Curcumin-Loaded Nanoparticles Impair the Pro-Tumor Activity of Acid-Stressed MSC in an In Vitro Model of Osteosarcoma
Abstract
1. Introduction
2. Results
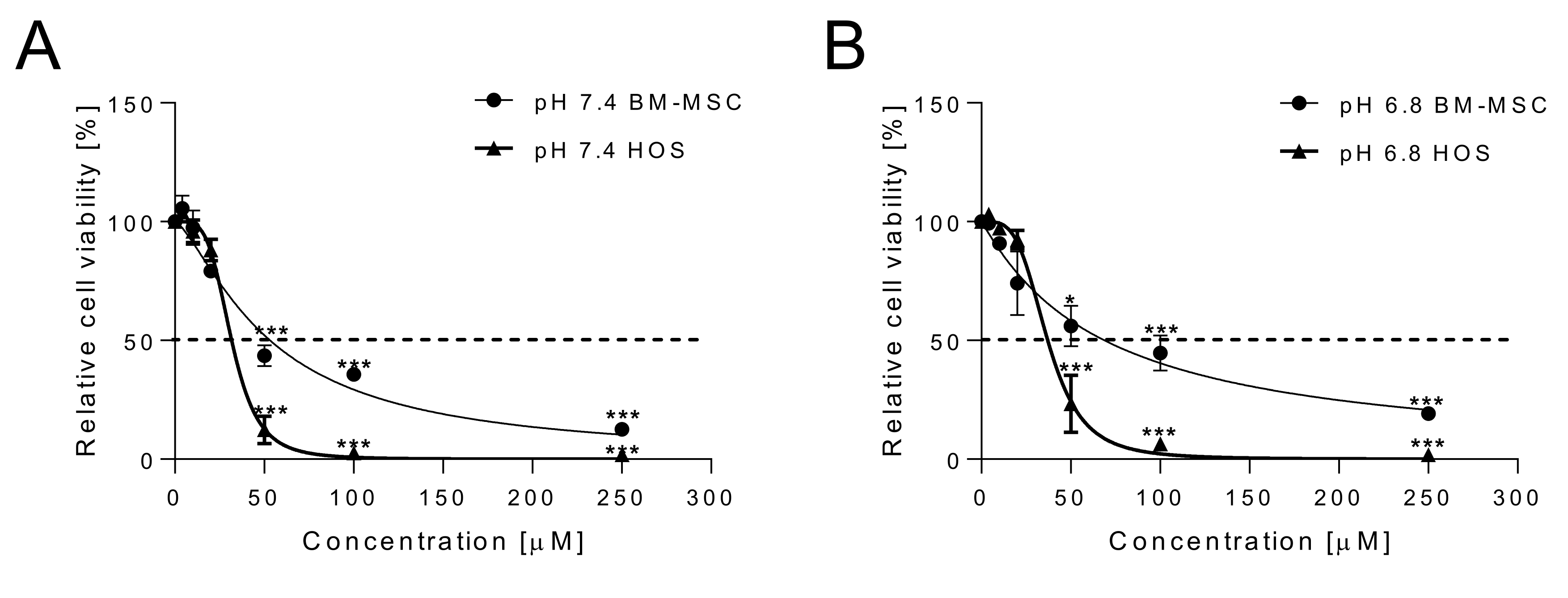
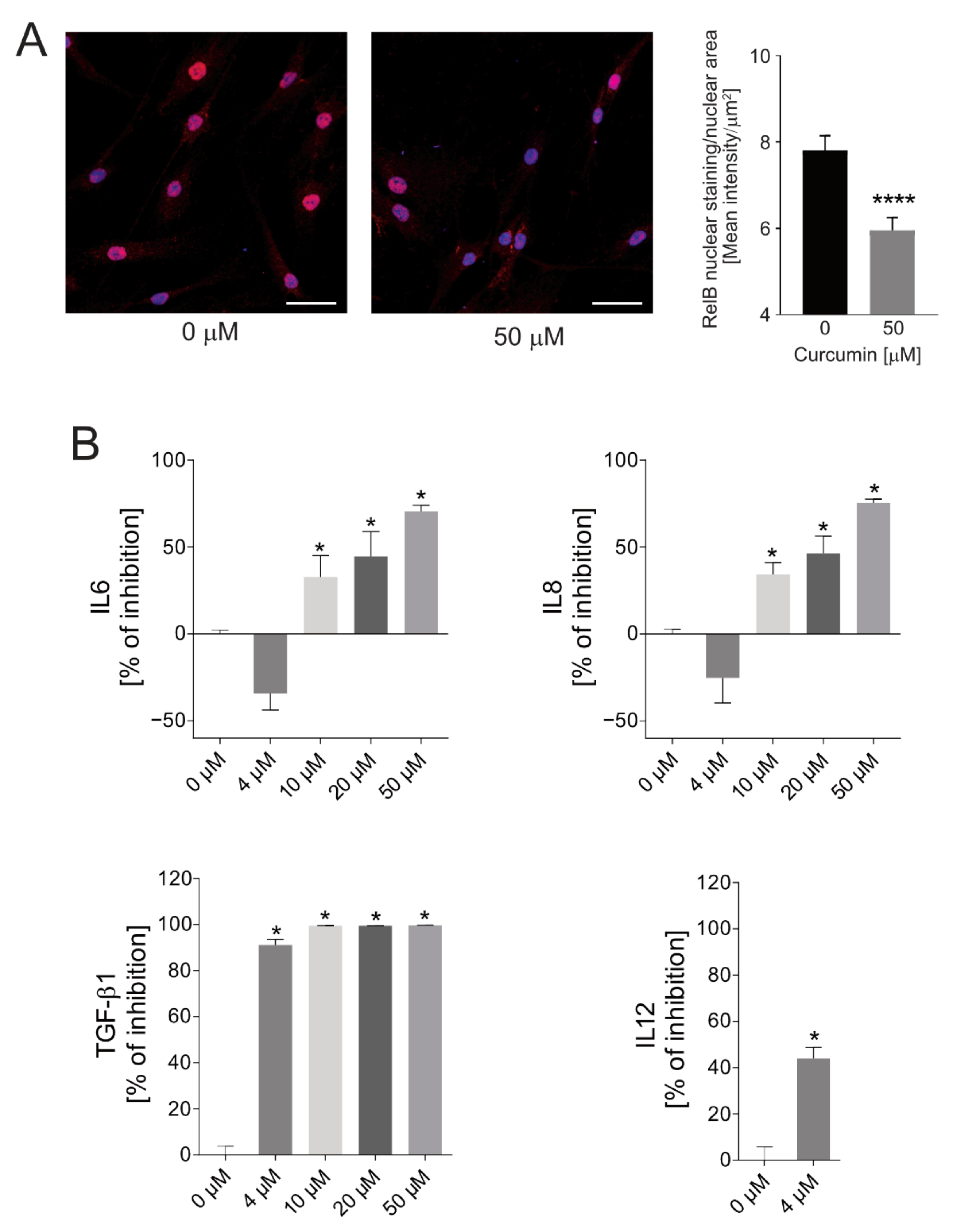
2.1. Cytotoxicity of Curcumin against Osteosarcoma Cells and Anti-Inflammatory Effects on Normal MSCs
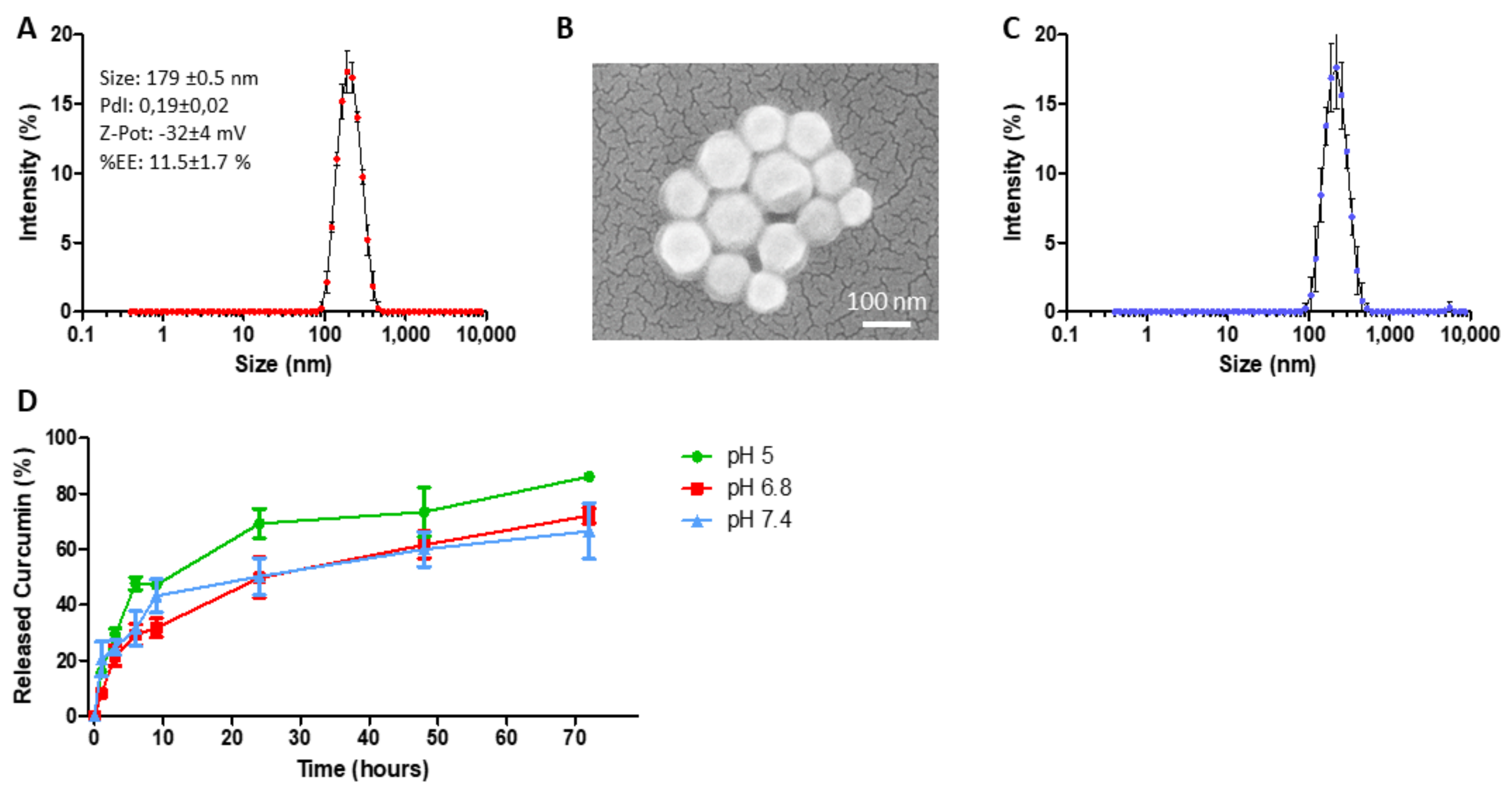
2.2. Curcumin-Loaded SPN (SPN-curc) Fabrication and Characterization
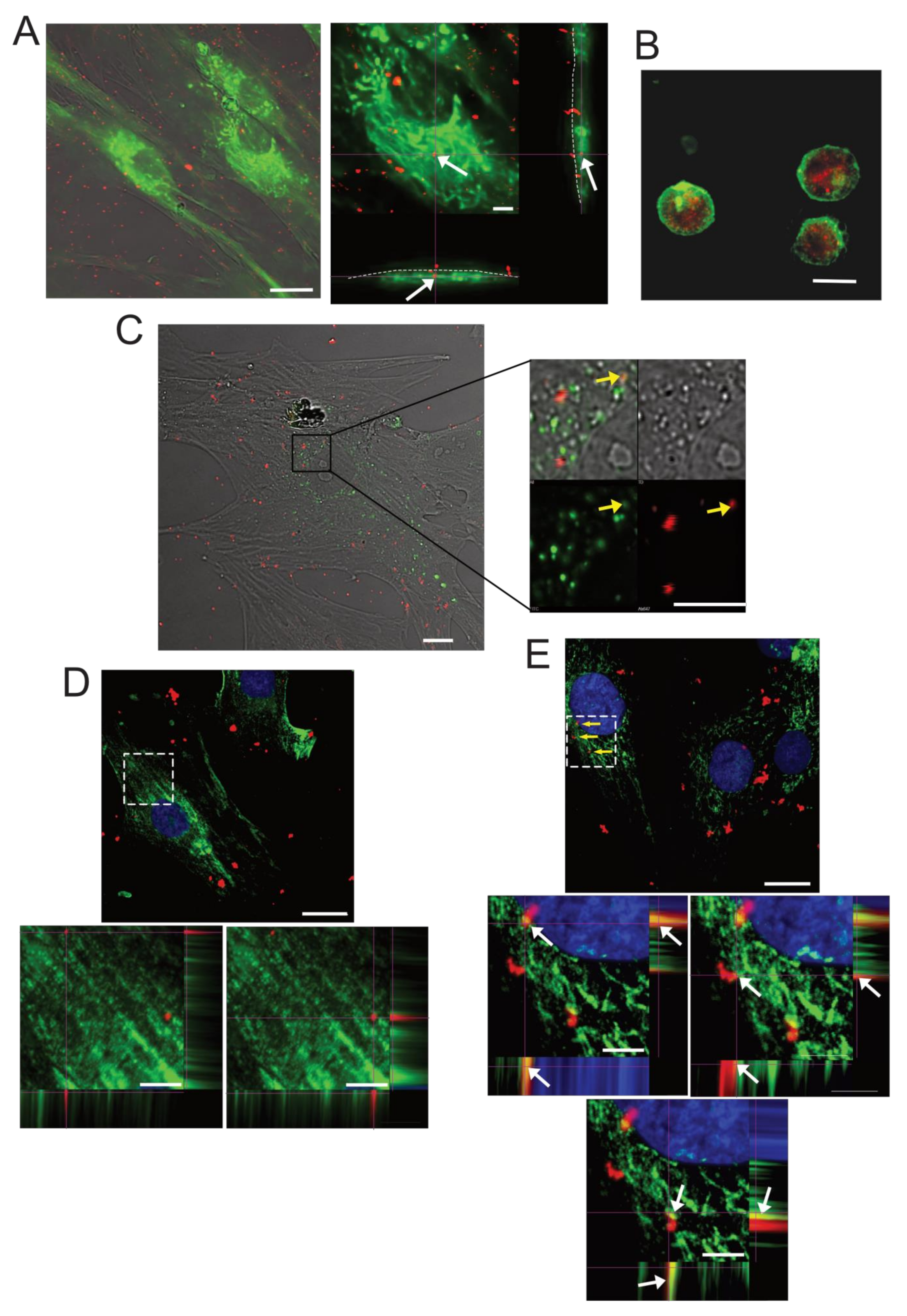
2.3. SPN-curc Internalization in BM-MSCs
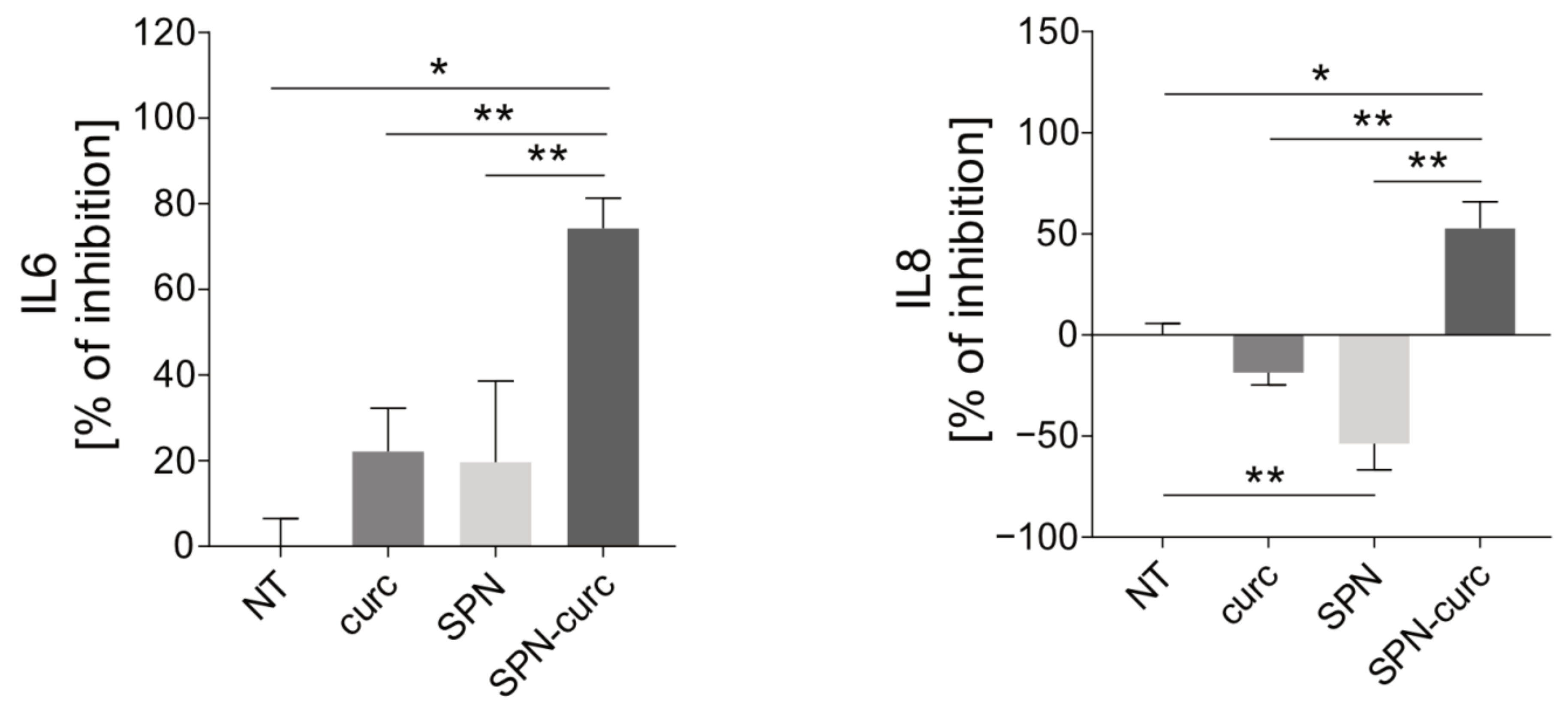
2.4. Anti-Inflammatory Efficacy of SPN-curc
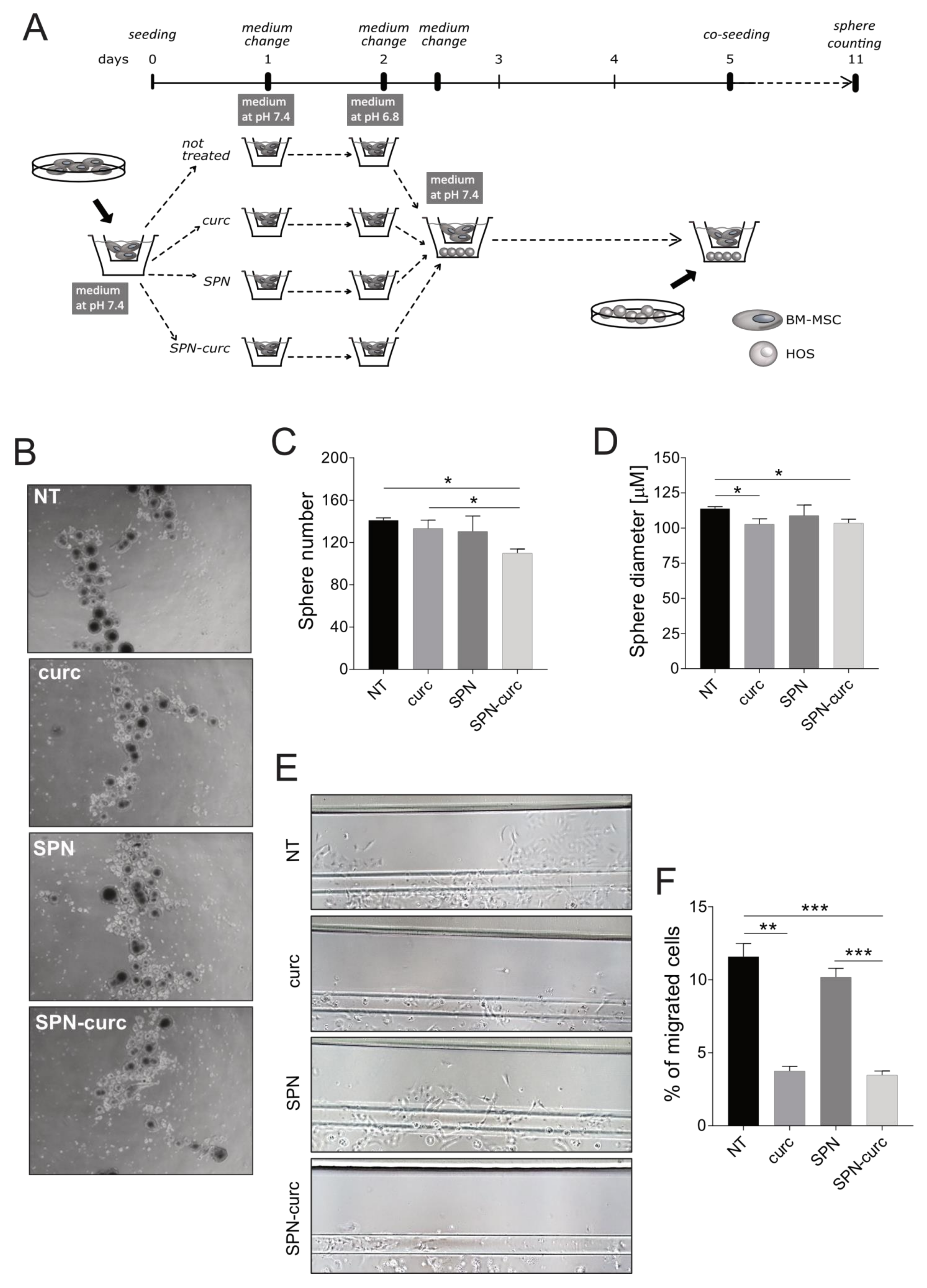
2.5. Curcumin-Loaded SPNs Revert the Acid-Mediated, Pro-Tumorigenic Phenotype of MSCs
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Nanoparticle Synthesis and Characterization
4.2.1. Materials
4.2.2. Synthesis of SPNs
4.2.3. Characterization of SPNs
4.2.4. Drug Loading and Release
4.3. Cellular Uptake
4.4. Lysotracker Staining
4.5. Immunofluorescence
4.6. Cytotoxicity Assay
4.7. ELISA
4.8. Sarcosphere-Forming Efficiency
4.9. Cell Invasion Assay in Microfluidic Devices
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TME | tumor microenvironment |
| CAFs | cancer-associated fibroblasts |
| MSCs | mesenchymal stromal cells |
| OS | osteosarcoma |
| RelA | RelA proto-oncogene NF-κB Subunit |
| RelB | RelB proto-oncogene NF-κB subunit |
| NFKB1 | nuclear factor kappa B subunit 1 |
| IκBα | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor alpha |
| SPN | spherical polymeric nanoparticles |
| SPN-curc | spherical polymeric nanoparticles encapsulating curcumin |
| CTCs | circulating tumor cells |
| BM-MSC | bone marrow-derived MSCs |
| IL6 | interleukin 6 |
| IL8 | interleukin 8 |
| IL-1β | interleukin 1beta |
| TNF-α | tumor necrosis factor alpha |
| TGF-β1 | transforming growth factor beta 1 |
| IL12 | interleukin 12 |
| PLGA | poly(lactic-co-glycolic acid) |
| DPPC | 1,2-dipalmitoyl-sn-glycero-3-phosphocholine |
| curc | curcumin |
| SPN-curc-Cy5 | spherical polymeric nanoparticles encapsulating curcumin and labelled with Cy5 fluorophore |
| CSCs | cancer stem cells |
References
- Bhome, R.; Bullock, M.D.; Al Saihati, H.A.; Goh, R.W.; Primrose, J.N.; Sayan, A.E.; Mirnezami, A.H. A top-down view of the tumor microenvironment: Structure, cells and signaling. Front. Cell Dev. Biol. 2015, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Hass, R.; Kasper, C.; Bohm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal. 2011, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Abe, R.; Fujita, Y.; Ando, S.; Inokuma, D.; Shimizu, H. Mesenchymal stem cells are recruited into wounded skin and contribute to wound repair by transdifferentiation into multiple skin cell type. J. Immunol. 2008, 180, 2581–2587. [Google Scholar] [CrossRef]
- Han, Y.; Li, X.; Zhang, Y.; Han, Y.; Chang, F.; Ding, J. Mesenchymal Stem Cells for Regenerative Medicine. Cells 2019, 8, 886. [Google Scholar] [CrossRef]
- Gao, F.; Chiu, S.M.; Motan, D.A.; Zhang, Z.; Chen, L.; Ji, H.L.; Tse, H.F.; Fu, Q.L.; Lian, Q. Mesenchymal stem cells and immunomodulation: Current status and future prospects. Cell Death Dis. 2016, 7, e2062. [Google Scholar] [CrossRef]
- Ridge, S.M.; Sullivan, F.J.; Glynn, S.A. Mesenchymal stem cells: Key players in cancer progression. Mol. Cancer 2017, 16, 31. [Google Scholar] [CrossRef]
- Shi, Y.; Du, L.; Lin, L.; Wang, Y. Tumour-associated mesenchymal stem/stromal cells: Emerging therapeutic targets. Nat. Rev. Drug Discov. 2017, 16, 35–52. [Google Scholar] [CrossRef]
- Alfranca, A.; Martinez-Cruzado, L.; Tornin, J.; Abarrategi, A.; Amaral, T.; de Alava, E.; Menendez, P.; Garcia-Castro, J.; Rodriguez, R. Bone microenvironment signals in osteosarcoma development. Cell. Mol. Life Sci. 2015, 72, 3097–3113. [Google Scholar] [CrossRef]
- Cortini, M.; Avnet, S.; Baldini, N. Mesenchymal stroma: Role in osteosarcoma progression. Cancer Lett. 2017, 405, 90–99. [Google Scholar] [CrossRef]
- Longhi, A.; Errani, C.; De Paolis, M.; Mercuri, M.; Bacci, G. Primary bone osteosarcoma in the pediatric age: State of the art. Cancer Treat. Rev. 2006, 32, 423–436. [Google Scholar] [CrossRef]
- Smeland, S.; Bielack, S.S.; Whelan, J.; Bernstein, M.; Hogendoorn, P.; Krailo, M.D.; Gorlick, R.; Janeway, K.A.; Ingleby, F.C.; Anninga, J.; et al. Survival and prognosis with osteosarcoma: Outcomes in more than 2000 patients in the EURAMOS-1 (European and American Osteosarcoma Study) cohort. Eur. J. Cancer 2019, 109, 36–50. [Google Scholar] [CrossRef]
- Bonuccelli, G.; Avnet, S.; Grisendi, G.; Salerno, M.; Granchi, D.; Dominici, M.; Kusuzaki, K.; Baldini, N. Role of mesenchymal stem cells in osteosarcoma and metabolic reprogramming of tumor cells. Oncotarget 2014, 5, 7575–7588. [Google Scholar] [CrossRef]
- Vallabhaneni, K.C.; Hassler, M.Y.; Abraham, A.; Whitt, J.; Mo, Y.Y.; Atfi, A.; Pochampally, R. Mesenchymal Stem/Stromal Cells under Stress Increase Osteosarcoma Migration and Apoptosis Resistance via Extracellular Vesicle Mediated Communication. PLoS ONE 2016, 11, e0166027. [Google Scholar] [CrossRef]
- Tu, B.; Du, L.; Fan, Q.M.; Tang, Z.; Tang, T.T. STAT3 activation by IL-6 from mesenchymal stem cells promotes the proliferation and metastasis of osteosarcoma. Cancer Lett. 2012, 325, 80–88. [Google Scholar] [CrossRef]
- Baglio, S.R.; Lagerweij, T.; Perez-Lanzon, M.; Ho, X.D.; Leveille, N.; Melo, S.A.; Cleton-Jansen, A.M.; Jordanova, E.S.; Roncuzzi, L.; Greco, M.; et al. Blocking Tumor-Educated MSC Paracrine Activity Halts Osteosarcoma Progression. Clin. Cancer Res. 2017, 23, 3721–3733. [Google Scholar] [CrossRef]
- Cortini, M.; Massa, A.; Avnet, S.; Bonuccelli, G.; Baldini, N. Tumor-Activated Mesenchymal Stromal Cells Promote Osteosarcoma Stemness and Migratory Potential via IL-6 Secretion. PLoS ONE 2016, 11, e0166500. [Google Scholar] [CrossRef]
- Kawano, M.; Tanaka, K.; Itonaga, I.; Iwasaki, T.; Tsumura, H. Interaction between human osteosarcoma and mesenchymal stem cells via an interleukin-8 signaling loop in the tumor microenvironment. Cell. Commun. Signal. 2018, 16, 13. [Google Scholar] [CrossRef]
- Xu, W.T.; Bian, Z.Y.; Fan, Q.M.; Li, G.; Tang, T.T. Human mesenchymal stem cells (hMSCs) target osteosarcoma and promote its growth and pulmonary metastasis. Cancer Lett. 2009, 281, 32–41. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, G.; Chen, R.; Hua, Y.; Cai, Z. Mesenchymal stem cells in the osteosarcoma microenvironment: Their biological properties, influence on tumor growth, and therapeutic implications. Stem Cell Res. Ther. 2018, 9, 22. [Google Scholar] [CrossRef]
- Avnet, S.; Di Pompo, G.; Chano, T.; Errani, C.; Ibrahim-Hashim, A.; Gillies, R.J.; Donati, D.M.; Baldini, N. Cancer-associated mesenchymal stroma fosters the stemness of osteosarcoma cells in response to intratumoral acidosis via NF-kappaB activation. Int. J. Cancer 2017, 140, 1331–1345. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Massa, A.; Perut, F.; Chano, T.; Woloszyk, A.; Mitsiadis, T.A.; Avnet, S.; Baldini, N. The effect of extracellular acidosis on the behaviour of mesenchymal stem cells in vitro. Eur. Cells Mater. 2017, 33, 252–267. [Google Scholar] [CrossRef]
- Peppicelli, S.; Bianchini, F.; Toti, A.; Laurenzana, A.; Fibbi, G.; Calorini, L. Extracellular acidity strengthens mesenchymal stem cells to promote melanoma progression. Cell Cycle 2015, 14, 3088–3100. [Google Scholar] [CrossRef]
- Bischoff, D.S.; Zhu, J.H.; Makhijani, N.S.; Yamaguchi, D.T. Acidic pH stimulates the production of the angiogenic CXC chemokine, CXCL8 (interleukin-8), in human adult mesenchymal stem cells via the extracellular signal-regulated kinase, p38 mitogen-activated protein kinase, and NF-kappaB pathways. J. Cell Biochem. 2008, 104, 1378–1392. [Google Scholar] [CrossRef]
- Perut, F.; Avnet, S.; Fotia, C.; Baglio, S.R.; Salerno, M.; Hosogi, S.; Kusuzaki, K.; Baldini, N. V-ATPase as an effective therapeutic target for sarcomas. Exp. Cell Res. 2014, 320, 21–32. [Google Scholar] [CrossRef]
- Kolosenko, I.; Avnet, S.; Baldini, N.; Viklund, J.; De Milito, A. Therapeutic implications of tumor interstitial acidification. Semin. Cancer Biol. 2017, 43, 119–133. [Google Scholar] [CrossRef]
- Avnet, S.; Lemma, S.; Cortini, M.; Pellegrini, P.; Perut, F.; Zini, N.; Kusuzaki, K.; Chano, T.; Grisendi, G.; Dominici, M.; et al. Altered pH gradient at the plasma membrane of osteosarcoma cells is a key mechanism of drug resistance. Oncotarget 2016, 7, 63408–63423. [Google Scholar] [CrossRef] [PubMed]
- Chano, T.; Avnet, S.; Kusuzaki, K.; Bonuccelli, G.; Sonveaux, P.; Rotili, D.; Mai, A.; Baldini, N. Tumour-specific metabolic adaptation to acidosis is coupled to epigenetic stability in osteosarcoma cells. Am. J. Cancer Res. 2016, 6, 859–875. [Google Scholar] [PubMed]
- Cortini, M.; Armirotti, A.; Columbaro, M.; Longo, D.L.; Di Pompo, G.; Cannas, E.; Maresca, A.; Errani, C.; Longhi, A.; Righi, A.; et al. Exploring Metabolic Adaptations to the Acidic Microenvironment of Osteosarcoma Cells Unveils Sphingosine 1-Phosphate as a Valuable Therapeutic Target. Cancers 2021, 13, 311. [Google Scholar] [CrossRef] [PubMed]
- Giordano, A.; Tommonaro, G. Curcumin and Cancer. Nutrients 2019, 11, 2376. [Google Scholar] [CrossRef]
- Karunaweera, N.; Raju, R.; Gyengesi, E.; Munch, G. Plant polyphenols as inhibitors of NF-kappaB induced cytokine production-a potential anti-inflammatory treatment for Alzheimer′s disease? Front. Mol. Neurosci. 2015, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Wilken, R.; Veena, M.S.; Wang, M.B.; Srivatsan, E.S. Curcumin: A review of anti-cancer properties and therapeutic activity in head and neck squamous cell carcinoma. Mol. Cancer 2011, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.F.; Takeda, T.; Li, B.; Tsuiji, K.; Kondo, A.; Tadakawa, M.; Nagase, S.; Yaegashi, N. Curcumin targets the AKT-mTOR pathway for uterine leiomyosarcoma tumor growth suppression. Int. J. Clin. Oncol. 2014, 19, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Orr, W.S.; Denbo, J.W.; Saab, K.R.; Ng, C.Y.; Wu, J.; Li, K.; Garner, J.M.; Morton, C.L.; Du, Z.; Pfeffer, L.M.; et al. Curcumin potentiates rhabdomyosarcoma radiosensitivity by suppressing NF-kappaB activity. PLoS ONE 2013, 8, e51309. [Google Scholar] [CrossRef]
- Lee, H.P.; Li, T.M.; Tsao, J.Y.; Fong, Y.C.; Tang, C.H. Curcumin induces cell apoptosis in human chondrosarcoma through extrinsic death receptor pathway. Int. Immunopharmacol. 2012, 13, 163–169. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.; Song, R.; Shen, Y.; Sun, Y.; Gu, Y.; Shu, Y.; Xu, Q. Targeting sarcoplasmic/endoplasmic reticulum Ca(2)+-ATPase 2 by curcumin induces ER stress-associated apoptosis for treating human liposarcoma. Mol. Cancer Ther. 2011, 10, 461–471. [Google Scholar] [CrossRef]
- Singh, M.; Pandey, A.; Karikari, C.A.; Singh, G.; Rakheja, D. Cell cycle inhibition and apoptosis induced by curcumin in Ewing sarcoma cell line SK-NEP-1. Med. Oncol 2010, 27, 1096–1101. [Google Scholar] [CrossRef]
- Chang, R.; Sun, L.; Webster, T.J. Short communication: Selective cytotoxicity of curcumin on osteosarcoma cells compared to healthy osteoblasts. Int. J. Nanomed. 2014, 9, 461–465. [Google Scholar]
- Chang, Z.; Xing, J.; Yu, X. Curcumin induces osteosarcoma MG63 cells apoptosis via ROS/Cyto-C/Caspase-3 pathway. Tumour Biol. 2014, 35, 753–758. [Google Scholar] [CrossRef]
- Walters, D.K.; Muff, R.; Langsam, B.; Born, W.; Fuchs, B. Cytotoxic effects of curcumin on osteosarcoma cell lines. Investig. New Drugs 2008, 26, 289–297. [Google Scholar] [CrossRef]
- Lee, D.S.; Lee, M.K.; Kim, J.H. Curcumin induces cell cycle arrest and apoptosis in human osteosarcoma (HOS) cells. Anticancer Res. 2009, 29, 5039–5044. [Google Scholar]
- Li, Y.; Zhang, J.; Ma, D.; Zhang, L.; Si, M.; Yin, H.; Li, J. Curcumin inhibits proliferation and invasion of osteosarcoma cells through inactivation of Notch-1 signaling. FEBS J. 2012, 279, 2247–2259. [Google Scholar] [CrossRef]
- Walker, B.C.; Mittal, S. Antitumor Activity of Curcumin in Glioblastoma. Int. J. Mol. Sci. 2020, 21, 9435. [Google Scholar] [CrossRef]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef]
- Feng, T.; Wei, Y.; Lee, R.J.; Zhao, L. Liposomal curcumin and its application in cancer. Int. J. Nanomed. 2017, 12, 6027–6044. [Google Scholar] [CrossRef]
- Jyoti, K.; Bhatia, R.K.; Martis, E.A.F.; Coutinho, E.C.; Jain, U.K.; Chandra, R.; Madan, J. Soluble curcumin amalgamated chitosan microspheres augmented drug delivery and cytotoxicity in colon cancer cells: In vitro and in vivo study. Colloids Surf. B Biointerfaces 2016, 148, 674–683. [Google Scholar] [CrossRef]
- Shinde, R.L.; Devarajan, P.V. Docosahexaenoic acid-mediated, targeted and sustained brain delivery of curcumin microemulsion. Drug Deliv. 2017, 24, 152–161. [Google Scholar] [CrossRef]
- Gera, M.; Sharma, N.; Ghosh, M.; Huynh, D.L.; Lee, S.J.; Min, T.; Kwon, T.; Jeong, D.K. Nanoformulations of curcumin: An emerging paradigm for improved remedial application. Oncotarget 2017, 8, 66680–66698. [Google Scholar] [CrossRef]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef]
- Palange, A.L.; Di Mascolo, D.; Carallo, C.; Gnasso, A.; Decuzzi, P. Lipid-polymer nanoparticles encapsulating curcumin for modulating the vascular deposition of breast cancer cells. Nanomedicine 2014, 10, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Stigliano, C.; Key, J.; Ramirez, M.; Aryal, S.; Decuzzi, P. Radiolabeled Polymeric Nanoconstructs Loaded with Docetaxel and Curcumin for Cancer Combinatorial Therapy and Nuclear Imaging. Adv. Funct. Mater. 2015, 25, 3371–3379. [Google Scholar] [CrossRef]
- Ameruoso, A.; Palomba, R.; Palange, A.L.; Cervadoro, A.; Lee, A.; Di Mascolo, D.; Decuzzi, P. Ameliorating Amyloid-beta Fibrils Triggered Inflammation via Curcumin-Loaded Polymeric Nanoconstructs. Front. Immunol. 2017, 8, 1411. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Yao, Y.; Wu, J.; Liu, Q.; Zhang, J.; Pu, X.; Zhang, Q.; Xia, L. Curcumin inhibits gastric cancer-derived mesenchymal stem cells mediated angiogenesis by regulating NF-kappaB/VEGF signaling. Am. J. Transl. Res. 2017, 9, 5538–5547. [Google Scholar] [PubMed]
- Abdel Aziz, M.T.; Khaled, H.M.; El Hindawi, A.; Roshdy, N.K.; Rashed, L.A.; Sabry, D.; Hassouna, A.A.; Taha, F.; Ali, W.I. Effect of mesenchymal stem cells and a novel curcumin derivative on Notch1 signaling in hepatoma cell line. Biomed. Res. Int. 2013, 2013, 129629. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, J.; Prasad, S.; Aggarwal, B.B. Curcumin and cancer cells: How many ways can curry kill tumor cells selectively? AAPS J. 2009, 11, 495–510. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.; Sun, L.; Webster, T.J. Selective inhibition of MG-63 osteosarcoma cell proliferation induced by curcumin-loaded self-assembled arginine-rich-RGD nanospheres. Int. J. Nanomed. 2015, 10, 3351–3365. [Google Scholar]
- Di Francesco, V.; Gurgone, D.; Palomba, R.; Ferreira, M.F.M.M.; Catelani, T.; Cervadoro, A.; Maffia, P.; Decuzzi, P. Modulating Lipoprotein Transcellular Transport and Atherosclerotic Plaque Formation in ApoE–/–Mice via Nanoformulated Lipid–Methotrexate Conjugates. ACS Appl. Mater. Interfaces 2020, 12, 37943–37956. [Google Scholar] [CrossRef]
- Maeda, H. The enhanced permeability and retention (EPR) effect in tumor vasculature: The key role of tumor-selective macromolecular drug targeting. Adv. Enzym. Regul. 2001, 41, 189–207. [Google Scholar] [CrossRef]
- Pelkmans, L.; Burli, T.; Zerial, M.; Helenius, A. Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic. Cell 2004, 118, 767–780. [Google Scholar] [CrossRef]
- Pols, M.S.; Klumperman, J. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 2009, 315, 1584–1592. [Google Scholar] [CrossRef]
- Mertins, S.D. Cancer stem cells: A systems biology view of their role in prognosis and therapy. Anticancer Drugs 2014, 25, 353–367. [Google Scholar] [CrossRef]
- Clarke, M.F.; Dick, J.E.; Dirks, P.B.; Eaves, C.J.; Jamieson, C.H.; Jones, D.L.; Visvader, J.; Weissman, I.L.; Wahl, G.M. Cancer stem cells—Perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006, 66, 9339–9344. [Google Scholar] [CrossRef]
- Salerno, M.; Avnet, S.; Bonuccelli, G.; Eramo, A.; De Maria, R.; Gambarotti, M.; Gamberi, G.; Baldini, N. Sphere-forming cell subsets with cancer stem cell properties in human musculoskeletal sarcomas. Int. J. Oncol. 2013, 43, 95–102. [Google Scholar] [CrossRef][Green Version]
- Avnet, S.; Lemma, S.; Cortini, M.; Di Pompo, G.; Perut, F.; Baldini, N. Pre-clinical Models for Studying the Interaction Between Mesenchymal Stromal Cells and Cancer Cells and the Induction of Stemness. Front. Oncol. 2019, 9, 305. [Google Scholar] [CrossRef]
- Cortini, M.; Baldini, N.; Avnet, S. New Advances in the Study of Bone Tumors: A Lesson From the 3D Environment. Front. Physiol. 2019, 10, 814. [Google Scholar] [CrossRef]
- Karunagaran, D.; Rashmi, R.; Kumar, T.R. Induction of apoptosis by curcumin and its implications for cancer therapy. Curr. Cancer Drug Targets 2005, 5, 117–129. [Google Scholar] [CrossRef]
- Kunwar, A.; Barik, A.; Mishra, B.; Rathinasamy, K.; Pandey, R.; Priyadarsini, K.I. Quantitative cellular uptake, localization and cytotoxicity of curcumin in normal and tumor cells. Biochim. Biophys. Acta 2008, 1780, 673–679. [Google Scholar] [CrossRef]
- Mutsaers, A.J.; Walkley, C.R. Cells of origin in osteosarcoma: Mesenchymal stem cells or osteoblast committed cells? Bone 2014, 62, 56–63. [Google Scholar] [CrossRef]
- Yin, J.; Chen, Y.; Zhang, Z.H.; Han, X. Stimuli-Responsive Block Copolymer-Based Assemblies for Cargo Delivery and Theranostic Applications. Polymers 2016, 8, 268. [Google Scholar] [CrossRef]
- Ivey, J.W.; Bonakdar, M.; Kanitkar, A.; Davalos, R.V.; Verbridge, S.S. Improving cancer therapies by targeting the physical and chemical hallmarks of the tumor microenvironment. Cancer Lett. 2016, 380, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Avnet, S.; Chano, T.; Massa, A.; Bonuccelli, G.; Lemma, S.; Falzetti, L.; Grisendi, G.; Dominici, M.; Baldini, N. Acid microenvironment promotes cell survival of human bone sarcoma through the activation of cIAP proteins and NF-kappaB pathway. Am. J. Cancer Res. 2019, 9, 1127–1144. [Google Scholar] [PubMed]
- Pires, B.R.B.; Silva, R.; Ferreira, G.M.; Abdelhay, E. NF-kappaB: Two Sides of the Same Coin. Genes 2018, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Setrerrahmane, S.; Xu, H. Tumor-related interleukins: Old validated targets for new anti-cancer drug development. Mol. Cancer 2017, 16, 153. [Google Scholar] [CrossRef]
- Waugh, D.J.; Wilson, C. The interleukin-8 pathway in cancer. Clin. Cancer Res. 2008, 14, 6735–6741. [Google Scholar] [CrossRef]
- Di Mascolo, D.; Christopher, J.L.; Aryal, S.; Ramirez, M.R.; Wang, J.; Candeloro, P.; Guindani, M.; Hsueh, W.A.; Decuzzi, P. Rosiglitazone-loaded nanospheres for modulating macrophage-specific inflammation in obesity. J. Control. Release 2013, 170, 460–468. [Google Scholar] [CrossRef]
- Stigliano, C.; Ramirez, M.R.; Singh, J.V.; Aryal, S.; Key, J.; Blanco, E.; Decuzzi, P. Methotraxate-Loaded Hybrid Nanoconstructs Target Vascular Lesions and Inhibit Atherosclerosis Progression in ApoE(−/−) Mice. Adv. Healthc. Mater. 2017, 6, 1601286. [Google Scholar] [CrossRef]
- Lee, A.; De Mei, C.; Fereira, M.; Marotta, R.; Yoon, H.Y.; Kim, K.; Kwon, I.C.; Decuzzi, P. Dexamethasone-loaded Polymeric Nanoconstructs for Monitoring and Treating Inflammatory Bowel Disease. Theranostics 2017, 7, 3653–3666. [Google Scholar] [CrossRef]
- Brown, H.K.; Tellez-Gabriel, M.; Heymann, D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017, 386, 189–195. [Google Scholar] [CrossRef]
- Van Duinen, V.; Trietsch, S.J.; Joore, J.; Vulto, P.; Hankemeier, T. Microfluidic 3D cell culture: From tools to tissue models. Curr. Opin. Biotechnol. 2015, 35, 118–126. [Google Scholar] [CrossRef]
- Aryal, S.; Hu, C.M.; Zhang, L. Polymeric nanoparticles with precise ratiometric control over drug loading for combination therapy. Mol. Pharm. 2011, 8, 1401–1407. [Google Scholar] [CrossRef]
- Aryal, S.; Key, J.; Stigliano, C.; Ananta, J.S.; Zhong, M.; Decuzzi, P. Engineered magnetic hybrid nanoparticles with enhanced relaxivity for tumor imaging. Biomaterials 2013, 34, 7725–7732. [Google Scholar] [CrossRef]
- Avnet, S.; Di Pompo, G.; Lemma, S.; Salerno, M.; Perut, F.; Bonuccelli, G.; Granchi, D.; Zini, N.; Baldini, N. V-ATPase is a candidate therapeutic target for Ewing sarcoma. Biochim. Biophys. Acta 2013, 1832, 1105–1116. [Google Scholar] [CrossRef][Green Version]
- Ferreira, M.; Rizzuti, I.F.; Palange, A.L.; Barbato, M.G.; Di Francesco, V.; Di Francesco, M.; Decuzzi, P. Optimizing the Pharmacological Properties of Discoidal Polymeric Nanoconstructs Against Triple-Negative Breast Cancer Cells. Front. Bioeng. Biotechnol. 2020, 8, 5. [Google Scholar] [CrossRef]
- Yang, T.T.; Sinai, P.; Kain, S.R. An acid phosphatase assay for quantifying the growth of adherent and nonadherent cells. Anal. Biochem. 1996, 241, 103–108. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Pompo, G.; Cortini, M.; Palomba, R.; Di Francesco, V.; Bellotti, E.; Decuzzi, P.; Baldini, N.; Avnet, S. Curcumin-Loaded Nanoparticles Impair the Pro-Tumor Activity of Acid-Stressed MSC in an In Vitro Model of Osteosarcoma. Int. J. Mol. Sci. 2021, 22, 5760. https://doi.org/10.3390/ijms22115760
Di Pompo G, Cortini M, Palomba R, Di Francesco V, Bellotti E, Decuzzi P, Baldini N, Avnet S. Curcumin-Loaded Nanoparticles Impair the Pro-Tumor Activity of Acid-Stressed MSC in an In Vitro Model of Osteosarcoma. International Journal of Molecular Sciences. 2021; 22(11):5760. https://doi.org/10.3390/ijms22115760
Chicago/Turabian StyleDi Pompo, Gemma, Margherita Cortini, Roberto Palomba, Valentina Di Francesco, Elena Bellotti, Paolo Decuzzi, Nicola Baldini, and Sofia Avnet. 2021. "Curcumin-Loaded Nanoparticles Impair the Pro-Tumor Activity of Acid-Stressed MSC in an In Vitro Model of Osteosarcoma" International Journal of Molecular Sciences 22, no. 11: 5760. https://doi.org/10.3390/ijms22115760
APA StyleDi Pompo, G., Cortini, M., Palomba, R., Di Francesco, V., Bellotti, E., Decuzzi, P., Baldini, N., & Avnet, S. (2021). Curcumin-Loaded Nanoparticles Impair the Pro-Tumor Activity of Acid-Stressed MSC in an In Vitro Model of Osteosarcoma. International Journal of Molecular Sciences, 22(11), 5760. https://doi.org/10.3390/ijms22115760









