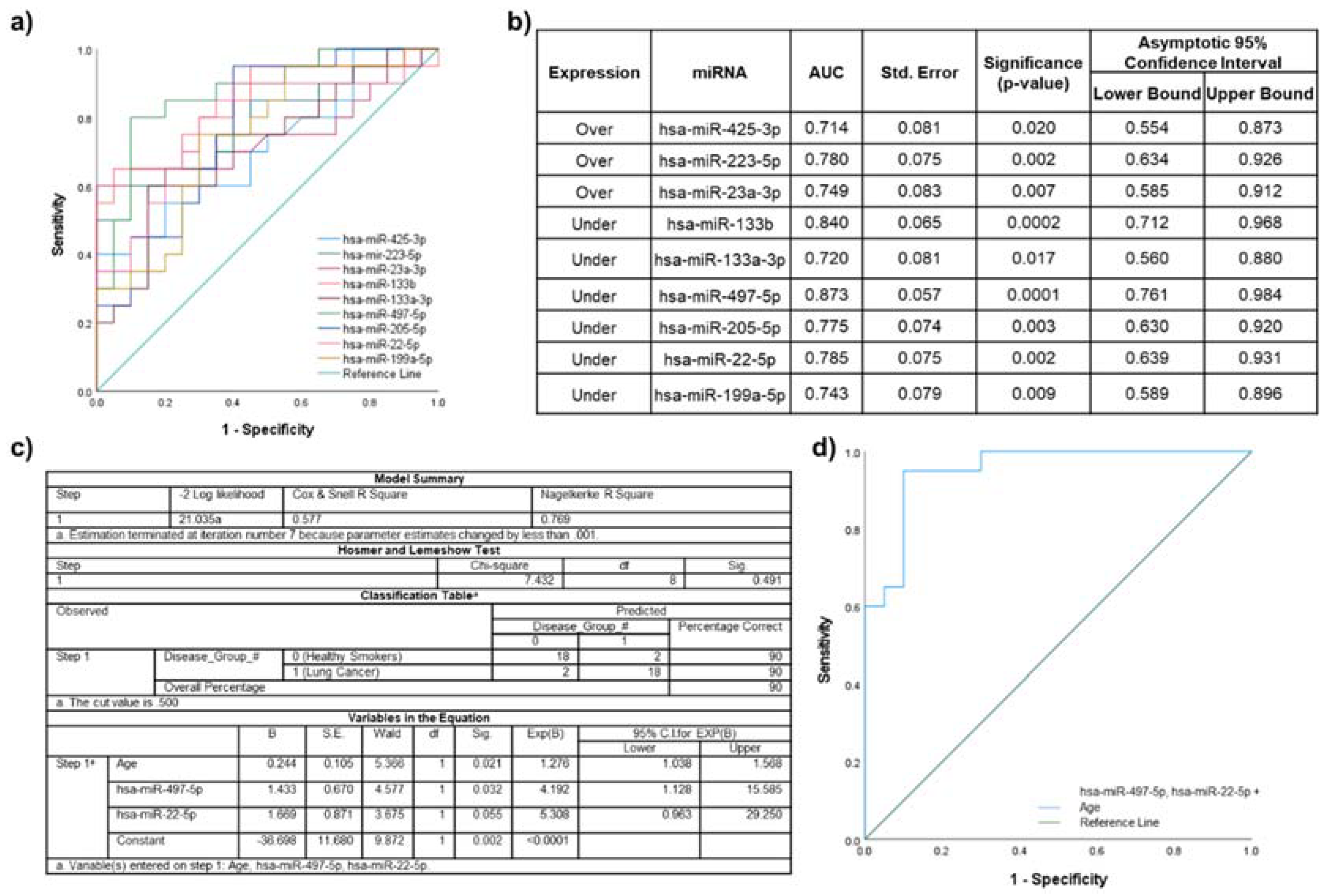
2.5. Biomarker Potential of Plasma EV miRNAs Assessed by ROC Curve Analyses
2.6. Lung Cancer Participants vs. Healthy Non-Smokers
2.7. Lung Cancer Participants vs. Healthy Smokers
2.8. Lung Cancer Participants vs. Stable COPD Participants
2.9. Biological Pathways Associated with miRNAs Differentially Expressed between Lung Cancer Participants and Healthy Non-Smoking Participants
3. Discussion
3.1. Main Results
3.2. Lung Disease Biomarker Potential of Plasma EV miRNAs
3.2.1. Lung Cancer Participants vs. Healthy Non-Smokers
3.2.2. Lung Disease Participant Cohorts
3.3. Identified miRNA-Regulated Target Genes in Each Patient Cohort and Their Involvement in Relevant Biological Pathways
3.4. Limitations
4. Materials and Methods
4.1. Statement of Ethics Approval
4.2. Blood Plasma Sample Collection and Processing
4.3. Plasma EV Isolation
4.4. Plasma EV Characterization
4.4.1. Nanoparticle Tracking Analysis
4.4.2. Western Blot of EV Markers
4.5. Plasma EV RNA Extraction and Purification
4.6. Plasma EV miRNA Profiling
4.6.1. Reverse Transcription
4.6.2. Quantitative PCR Using miRCURY LNA Serum/Plasma Focus PCR Panels
4.7. miRNA Function Enrichment Analysis
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jørgen Vestbo, S.S.H.; Agusti, A.G.; Jones, P.W.; Vogelmeier, C.; Anzueto, A.; Barnes, P.J.; Fabbri, L.M.; Martinez, F.J.; Nishimura, M.; Stockley, R.A.; et al. Global Strategy for the Diagnosis, Management, Prevention of Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2013, 187, 347–365. [Google Scholar] [CrossRef]
- WHO. COPD Predicted to Be Thrid Leading Cause of Death in 2030. Available online: http://www.who.int/respiratory/copd/World_Health_Statistics_2008/en/ (accessed on 10 January 2019).
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer statistics, 2009. CA Cancer J. Clin. 2009, 59, 225–249. [Google Scholar] [CrossRef]
- Houghton, A.M.; Mouded, M.; Shapiro, S.D. Common origins of lung cancer and COPD. Nat. Med. 2008, 14, 1023–1024. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.C.; Chen, Y.; Krewski, D.; Calle, E.E.; Thun, M.J. Chronic obstructive pulmonary disease is associated with lung cancer mortality in a prospective study of never smokers. Am. J. Respir. Crit. Care Med. 2007, 176, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Young, R.P.; Hopkins, R.J. Link between COPD and lung cancer. Respir. Med. 2010, 104, 758–759. [Google Scholar] [CrossRef]
- Sekine, Y.; Katsura, H.; Koh, E.; Hiroshima, K.; Fujisawa, T. Early detection of COPD is important for lung cancer surveillance. Eur. Respir. J. 2012, 39, 1230–1240. [Google Scholar] [CrossRef]
- Durham, A.L.; Adcock, I.M. The relationship between COPD and lung cancer. Lung Cancer 2015, 90, 121–127. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Parris, B.A.; O’Farrell, H.E.; Fong, K.M.; Yang, I.A. Chronic obstructive pulmonary disease (COPD) and lung cancer: Common pathways for pathogenesis. J. Thorac. Dis. 2019, 11, S2155–S2172. [Google Scholar] [CrossRef]
- Shivdasani, R.A. MicroRNAs: Regulators of gene expression and cell differentiation. Blood 2006, 108, 3646–3653. [Google Scholar] [CrossRef]
- Rebane, A.; Akdis, C.A. MicroRNAs: Essential players in the regulation of inflammation. J. Allergy Clin. Immunol. 2013, 132, 15–26. [Google Scholar] [CrossRef]
- Muth, D.C.; Powell, B.H.; Zhao, Z.; Witwer, K.W. miRNAs in platelet-poor blood plasma and purified RNA are highly stable: A confirmatory study. BMC Res. Notes 2018, 11, 273. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Z.; Zhou, S.; Wen, J.; Geng, B.; Yang, J.; Cui, Q. Genome-wide analysis of human microRNA stability. BioMed Res. Int. 2013, 2013, 368975. [Google Scholar] [CrossRef]
- Bail, S.; Swerdel, M.; Liu, H.; Jiao, X.; Goff, L.A.; Hart, R.P.; Kiledjian, M. Differential regulation of microRNA stability. RNA 2010, 16, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Baudrimont, A.; Voegeli, S.; Viloria, E.C.; Stritt, F.; Lenon, M.; Wada, T.; Jaquet, V.; Becskei, A. Multiplexed gene control reveals rapid mRNA turnover. Sci. Adv. 2017, 3, e1700006. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neuro-Oncol. 2013, 113, 1–11. [Google Scholar] [CrossRef]
- Nolte-’t Hoen, E.N.; Buermans, H.P.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.; t Hoen, P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef]
- Pattarayan, D.; Thimmulappa, R.K.; Ravikumar, V.; Rajasekaran, S. Diagnostic Potential of Extracellular MicroRNA in Respiratory Diseases. Clin. Rev. Allergy Immunol. 2018, 54, 480–492. [Google Scholar] [CrossRef]
- Benedikter, B.J.; Wouters, E.F.M.; Savelkoul, P.H.M.; Rohde, G.G.U.; Stassen, F.R.M. Extracellular vesicles released in response to respiratory exposures: Implications for chronic disease. J. Toxicol. Environ. Health Part B 2018, 21, 142–160. [Google Scholar] [CrossRef]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347. [Google Scholar] [CrossRef]
- Anderson, N.L.; Anderson, N.G. The human plasma proteome: History, character, and diagnostic prospects. Mol. Cell. Proteom. MCP 2002, 1, 845–867. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 20 May 2021).
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef] [PubMed]
- O’Farrell, H.E.; Yang, I.A. Extracellular vesicles in chronic obstructive pulmonary disease (COPD). J. Thorac. Dis. 2019, 11, S2141–S2154. [Google Scholar] [CrossRef]
- Young, R.P.; Whittington, C.F.; Hopkins, R.J.; Hay, B.A.; Epton, M.J.; Black, P.N.; Gamble, G.D. Chromosome 4q31 locus in COPD is also associated with lung cancer. Eur. Respir. J. 2010, 36, 1375–1382. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-H.; Sun, S.-S.; Li, N.; Lv, P.; Xie, S.-Y.; Wang, P.-Y. MiR-205 as a promising biomarker in the diagnosis and prognosis of lung cancer. Oncotarget 2017, 8, 91938–91949. [Google Scholar] [CrossRef]
- Larzabal, L.; de Aberasturi, A.L.; Redrado, M.; Rueda, P.; Rodriguez, M.J.; Bodegas, M.E.; Montuenga, L.M.; Calvo, A. TMPRSS4 regulates levels of integrin α5 in NSCLC through miR-205 activity to promote metastasis. Br. J. Cancer 2014, 110, 764–774. [Google Scholar] [CrossRef]
- Chen, J.; Hu, C.; Pan, P. Extracellular Vesicle MicroRNA Transfer in Lung Diseases. Front. Physiol. 2017, 8, 1028. [Google Scholar] [CrossRef]
- Xie, L.; Wu, M.; Lin, H.; Liu, C.; Yang, H.; Zhan, J.; Sun, S. An increased ratio of serum miR-21 to miR-181a levels is associated with the early pathogenic process of chronic obstructive pulmonary disease in asymptomatic heavy smokers. Mol. Biosyst. 2014, 10, 1072–1081. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Gadalla, R.; El-Ghonaimy, E.A.; Samir, O.; Mohamed, H.T.; Hassan, H.; Greve, B.; El-Shinawi, M.; Mohamed, M.M.; Götte, M. Syndecan-1 is a novel molecular marker for triple negative inflammatory breast cancer and modulates the cancer stem cell phenotype via the IL-6/STAT3, Notch and EGFR signaling pathways. Mol. Cancer 2017, 16, 57. [Google Scholar] [CrossRef]
- Parimon, T.; Brauer, R.; Schlesinger, S.Y.; Xie, T.; Jiang, D.; Ge, L.; Huang, Y.; Birkland, T.P.; Parks, W.C.; Habiel, D.M.; et al. Syndecan-1 Controls Lung Tumorigenesis by Regulating miRNAs Packaged in Exosomes. Am. J. Pathol. 2018, 188, 1094–1103. [Google Scholar] [CrossRef]
- Lou, C.; Xiao, M.; Cheng, S.; Lu, X.; Jia, S.; Ren, Y.; Li, Z. MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1α expression. Cell Death Dis. 2016, 7, e2159. [Google Scholar] [CrossRef]
- Aushev, V.N.; Zborovskaya, I.B.; Laktionov, K.K.; Girard, N.; Cros, M.P.; Herceg, Z.; Krutovskikh, V. Comparisons of microRNA patterns in plasma before and after tumor removal reveal new biomarkers of lung squamous cell carcinoma. PLoS ONE 2013, 8, e78649. [Google Scholar] [CrossRef]
- Del Vescovo, V.; Cantaloni, C.; Cucino, A.; Girlando, S.; Silvestri, M.; Bragantini, E.; Fasanella, S.; Cuorvo, L.V.; Palma, P.D.; Rossi, G.; et al. miR-205 Expression levels in nonsmall cell lung cancer do not always distinguish adenocarcinomas from squamous cell carcinomas. Am. J. Surg. Pathol. 2011, 35, 268–275. [Google Scholar] [CrossRef]
- Endzeliņš, E.; Berger, A.; Melne, V.; Bajo-Santos, C.; Soboļevska, K.; Ābols, A.; Rodriguez, M.; Šantare, D.; Rudņickiha, A.; Lietuvietis, V.; et al. Detection of circulating miRNAs: Comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 2017, 17, 730. [Google Scholar] [CrossRef]
- Leng, Q.; Lin, Y.; Jiang, F.; Lee, C.-J.; Zhan, M.; Fang, H.; Wang, Y.; Jiang, F. A plasma miRNA signature for lung cancer early detection. Oncotarget 2017, 8, 111902–111911. [Google Scholar] [CrossRef]
- Ma, J.; Mannoor, K.; Gao, L.; Tan, A.; Guarnera, M.A.; Zhan, M.; Shetty, A.; Stass, S.A.; Xing, L.; Jiang, F. Characterization of microRNA transcriptome in lung cancer by next-generation deep sequencing. Mol. Oncol. 2014, 8, 1208–1219. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.; Yang, Z.; Li, Y.; Han, B.; Chen, L.A. miR-339-5p inhibits metastasis of non-small cell lung cancer by regulating the epithelial-to-mesenchymal transition. Oncol. Lett. 2018, 15, 2508–2514. [Google Scholar] [CrossRef]
- Sun, Y.; Mei, H.; Xu, C.; Tang, H.; Wei, W. Circulating microRNA-339-5p and -21 in plasma as an early detection predictors of lung adenocarcinoma. Pathol. Res. Pract. 2018, 214, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Sanfiorenzo, C.; Ilie, M.I.; Belaid, A.; Barlési, F.; Mouroux, J.; Marquette, C.H.; Brest, P.; Hofman, P. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS ONE 2013, 8, e54596. [Google Scholar] [CrossRef]
- Xu, J.; Wang, T.; Cao, Z.; Huang, H.; Li, J.; Liu, W.; Liu, S.; You, L.; Zhou, L.; Zhang, T.; et al. MiR-497 downregulation contributes to the malignancy of pancreatic cancer and associates with a poor prognosis. Oncotarget 2014, 5, 6983–6993. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.T.; Jiang, C.C.; Wang, G.P.; Li, Y.P.; Wang, C.Y.; Guo, X.Y.; Yang, R.H.; Feng, Y.; Wang, F.H.; Tseng, H.Y.; et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene 2013, 32, 1910–1920. [Google Scholar] [CrossRef]
- Qu, F.; Ye, J.; Pan, X.; Wang, J.; Gan, S.; Chu, C.; Chu, J.; Zhang, X.; Liu, M.; He, H.; et al. MicroRNA-497-5p down-regulation increases PD-L1 expression in clear cell renal cell carcinoma. J. Drug Target. 2019, 27, 67–74. [Google Scholar] [CrossRef]
- Zheng, D.; Haddadin, S.; Wang, Y.; Gu, L.Q.; Perry, M.C.; Freter, C.E.; Wang, M.X. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int. J. Clin. Exp. Pathol. 2011, 4, 575–586. [Google Scholar]
- Mavridis, K.; Gueugnon, F.; Petit-Courty, A.; Courty, Y.; Barascu, A.; Guyetant, S.; Scorilas, A. The oncomiR miR-197 is a novel prognostic indicator for non-small cell lung cancer patients. Br. J. Cancer 2015, 112, 1527–1535. [Google Scholar] [CrossRef]
- Yin, Q.; Han, Y.; Zhu, D.; Li, Z.; Shan, S.; Jin, W.; Lu, Q.; Ren, T. miR-145 and miR-497 suppress TGF-β-induced epithelial-mesenchymal transition of non-small cell lung cancer by targeting MTDH. Cancer Cell Int. 2018, 18, 105. [Google Scholar] [CrossRef]
- Zhao, W.-y.; Wang, Y.; An, Z.-j.; Shi, C.-g.; Zhu, G.-a.; Wang, B.; Lu, M.-y.; Pan, C.-k.; Chen, P. Downregulation of miR-497 promotes tumor growth and angiogenesis by targeting HDGF in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2013, 435, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, L.; Liu, W.; Li, F. MicroRNA-497-5p inhibits proliferation and invasion of non-small cell lung cancer by regulating FGF2. Oncol. Lett. 2019, 17, 3425–3431. [Google Scholar] [CrossRef]
- Wang, W.-S.; Liu, L.-X.; Li, G.-P.; Chen, Y.; Li, C.-Y.; Jin, D.-Y.; Wang, X.-L. Combined Serum CA19-9 and miR-27a-3p in Peripheral Blood Mononuclear Cells to Diagnose Pancreatic Cancer. Cancer Prev. Res. 2013, 6, 331. [Google Scholar] [CrossRef] [PubMed]
- Zuo, D.; Chen, L.; Liu, X.; Wang, X.; Xi, Q.; Luo, Y.; Zhang, N.; Guo, H. Combination of miR-125b and miR-27a enhances sensitivity and specificity of AFP-based diagnosis of hepatocellular carcinoma. Tumor Biol. 2016, 37, 6539–6549. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Qian, K.; Wei, X.; Deng, H.; Zhao, B.; Chen, Q.; Zhang, J.; Liu, H. miR-27a promotes proliferation, migration, and invasion of colorectal cancer by targeting FAM172A and acts as a diagnostic and prognostic biomarker. Oncol. Rep. 2017, 37, 3554–3564. [Google Scholar] [CrossRef]
- Szczyrek, M.; Kuźnar-Kamińska, B.; Grenda, A.; Krawczyk, P.; Sawicki, M.; Głogowski, M.; Balicka, G.; Rolska-Kopińska, A.; Nicoś, M.; Jakimiec, M.; et al. Diagnostic value of plasma expression of microRNAs complementary to Drosha and Dicer in lung cancer patients. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3857–3866. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Pan, B.; Sun, L.; Chen, X.; Zeng, K.; Hu, X.; Xu, T.; Xu, M.; Wang, S. Circulating Exosomal miR-27a and miR-130a Act as Novel Diagnostic and Prognostic Biomarkers of Colorectal Cancer. Cancer Epidemiol. Prev. Biomark. 2018, 27, 746. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Chen, H.; Xu, C.; Zhang, Y.; Zhang, Q.; Chen, L.; Ding, Q.; Deng, Z. Exosomal miR-106b serves as a novel marker for lung cancer and promotes cancer metastasis via targeting PTEN. Life Sci. 2020, 244, 117297. [Google Scholar] [CrossRef] [PubMed]
- Soeda, S.; Ohyashiki, J.H.; Ohtsuki, K.; Umezu, T.; Setoguchi, Y.; Ohyashiki, K. Clinical relevance of plasma miR-106b levels in patients with chronic obstructive pulmonary disease. Int. J. Mol. Med. 2013, 31, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Xu, J.; Liu, H.; Zhao, Z. Peripheral leukocyte microRNAs as novel biomarkers for COPD. Int. J. Chron Obs. Pulmon Dis. 2017, 12, 1101–1112. [Google Scholar] [CrossRef]
- Wu, F.; Yin, Z.; Yang, L.; Fan, J.; Xu, J.; Jin, Y.; Yu, J.; Zhang, D.; Yang, G. Smoking Induced Extracellular Vesicles Release and Their Distinct Properties in Non-Small Cell Lung Cancer. J. Cancer 2019, 10, 3435–3443. [Google Scholar] [CrossRef]
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.-P.; Belting, M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380. [Google Scholar] [CrossRef]
- Zheng, Y. Study Design Considerations for Cancer Biomarker Discoveries. J. Appl. Lab. Med. 2018, 3, 282–289. [Google Scholar] [CrossRef]
- Diamandis, E.P. Cancer biomarkers: Can we turn recent failures into success? J. Natl. Cancer Inst. 2010, 102, 1462–1467. [Google Scholar] [CrossRef]
- Ding, M.; Wang, C.; Lu, X.; Zhang, C.; Zhou, Z.; Chen, X.; Zhang, C.-Y.; Zen, K.; Zhang, C. Comparison of commercial exosome isolation kits for circulating exosomal microRNA profiling. Anal. Bioanal. Chem. 2018, 410, 3805–3814. [Google Scholar] [CrossRef]
- Moon, S.; Shin, D.W.; Kim, S.; Lee, Y.-S.; Mankhong, S.; Yang, S.W.; Lee, P.H.; Park, D.-H.; Kwak, H.-B.; Lee, J.-S.; et al. Enrichment of Exosome-Like Extracellular Vesicles from Plasma Suitable for Clinical Vesicular miRNA Biomarker Research. J. Clin. Med. 2019, 8, 1995. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; da Silva, A.M.; Calin, G.; Pantel, K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef] [PubMed]


| Clinical Characteristic | Healthy Non-Smokers | Healthy Smokers | Lung Cancer | COPD (Stable) | Total | Significance |
|---|---|---|---|---|---|---|
| Gender (n, %) | ||||||
| Males | 7 (35) | 13 (65) | 10 (50) | 12 (60) | 42 (52.5) | p = 0.240 " 1 |
| Females | 13 (65) | 7 (35) | 10 (50) | 8 (40) | 38 (47.5) | |
| Age (mean, SD) | 55.1 (7.0) | 60.4 (4.2) | 66.7 (7.3) | 70.3 (7.6) | 63.2 (8.6) | p < 0.0001 * " 2 |
| Smoking history (n, %) | ||||||
| Never | 20 (100) | 0 | 0 | 1 (5) | 21 (26.3) | p = 0.02 * " 2 |
| Former | - | 8 (40) | 14 (70) | 15 (75) | 37 (46.3) | |
| Current | - | 12 (60) | 5 (25) | 4 (20) | 22 (27.5) | |
| Pack years (mean, SD) | - | 46.2 (15.8) | 39.8 (23.2) | 58.2 (23.2) | 48.3 (21.8) | p = 0.058 " 2 |
| TNM (n, %) | ||||||
| IA or B | - | - | 9 (45) | - | - | - |
| IIA or B | - | - | 2 (10) | - | - | - |
| IIIA or B | - | - | 1 (5) | - | - | - |
| IVA or B | - | - | 6 (30) | - | - | - |
| NSCLC subtype | ||||||
| Adenocarcinoma | 14 (70) | |||||
| Squamous cell carcinoma | 2 (10) | |||||
| Non-small cell carcinoma | 4 (20) | |||||
| GOLD stage (n, %) | ||||||
| GOLD 0 | - | - | - | 3 (15) | - | - |
| GOLD 1 | - | - | - | 0 | - | - |
| GOLD 2 | - | - | - | 8 (40) | - | - |
| GOLD 3 | - | - | - | 5 (25) | - | - |
| GOLD 4 | - | - | - | 4 (20) | - | - |
| Healthy Non-Smokers | Healthy Smokers | COPD (Stable) | |
|---|---|---|---|
| Lung cancer | miRNAs underexpressed (fold regulation; significance) | ||
| hsa-miR-221-3p (-2.2; p = 0.001 1) hsa-miR-200a-3p (-3.43; p = 0.013 1) hsa-miR-133b (-9.08; p < 0.0001 1) hsa-miR-497-5p (-4.76; p = 0.001 2) hsa-miR-136-5p (-3.6; p = 0.01 2) hsa-miR-326 (-4.47; p = 0.011 2) hsa-miR-107 (-2.27; p < 0.0001 1) hsa-miR-205-5p (-4.45; p = 0.0001 2) hsa-miR-339-5p (-2.62; p = 0.002 1) hsa-miR-122-5p (-2.6; p = 0.032 2) hsa-miR-103a-3p (-2.14; p < 0.0001 2) hsa-miR-210-3p (-2.37; p = 0.0002 2) hsa-miR-199a-5p (-8.17; p = 0.0005 2) | hsa-miR-133b (-6.69; p = 0.0001 1) hsa-miR-133a-3p (-4.9; p = 0.017 2) hsa-miR-451a (-2.18; p < 0.0001 2) hsa-miR-497-5p (-5.12; p = 0.0001 2) hsa-miR-107 (-2.19; p < 0.0001 1) hsa-miR-205-5p (-3.74; p = 0.003 2) hsa-miR-22-5p (-2.07; p = 0.002 2) hsa-miR-103a-3p (-2.02; p < 0.0001 1) hsa-miR-210-3p (-2.14; p = 0.001 2) hsa-miR-199a-5p (-4.56; p = 0.009 2) hsa-miR-144-3p (-2.17; p < 0.0001 1) hsa-miR-7-5p (-2.85; p = 0.014 2) | hsa-miR-133b (-4.75; p = 0.001 2) hsa-miR-130a-3p (-2.09; p < 0.0001 1) hsa-miR-497-5p (-3.15; p = 0.008 2) hsa-miR-1260a (-2.53; p < 0.0001 1) hsa-miR-210-3p (-2.29; p = 0.0003 2) hsa-miR-543 (-2.36; p = 0.027 2) | |
| miRNAs overexpressed (fold regulation; significance) | |||
| hsa-miR-197-3p (2.45; p = 0.019 2) hsa-miR-223-3p (2.49; p = 0.017 2) | hsa-miR-505-3p (2.27; p = 0.022 2) hsa-miR-23b-3p (2.12; p = 0.002 2) hsa-miR-361-5p (2.18; p = 0.001 2) hsa-miR-27a-3p (2.06; p = 0.0002 1) hsa-miR-328-3p (2.55; p = 0.007 2) hsa-let-7b-3p (2.05; p = 0.002 2) hsa-miR-425-3p (2.12; p = 0.021 2) hsa-miR-197-3p (3.31; p < 0.0001 1) hsa-miR-223-5p (3.75; p = 0.002 2) hsa-miR-191-5p (2.11; p < 0.0001 1) hsa-miR-23a-3p (2.01; p = 0.007 2) hsa-miR-423-3p (2.26; p = 0.007 1) hsa-miR-223-3p (4.52; p < 0.0001 2) hsa-miR-24-3p (2.12; p = 0.0002 1) | hsa-miR-361-5p (2.08; p = 0.002 1) hsa-miR-27a-3p (2.06; p = 0.0005 1) hsa-miR-197-3p (2.26; p = 0.004 1) hsa-miR-223-5p (3; p = 0.042 2) hsa-miR-193a-5p (2.18; p = 0.007 2) hsa-miR-423-3p (2.23; p = 0.01 1) hsa-miR-223-3p (3.54; p = 0.001 2) hsa-miR-106b-3p (3.37; p = 0.004 2) | |
| Patient Groups | KEGG Pathway | Significance | # miRNAs | List of Identified Target Genes | # Genes Validated from GeneGlobe Analysis |
|---|---|---|---|---|---|
| Lung cancer vs. healthy non-smokers | Fatty acid biosynthesis (hsa00061) | p < 0.0001 1 | 2 | FASN ACSL3 | 2 |
| Fatty acid metabolism (hsa01212) | p < 0.0001 1 | 10 | FASN ACSL3 PTPLB SCD5 ACAT2 ELOVL2 ELOVL6 ACAT1 ACADM MECR | 4 | |
| Proteoglycans in cancer (hsa05205) | p = 0.009 1 | 12 | BRAF WNT16 WNT7A SOS2 CBL ROCK2 FZD6 ITGA5 MRAS ANK2 COL21A1 ANK3 SLC9A1 FZD4 FZD10 IGF2 EIF4B DDX5 PLAUR FGF2 TGFB2 ANK1 AKT3 WNT3A SMO VEGFA FGFR1 KDR GRB2 WNT7B MDM2 ELK1 PRKACB | 21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Farrell, H.E.; Bowman, R.V.; Fong, K.M.; Yang, I.A. Plasma Extracellular Vesicle miRNAs Can Identify Lung Cancer, Current Smoking Status, and Stable COPD. Int. J. Mol. Sci. 2021, 22, 5803. https://doi.org/10.3390/ijms22115803
O’Farrell HE, Bowman RV, Fong KM, Yang IA. Plasma Extracellular Vesicle miRNAs Can Identify Lung Cancer, Current Smoking Status, and Stable COPD. International Journal of Molecular Sciences. 2021; 22(11):5803. https://doi.org/10.3390/ijms22115803
Chicago/Turabian StyleO’Farrell, Hannah E., Rayleen V. Bowman, Kwun M. Fong, and Ian A. Yang. 2021. "Plasma Extracellular Vesicle miRNAs Can Identify Lung Cancer, Current Smoking Status, and Stable COPD" International Journal of Molecular Sciences 22, no. 11: 5803. https://doi.org/10.3390/ijms22115803
APA StyleO’Farrell, H. E., Bowman, R. V., Fong, K. M., & Yang, I. A. (2021). Plasma Extracellular Vesicle miRNAs Can Identify Lung Cancer, Current Smoking Status, and Stable COPD. International Journal of Molecular Sciences, 22(11), 5803. https://doi.org/10.3390/ijms22115803






