Genome-Wide Role of HSF1 in Transcriptional Regulation of Desiccation Tolerance in the Anhydrobiotic Cell Line, Pv11
Abstract
1. Introduction
2. Results
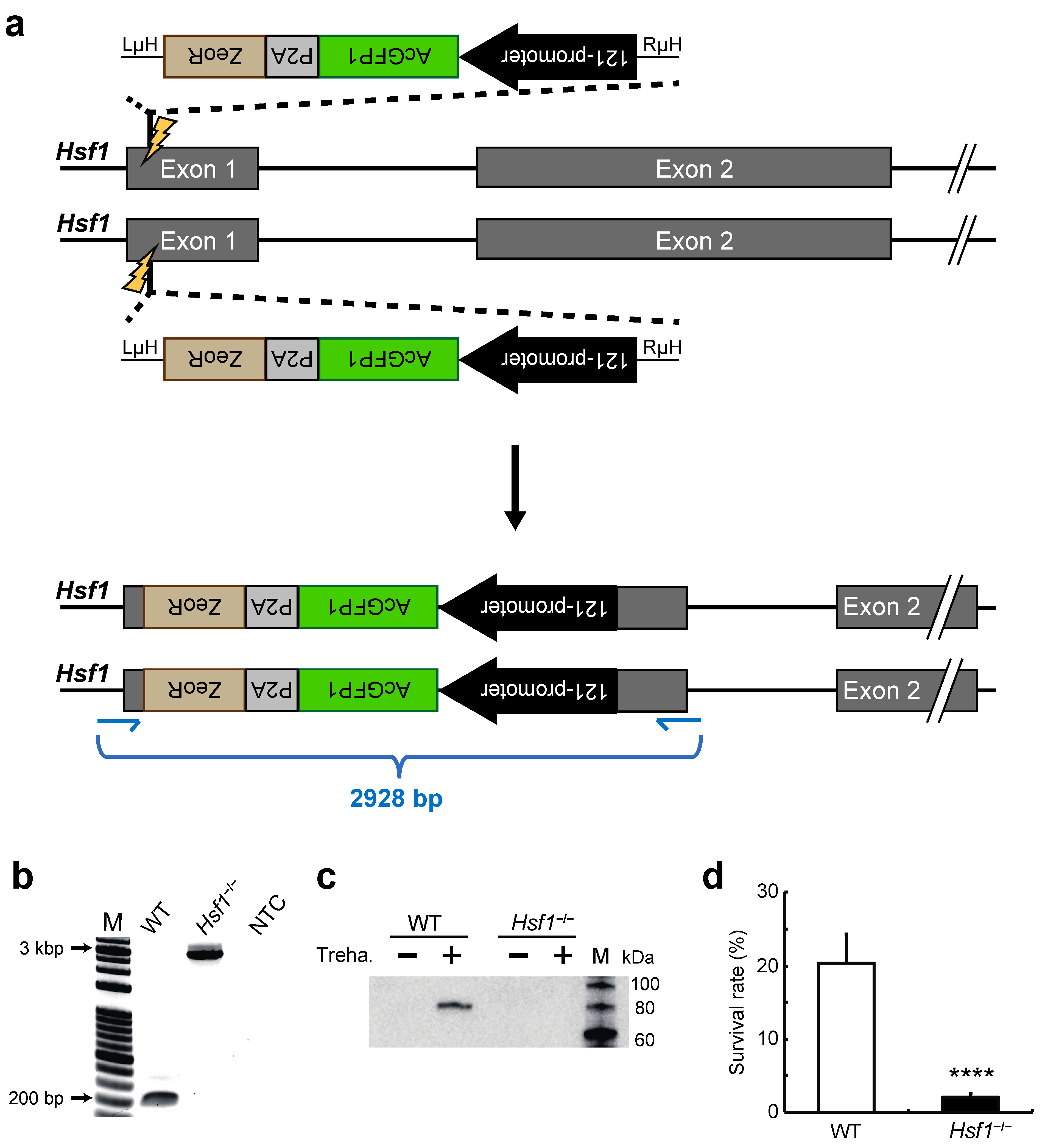
2.1. Establishment of a HSF1-Knockout Clonal Cell Line Using the CRIS-PITCh Technique
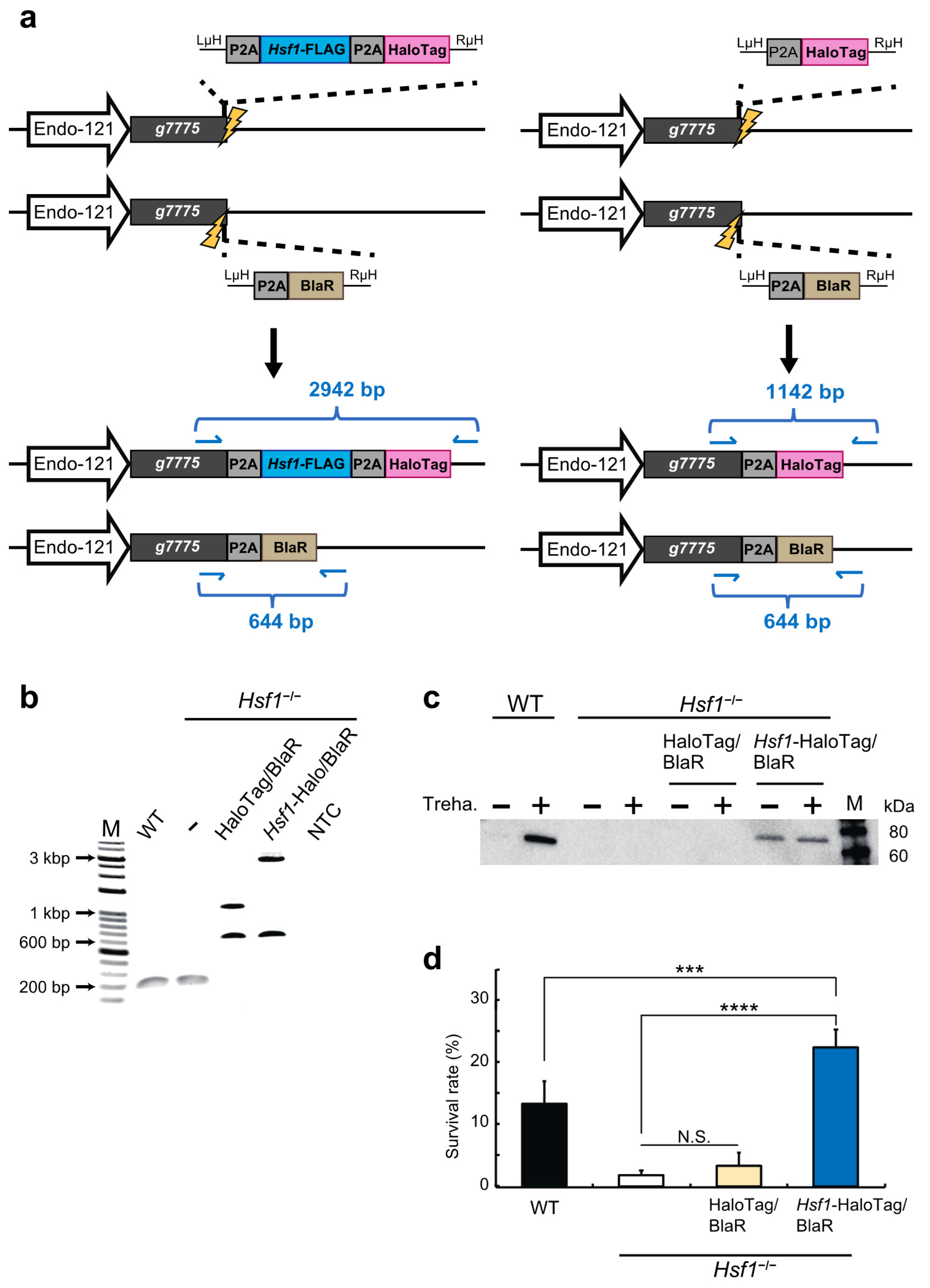
2.2. HSF1 Rescue of the HSF1-Knockout Cell Line Using the CRIS-PITCh Technique
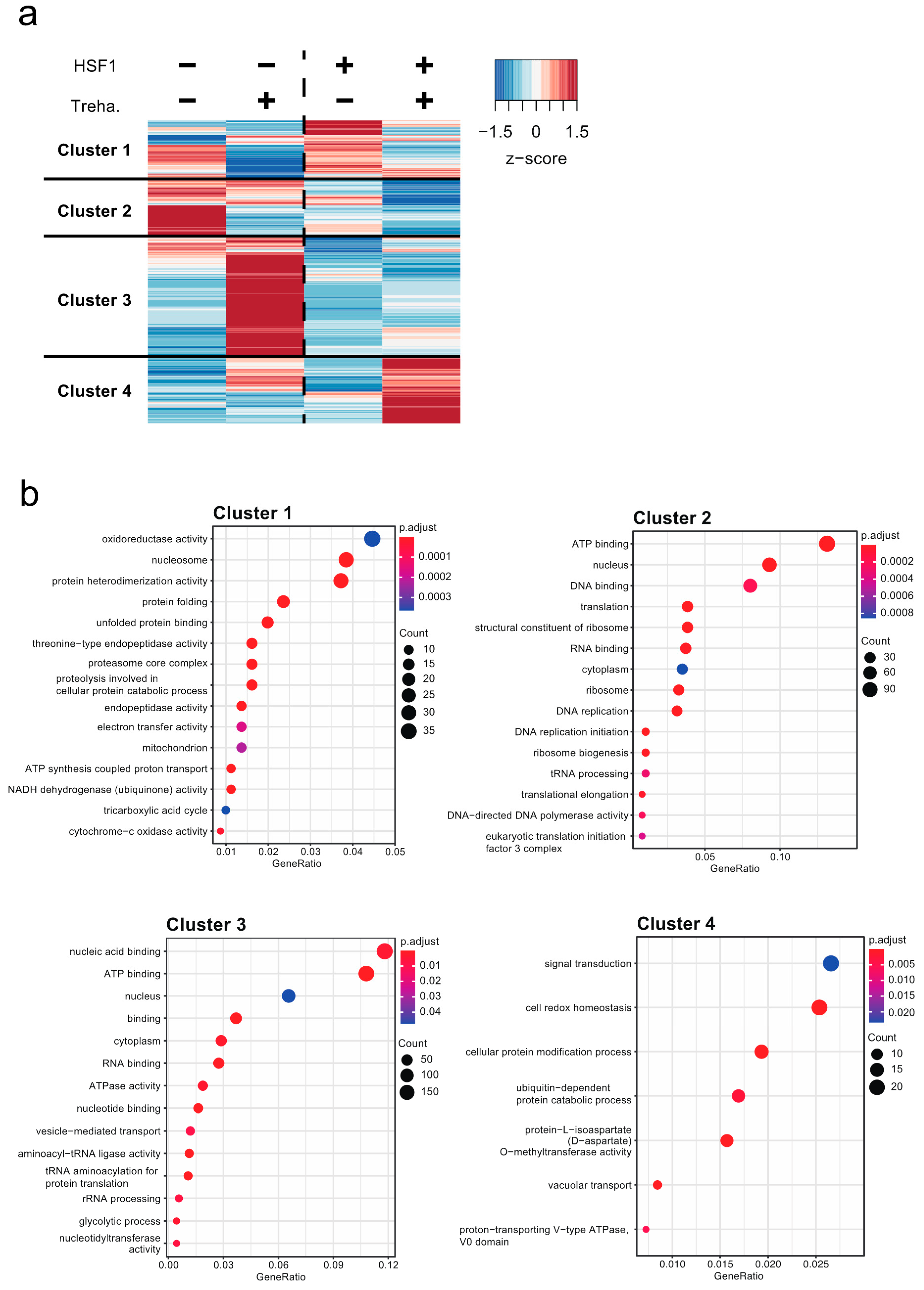
2.3. Contribution of HSF1 to the Regulation of Genes Crucial for Anhydrobiosis
2.4. Direct Transcriptional Regulation of Cluster 4 Genes by HSF1
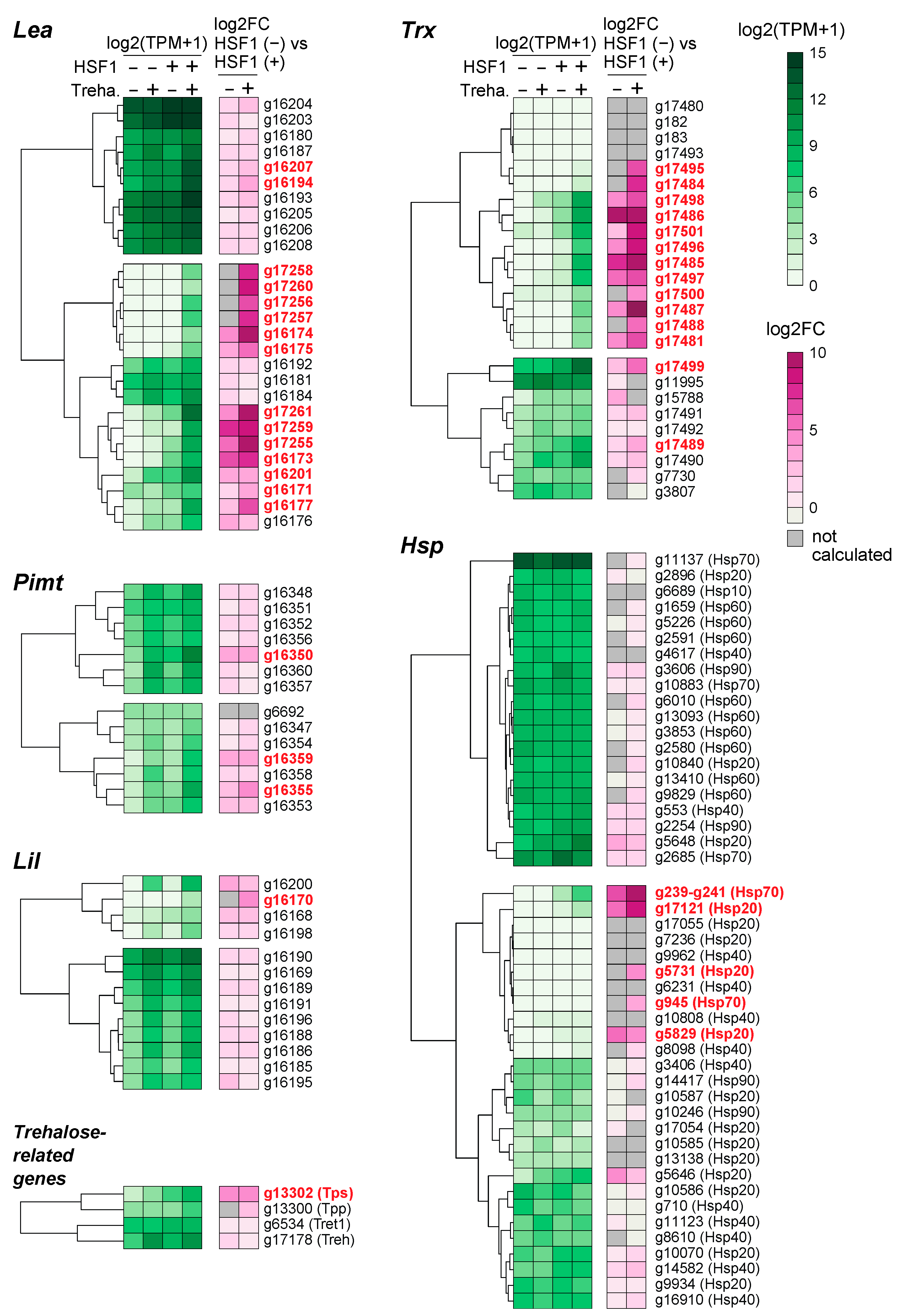
2.5. Differential Contribution of HSF1 to the Expression of Anhydrobiosis-Related Genes
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Vector Construction
4.3. Transfection and Cell Sorting
4.4. Genomic PCR and Western Blotting
4.5. Desiccation-Rehydration Experiment and Calculation of Survival Rate
4.6. HSP Gene Prediction
4.7. Library Preparation for Illumina Sequencing
4.8. Quality Control and Trimming
4.9. CAGE Libraries Processing and Gene Model Update
4.10. mRNA-Seq Library Processing and Analysis
4.11. Motif Enrichment Analysis
4.12. ChIP-qPCR
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rothschild, L.J.; Mancinelli, R.L. Life in extreme environments. Nature 2001, 409, 1092–1101. [Google Scholar] [CrossRef]
- Kaczmarek, L.; Roszkowska, M.; Fontaneto, D.; Jezierska, M.; Pietrzak, B.; Wieczorek, R.; Poprawa, I.; Kosicki, J.Z.; Karachitos, A.; Kmita, H. Staying young and fit? Ontogenetic and phylogenetic consequences of animal anhydrobiosis. J. Zool. 2019, 309, 1–11. [Google Scholar] [CrossRef]
- Koshland, D.; Tapia, H. Desiccation tolerance: An unusual window into stress biology. Mol. Biol. Cell 2019, 30, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M. Anhydrobiosis in invertebrates. Appl. Entomol. Zool. 2006, 41, 15–31. [Google Scholar] [CrossRef]
- Alpert, P. Constraints of tolerance: Why are desiccation-tolerant organisms so small or rare? J. Exp. Biol. 2006, 209, 1575–1584. [Google Scholar] [CrossRef]
- Erkut, C.; Kurzchalia, T.V. The C. elegans dauer larva as a paradigm to study metabolic suppression and desiccation tolerance. Planta 2015, 242, 389–396. [Google Scholar] [CrossRef]
- Tunnacliffe, A.; Lapinski, J. Resurrecting Van Leeuwenhoek’s rotifers: A reappraisal of the role of disaccharides in anhydrobiosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003, 358, 1755–1771. [Google Scholar] [CrossRef]
- Welnicz, W.; Grohme, M.A.; Kaczmarek, L.; Schill, R.O.; Frohme, M. Anhydrobiosis in tardigrades-the last decade. J. Insect Physiol. 2011, 57, 577–583. [Google Scholar] [CrossRef]
- Kikuta, S.; Watanabe, S.J.; Sato, R.; Gusev, O.; Nesmelov, A.; Sogame, Y.; Cornette, R.; Kikawada, T. Towards water-free biobanks: Long-term dry-preservation at room temperature of desiccation-sensitive enzyme luciferase in air-dried insect cells. Sci. Rep. 2017, 7, 6540. [Google Scholar] [CrossRef] [PubMed]
- Hinton, H.E. A new chironomid from Africa, the larva of which can be dehydrated without injury. Proc. Zool. Soc. Lond. 1951, 121, 371–380. [Google Scholar] [CrossRef]
- Hinton, H.E. Cryptobiosis in the larva of Polypedilum-vanderplanki Hint (Chironomidae). J. Insect Physiol. 1960, 5, 286–300. [Google Scholar] [CrossRef]
- Cornette, R.; Kikawada, T. The induction of anhydrobiosis in the sleeping chironomid: Current status of our knowledge. IUBMB Life 2011, 63, 419–429. [Google Scholar] [CrossRef]
- Sogame, Y.; Kikawada, T. Current findings on the molecular mechanisms underlying anhydrobiosis in Polypedilum vanderplanki. Curr. Opin. Insect Sci. 2017, 19, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, Y.; Imanishi, S.; Mitsumasu, K.; Kanamori, Y.; Iwata, K.; Watanabe, M.; Kikawada, T.; Okuda, T. Cells from an anhydrobiotic chironomid survive almost complete desiccation. Cryobiology 2010, 60, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Imanishi, S.; Akiduki, G.; Cornette, R.; Okuda, T. Air-dried cells from the anhydrobiotic insect, Polypedilum vanderplanki, can survive long term preservation at room temperature and retain proliferation potential after rehydration. Cryobiology 2016, 73, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Gusev, O.; Suetsugu, Y.; Cornette, R.; Kawashima, T.; Logacheva, M.D.; Kondrashov, A.S.; Penin, A.A.; Hatanaka, R.; Kikuta, S.; Shimura, S.; et al. Comparative genome sequencing reveals genomic signature of extreme desiccation tolerance in the anhydrobiotic midge. Nat. Commun. 2014, 5, 4784. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.G.; Hiki, Y.; Hiroi, N.F.; Shagimardanova, E.; Gusev, O.; Cornette, R.; Kikawada, T.; Funahashi, A. Identification of a master transcription factor and a regulatory mechanism for desiccation tolerance in the anhydrobiotic cell line Pv11. PLoS ONE 2020, 15, e0230218. [Google Scholar] [CrossRef]
- Miyata, Y.; Fuse, H.; Tokumoto, S.; Hiki, Y.; Deviatiiarov, R.; Yoshida, Y.; Yamada, T.G.; Cornette, R.; Gusev, O.; Shagimardanova, E.; et al. Cas9-mediated genome editing reveals a significant contribution of calcium signaling pathways to anhydrobiosis in Pv11. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hatanaka, R.; Gusev, O.; Cornette, R.; Shimura, S.; Kikuta, S.; Okada, J.; Okuda, T.; Kikawada, T. Diversity of the expression profiles of late embryogenesis abundant (LEA) protein encoding genes in the anhydrobiotic midge Polypedilum vanderplanki. Planta 2015, 242, 451–459. [Google Scholar] [CrossRef]
- Nesmelov, A.; Cornette, R.; Gusev, O.; Kikawada, T. The antioxidant system in the anhydrobiotic midge as an essential, adaptive mechanism for desiccation survival. In Survival Strategies in Extreme Cold and Desiccation. Advances in Experimental Medicine and Biology; Iwaya-Inoue, M., Sakurai, M., Uemura, M., Eds.; Springer Nature: Singapore, 2018; pp. 259–270. [Google Scholar] [CrossRef]
- Deviatiiarov, R.; Ayupov, R.; Laikov, A.; Shagimardanova, E.; Kikawada, T.; Gusev, O. Diversity and regulation of s-adenosylmethionine dependent methyltransferases in the anhydrobiotic midge. Insects 2020, 11, 634. [Google Scholar] [CrossRef]
- Voronina, T.A.; Nesmelov, A.A.; Kondratyeva, S.A.; Deviatiiarov, R.M.; Miyata, Y.; Tokumoto, S.; Cornette, R.; Gusev, O.A.; Kikawada, T.; Shagimardanova, E.I. New group of transmembrane proteins associated with desiccation tolerance in the anhydrobiotic midge Polypedilum vanderplanki. Sci. Rep. 2020, 10, 11633. [Google Scholar] [CrossRef] [PubMed]
- Goyal, K.; Walton, L.J.; Tunnacliffe, A. LEA proteins prevent protein aggregation due to water stress. Biochem. J. 2005, 388, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, R.; Hagiwara-Komoda, Y.; Furuki, T.; Kanamori, Y.; Fujita, M.; Cornette, R.; Sakurai, M.; Okuda, T.; Kikawada, T. An abundant LEA protein in the anhydrobiotic midge, PvLEA4, acts as a molecular shield by limiting growth of aggregating protein particles. Insect Biochem. Mol. Biol. 2013, 43, 1055–1067. [Google Scholar] [CrossRef]
- Furuki, T.; Sakurai, M. Group 3 LEA protein model peptides protect enzymes against desiccation stress. Biochim. Biophys. Acta 2016, 1864, 1237–1243. [Google Scholar] [CrossRef]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.K.K.; Mahawar, M. PIMT-mediated protein repair: Mechanism and implications. Biochemistry 2019, 84, 453–463. [Google Scholar] [CrossRef]
- Mazin, P.V.; Shagimardanova, E.; Kozlova, O.; Cherkasov, A.; Sutormin, R.; Stepanova, V.V.; Stupnikov, A.; Logacheva, M.; Penin, A.; Sogame, Y.; et al. Cooption of heat shock regulatory system for anhydrobiosis in the sleeping chironomid Polypedilum vanderplanki. Proc. Natl Acad Sci. USA 2018, 115, E2477–E2486. [Google Scholar] [CrossRef]
- Nakai, A. Heat Shock Factor; Springer: Tokyo, Japan, 2016; 301p. [Google Scholar]
- Brunquell, J.; Morris, S.; Lu, Y.; Cheng, F.; Westerheide, S.D. The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans. BMC Genom. 2016, 17, 559. [Google Scholar] [CrossRef] [PubMed]
- Sogame, Y.; Okada, J.; Kikuta, S.; Miyata, Y.; Cornette, R.; Gusev, O.; Kikawada, T. Establishment of gene transfer and gene silencing methods in a desiccation-tolerant cell line, Pv11. Extremophiles 2017, 21, 65–72. [Google Scholar] [CrossRef]
- Miyata, Y.; Tokumoto, S.; Sogame, Y.; Deviatiiarov, R.; Okada, J.; Cornette, R.; Gusev, O.; Shagimardanova, E.; Sakurai, M.; Kikawada, T. Identification of a novel strong promoter from the anhydrobiotic midge, Polypedilum vanderplanki, with conserved function in various insect cell lines. Sci. Rep. 2019, 9, 7004. [Google Scholar] [CrossRef]
- Tokumoto, S.; Miyata, Y.; Usui, K.; Deviatiiarov, R.; Ohkawa, T.; Kondratieva, S.; Shagimardanova, E.; Gusev, O.; Cornette, R.; Itoh, M.; et al. Development of a Tet-On inducible expression system for the anhydrobiotic cell line, Pv11. Insects 2020, 11, 781. [Google Scholar] [CrossRef]
- Sakuma, T.; Nakade, S.; Sakane, Y.; Suzuki, K.T.; Yamamoto, T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 2016, 11, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Nakamae, K.; Nishimura, Y.; Takenaga, M.; Nakade, S.; Sakamoto, N.; Ide, H.; Sakuma, T.; Yamamoto, T. Establishment of expanded and streamlined pipeline of PITCh knock-in—A web-based design tool for MMEJ-mediated gene knock-in, PITCh designer, and the variations of PITCh, PITCh-TG and PITCh-KIKO. Bioengineered 2017, 8, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Plowman, G.D.; Hunter, T.; Sudarsanam, S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002, 27, 514–520. [Google Scholar] [CrossRef]
- Patel, S.; Latterich, M. The AAA team: Related ATPases with diverse functions. Trends Cell Biol. 1998, 8, 65–71. [Google Scholar] [CrossRef]
- Neuwald, A.F.; Aravind, L.; Spouge, J.L.; Koonin, E.V. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999, 9, 27–43. [Google Scholar] [PubMed]
- Zhang, X.; Shaw, A.; Bates, P.A.; Newman, R.H.; Gowen, B.; Orlova, E.; Gorman, M.A.; Kondo, H.; Dokurno, P.; Lally, J.; et al. Structure of the AAA ATPase p97. Mol. Cell 2000, 6, 1473–1484. [Google Scholar] [CrossRef]
- Ye, Y. Diverse functions with a common regulator: Ubiquitin takes command of an AAA ATPase. J. Struct. Biol. 2006, 156, 29–40. [Google Scholar] [CrossRef]
- Klug, A.; Schwabe, J.W. Protein motifs 5. Zinc fingers. FASEB J. 1995, 9, 597–604. [Google Scholar] [CrossRef]
- Li, W.T.; He, M.; Wang, J.; Wang, Y.P. Zinc finger protein (ZFP) in plants-A review. Plant. Omics 2013, 6, 474–480. [Google Scholar]
- Vasiliou, V.; Vasiliou, K.; Nebert, D.W. Human ATP-binding cassette (ABC) transporter family. Hum. Genom. 2009, 3, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Theodoulou, F.L.; Kerr, I.D. ABC transporter research: Going strong 40 years on. Biochem. Soc. Trans. 2015, 43, 1033–1040. [Google Scholar] [CrossRef]
- Guertin, M.J.; Lis, J.T. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 2010, 6, e1001114. [Google Scholar] [CrossRef] [PubMed]
- Gusev, O.; Cornette, R.; Kikawada, T.; Okuda, T. Expression of heat shock protein-coding genes associated with anhydrobiosis in an African chironomid Polypedilum vanderplanki. Cell Stress Chaperones 2011, 16, 81–90. [Google Scholar] [CrossRef]
- Yamada, T.G.; Suetsugu, Y.; Deviatiiarov, R.; Gusev, O.; Cornette, R.; Nesmelov, A.; Hiroi, N.; Kikawada, T.; Funahashi, A. Transcriptome analysis of the anhydrobiotic cell line Pv11 infers the mechanism of desiccation tolerance and recovery. Sci. Rep. 2018, 8, 17941. [Google Scholar] [CrossRef]
- Savic, D.; Partridge, E.C.; Newberry, K.M.; Smith, S.B.; Meadows, S.K.; Roberts, B.S.; Mackiewicz, M.; Mendenhall, E.M.; Myers, R.M. CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins. Genome Res. 2015, 25, 1581–1589. [Google Scholar] [CrossRef] [PubMed]
- Partridge, E.C.; Watkins, T.A.; Mendenhall, E.M. Every transcription factor deserves its map: Scaling up epitope tagging of proteins to bypass antibody problems. Bioessays 2016, 38, 801–811. [Google Scholar] [CrossRef]
- Xiong, X.; Zhang, Y.; Yan, J.; Jain, S.; Chee, S.; Ren, B.; Zhao, H. A Scalable epitope tagging approach for high throughput ChIP-Seq analysis. ACS Synth. Biol. 2017, 6, 1034–1042. [Google Scholar] [CrossRef]
- Kidder, B.L.; Hu, G.; Zhao, K. ChIP-Seq: Technical considerations for obtaining high-quality data. Nat. Immunol. 2011, 12, 918–922. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; MacRae, T.H. Stress tolerance in diapausing embryos of Artemia franciscana is dependent on heat shock factor 1 (Hsf1). PLoS ONE 2018, 13, e0200153. [Google Scholar] [CrossRef]
- Hibshman, J.D.; Clegg, J.S.; Goldstein, B. Mechanisms of desiccation tolerance: Themes and variations in brine shrimp, roundworms, and tardigrades. Front. Physiol. 2020, 11, 592016. [Google Scholar] [CrossRef]
- Ryabova, A.; Cornette, R.; Cherkasov, A.; Watanabe, M.; Okuda, T.; Shagimardanova, E.; Kikawada, T.; Gusev, O. Combined metabolome and transcriptome analysis reveals key components of complete desiccation tolerance in an anhydrobiotic insect. Proc. Natl Acad Sci. USA 2020, 117, 19209–19220. [Google Scholar] [CrossRef]
- Muerdter, F.; Boryn, L.M.; Arnold, C.D. STARR-seq—Principles and Applications. Genomics 2015, 106, 145–150. [Google Scholar] [CrossRef]
- Chatterjee, S.; Ahituv, N. Gene regulatory elements, major drivers of human disease. Annu. Rev. Genom. Hum. Genet. 2017, 18, 45–63. [Google Scholar] [CrossRef]
- Parnas, O.; Jovanovic, M.; Eisenhaure, T.M.; Herbst, R.H.; Dixit, A.; Ye, C.J.; Przybylski, D.; Platt, R.J.; Tirosh, I.; Sanjana, N.E.; et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 2015, 162, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Sanjana, N.E.; Zheng, K.; Shalem, O.; Lee, K.; Shi, X.; Scott, D.A.; Song, J.; Pan, J.Q.; Weissleder, R.; et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 2015, 160, 1246–1260. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, A.; Fujimoto, M.; Tan, K.; Kurashima, A.; Srivastava, P.; Okada, M.; Takii, R.; Nakai, A. HSF1 is required for induction of mitochondrial chaperones during the mitochondrial unfolded protein response. FEBS Open Bio 2020, 10, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Kouno, T.; Moody, J.; Kwon, A.T.; Shibayama, Y.; Kato, S.; Huang, Y.; Bottcher, M.; Motakis, E.; Mendez, M.; Severin, J.; et al. C1 CAGE detects transcription start sites and enhancer activity at single-cell resolution. Nat. Commun. 2019, 10, 360. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokumoto, S.; Miyata, Y.; Deviatiiarov, R.; Yamada, T.G.; Hiki, Y.; Kozlova, O.; Yoshida, Y.; Cornette, R.; Funahashi, A.; Shagimardanova, E.; et al. Genome-Wide Role of HSF1 in Transcriptional Regulation of Desiccation Tolerance in the Anhydrobiotic Cell Line, Pv11. Int. J. Mol. Sci. 2021, 22, 5798. https://doi.org/10.3390/ijms22115798
Tokumoto S, Miyata Y, Deviatiiarov R, Yamada TG, Hiki Y, Kozlova O, Yoshida Y, Cornette R, Funahashi A, Shagimardanova E, et al. Genome-Wide Role of HSF1 in Transcriptional Regulation of Desiccation Tolerance in the Anhydrobiotic Cell Line, Pv11. International Journal of Molecular Sciences. 2021; 22(11):5798. https://doi.org/10.3390/ijms22115798
Chicago/Turabian StyleTokumoto, Shoko, Yugo Miyata, Ruslan Deviatiiarov, Takahiro G. Yamada, Yusuke Hiki, Olga Kozlova, Yuki Yoshida, Richard Cornette, Akira Funahashi, Elena Shagimardanova, and et al. 2021. "Genome-Wide Role of HSF1 in Transcriptional Regulation of Desiccation Tolerance in the Anhydrobiotic Cell Line, Pv11" International Journal of Molecular Sciences 22, no. 11: 5798. https://doi.org/10.3390/ijms22115798
APA StyleTokumoto, S., Miyata, Y., Deviatiiarov, R., Yamada, T. G., Hiki, Y., Kozlova, O., Yoshida, Y., Cornette, R., Funahashi, A., Shagimardanova, E., Gusev, O., & Kikawada, T. (2021). Genome-Wide Role of HSF1 in Transcriptional Regulation of Desiccation Tolerance in the Anhydrobiotic Cell Line, Pv11. International Journal of Molecular Sciences, 22(11), 5798. https://doi.org/10.3390/ijms22115798








