Protein Sumoylation Is Crucial for Phagocytosis in Entamoeba histolytica Trophozoites
Abstract
1. Introduction
2. Results
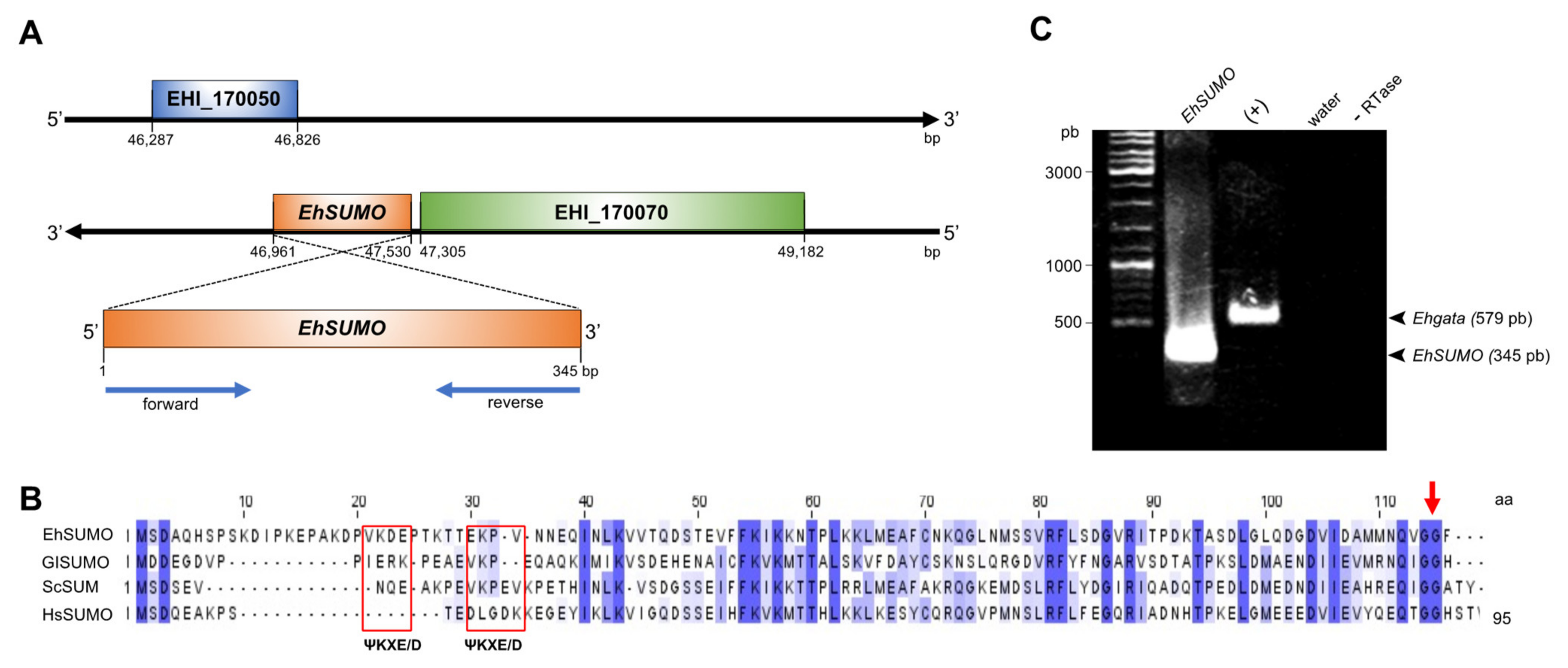
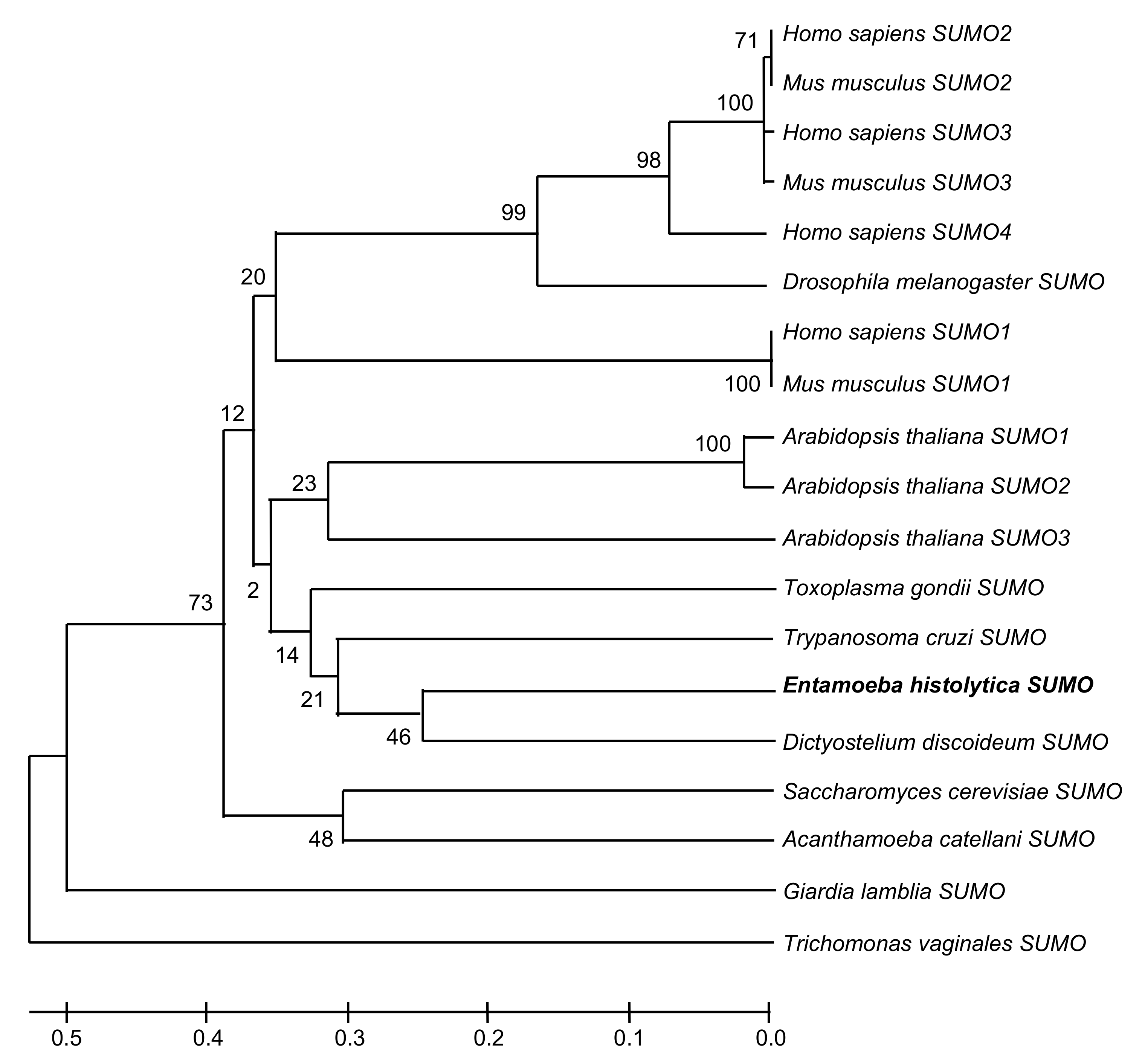
2.1. In Silico Analysis Predicts the Existence of SUMO and the Sumoylation-Desumoylation Machineries in E. histolytica
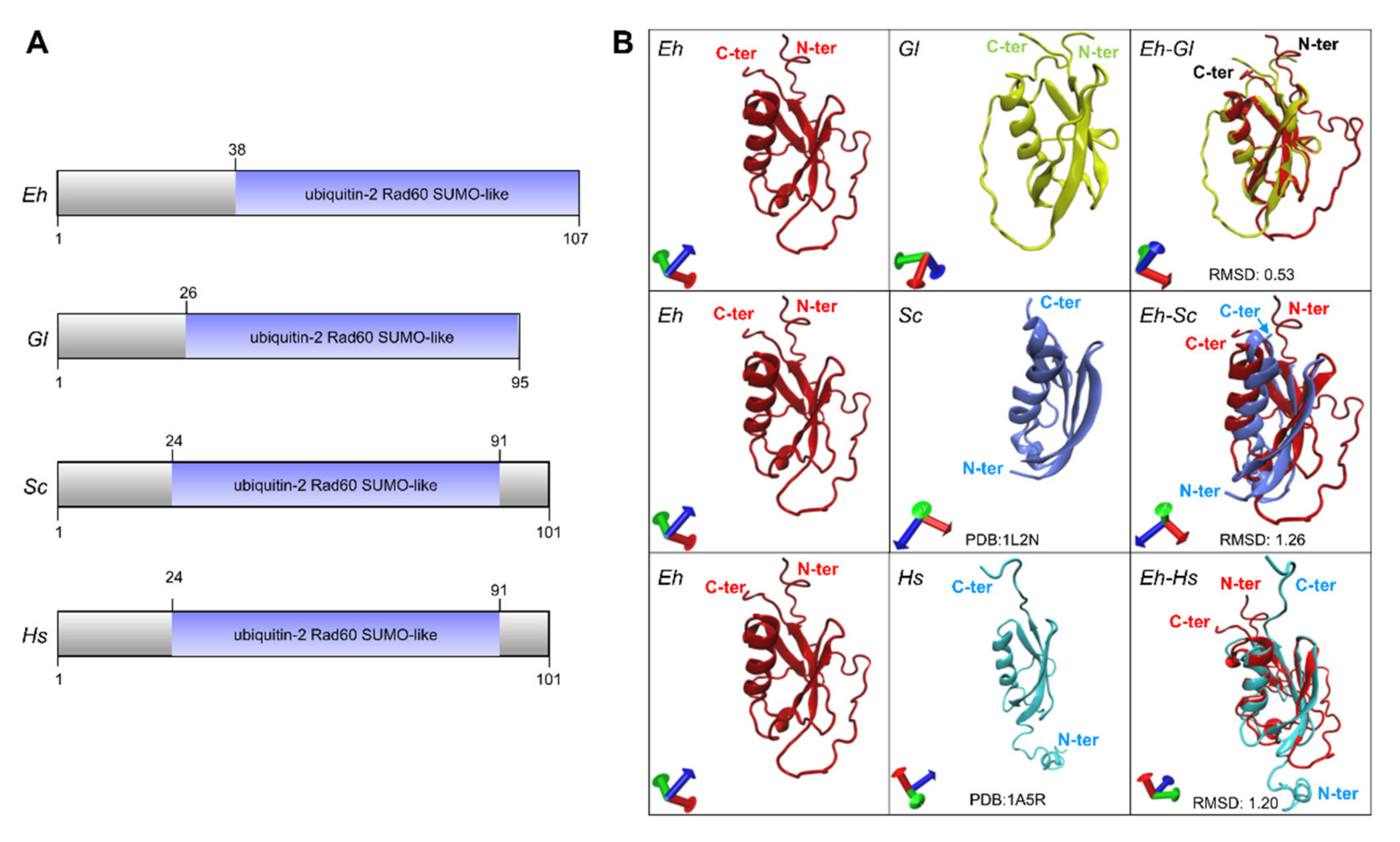
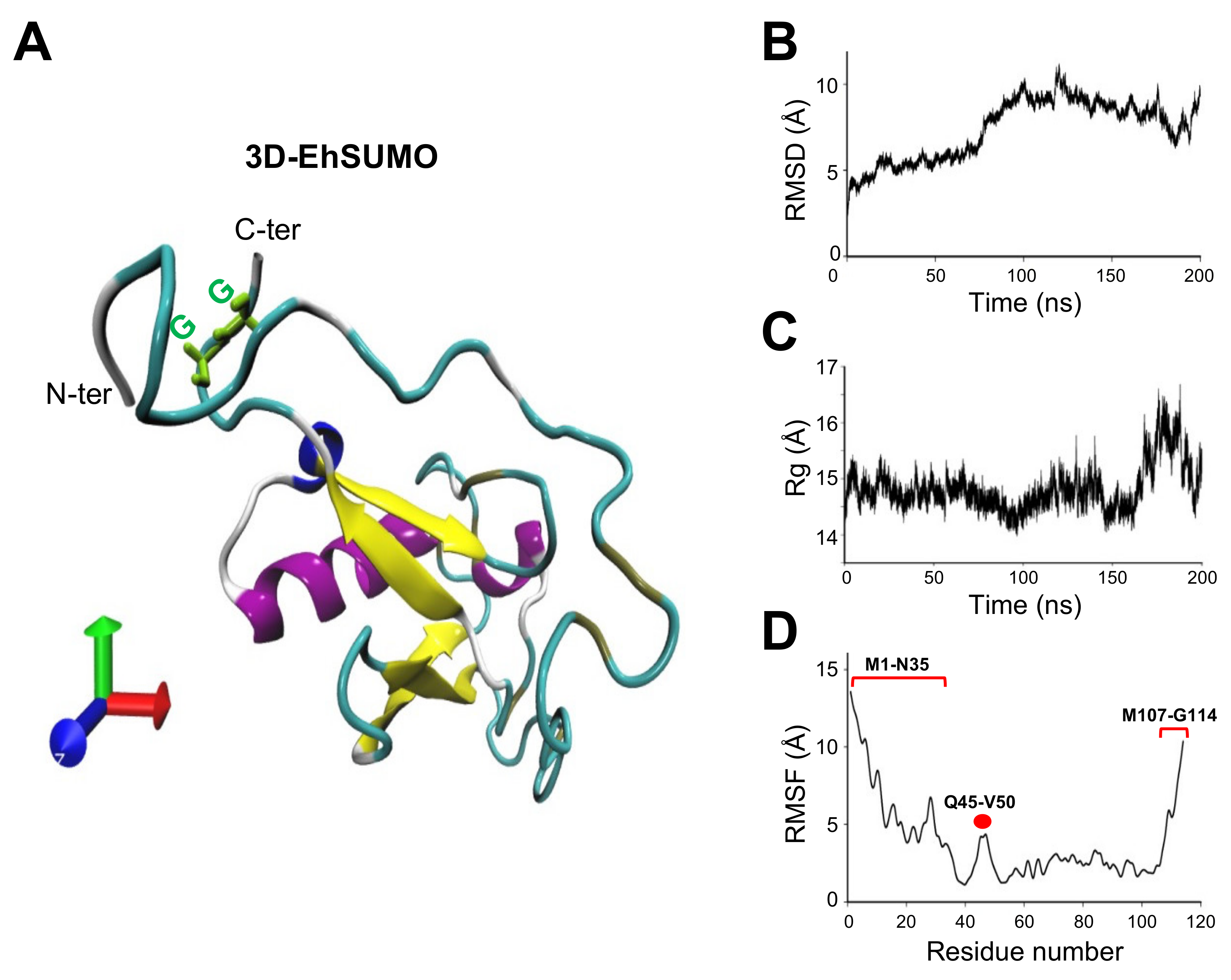
2.2. The Predicted 3D-Structure of EhSUMO Is Like Other Orthologues
2.3. Under the Stimulus of Erythrocytes, EhSUMO Moves from the Cytoplasm to the Target Cell Adherence Points, Phagocytic Cups, and Phagosomes
2.4. In Silico Analysis Reveals Sumoylation Sites in ESCRT-III and EhADH Proteins
2.5. EhADH Protein Interacts with EhSUMO
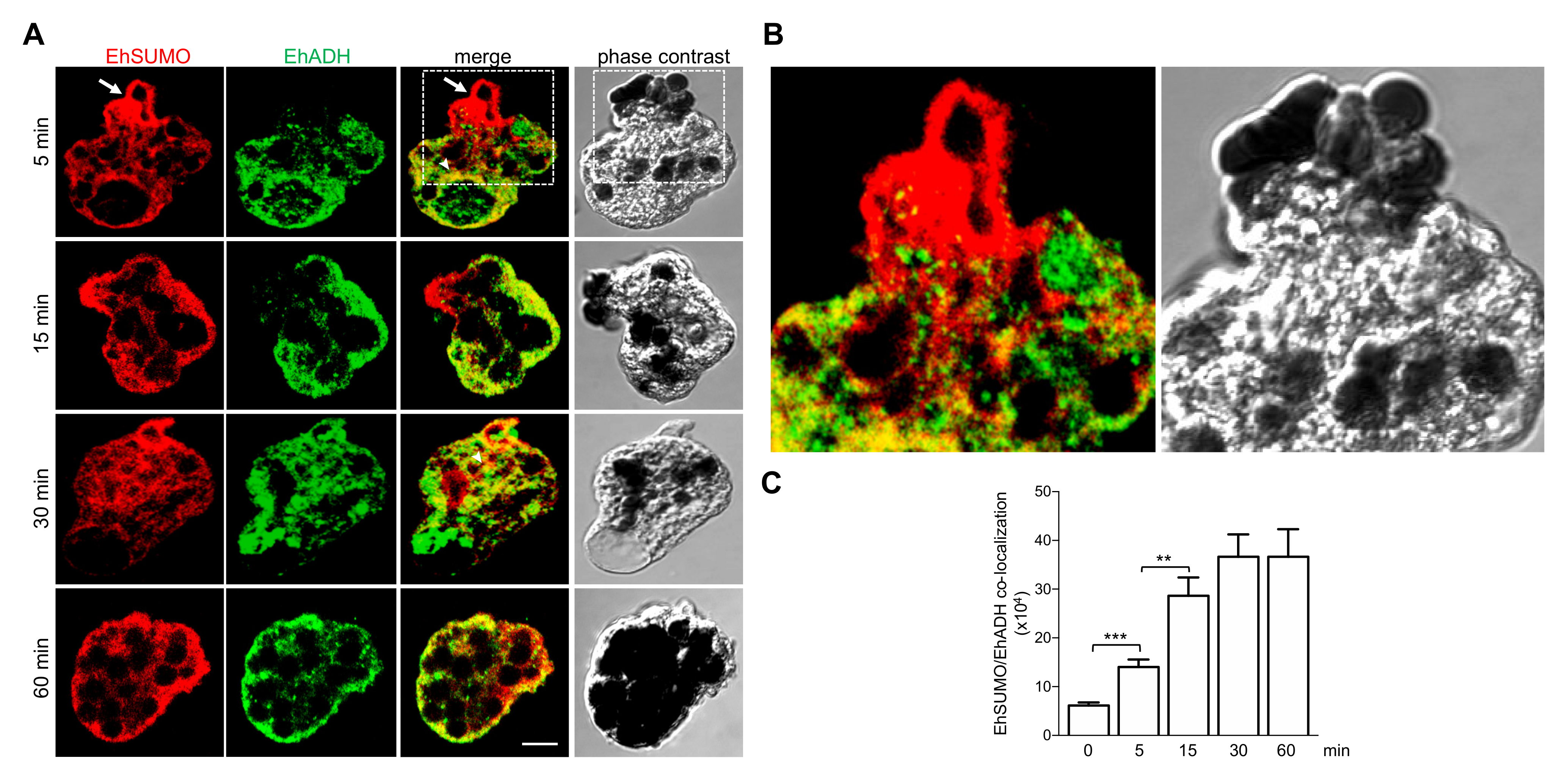
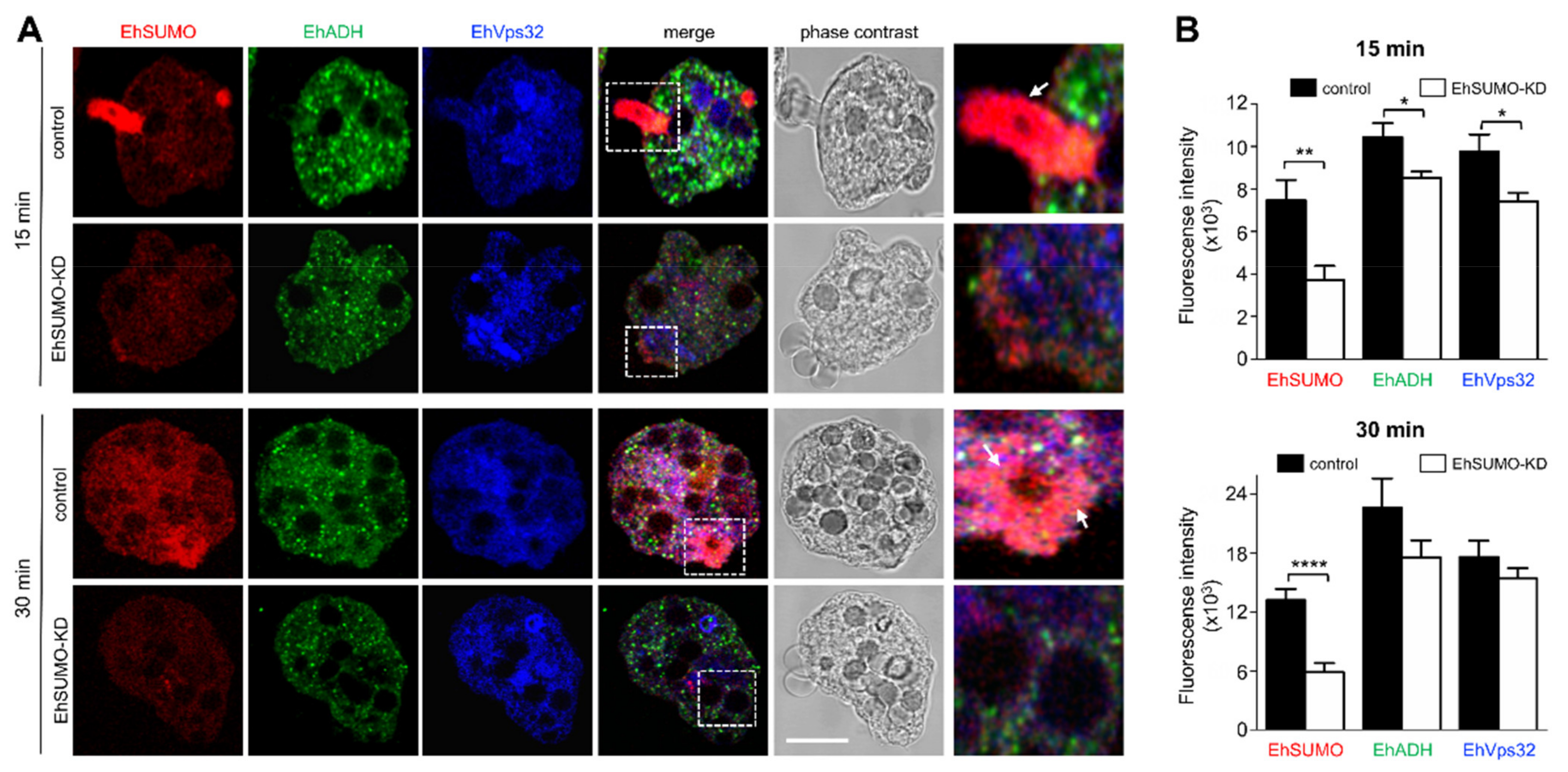
2.6. Colocation of EhSUMO and EhADH Increases during Phagocytosis
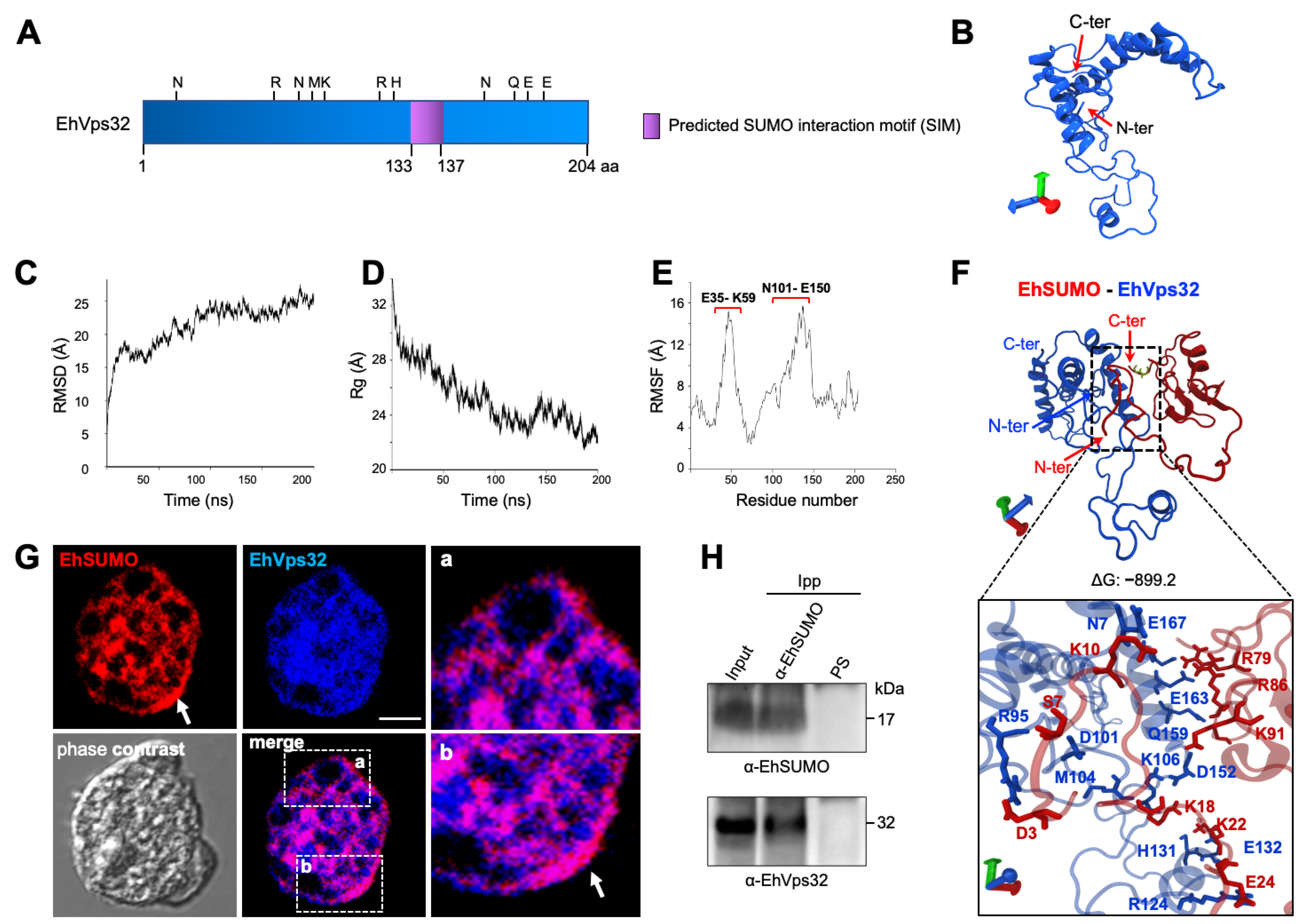
2.7. In Silico Analysis Discloses Interaction between EhVps32 and EhSUMO
2.8. Immunofluorescence and Immunoprecipitation Experiments Confirm the EhSUMO and EhVps32 Interaction
2.9. Interaction between EhVps32 and EhSUMO Continues through Phagocytosis
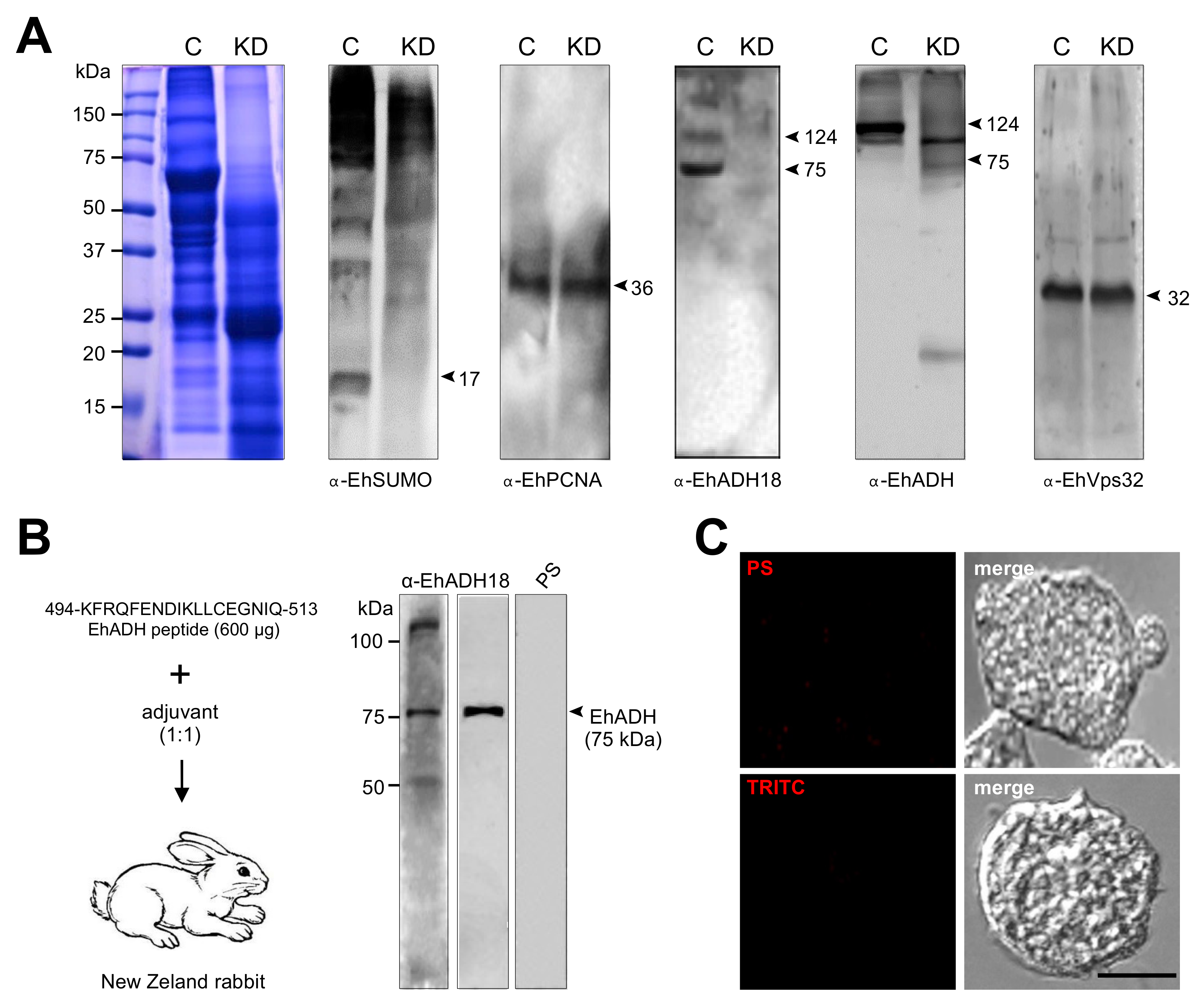
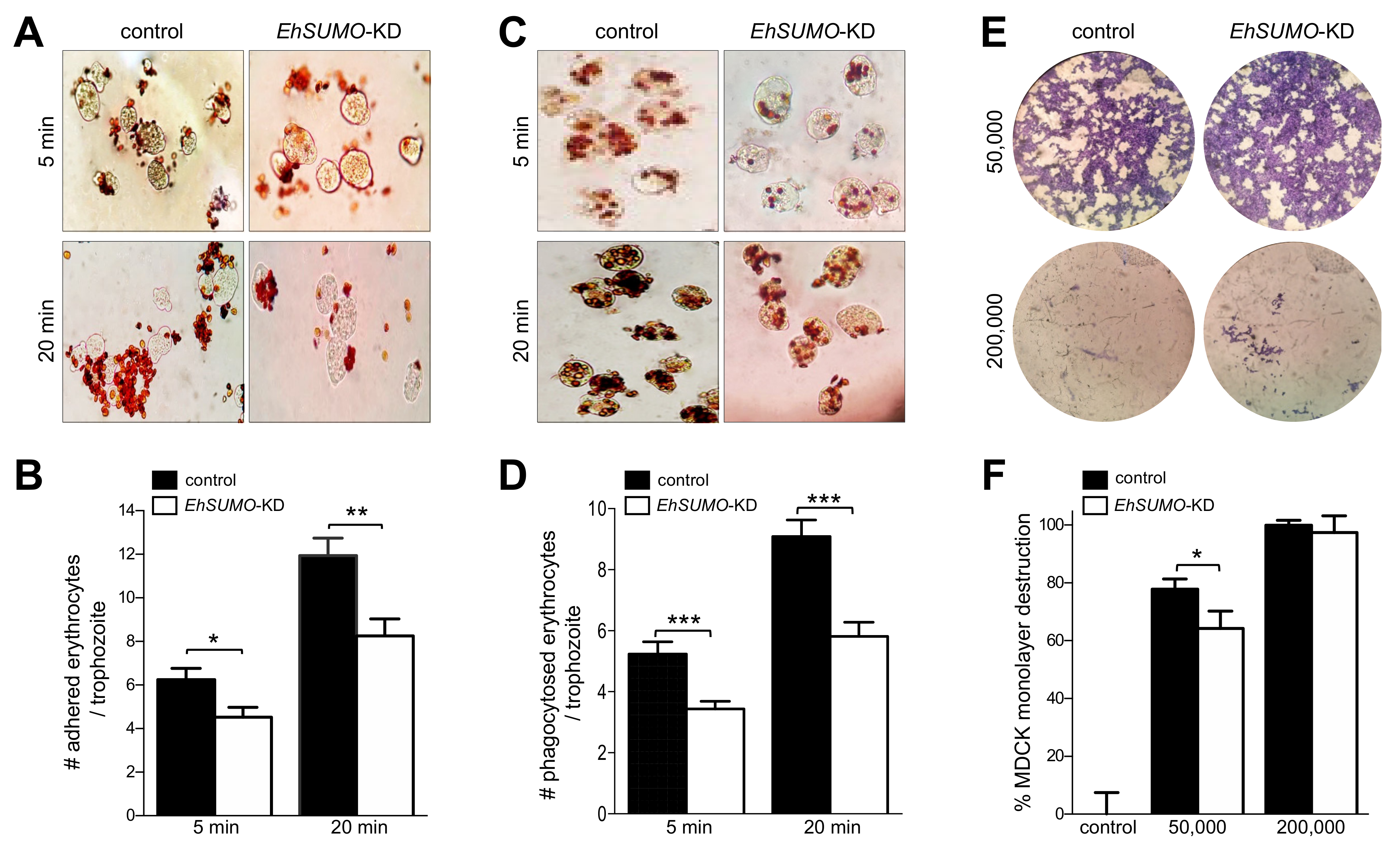
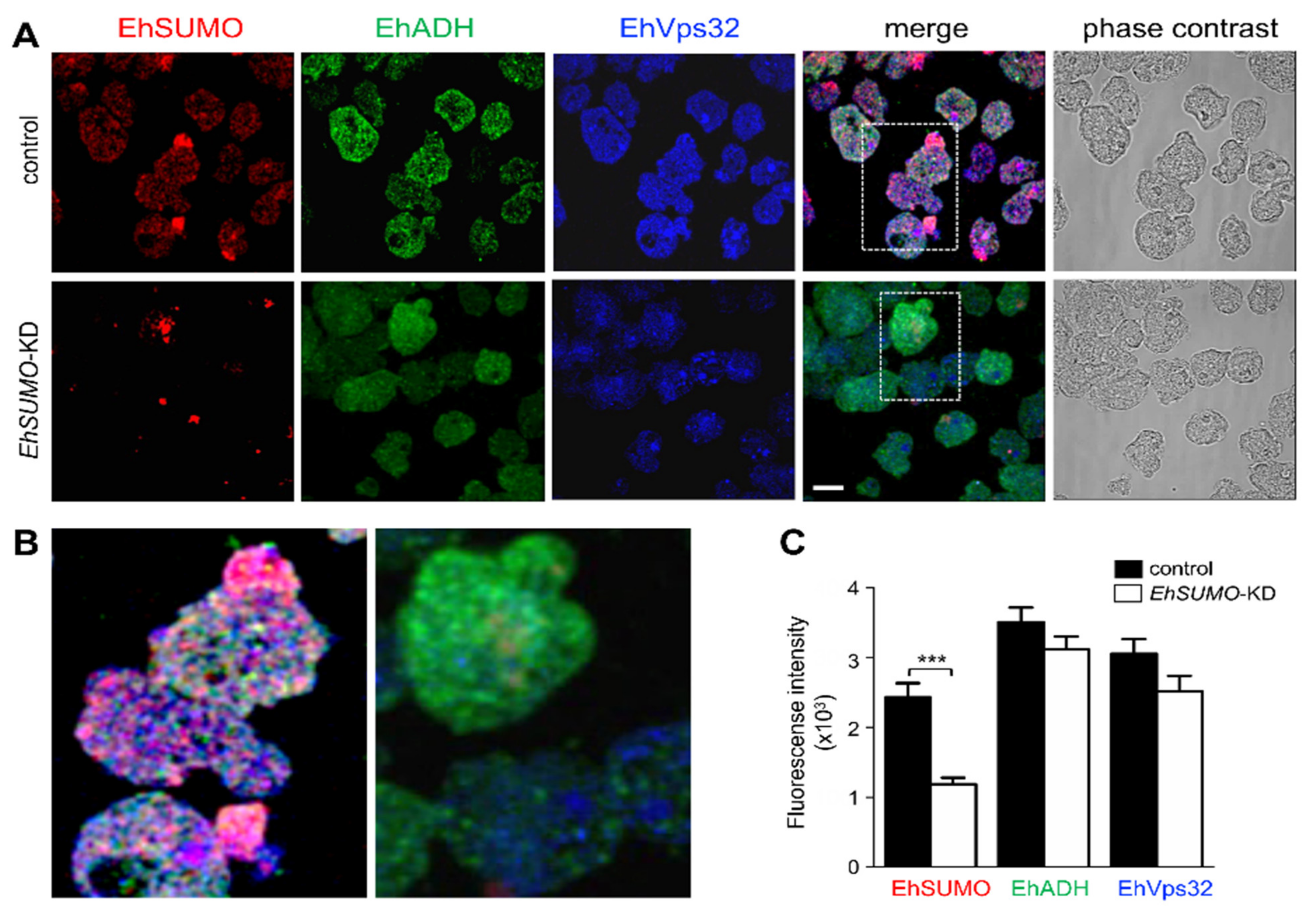
2.10. EhSUMO Knocked-Down Trophozoites Exhibited Diminished Adhesion, Erythrophagocytosis and Cytopathic Effect in Comparison to the Wild Type Strain
3. Discussion
4. Materials and Methods
4.1. Culture of Trophozoites and Epithelial Cells
4.2. SUMO Searching and Phylogenetic Analysis
4.3. Secondary and Tertiary Structure of EhSUMO
4.4. Molecular Dynamics Simulations (MDS)
4.5. Cloning of the E. histolytica SUMO Gene (EhSUMO)
4.6. Expression and Purification of Recombinant Protein, and Generation of Anti-Ehsumo Antibodies
4.7. Production of the α-EhADH18 Antibody
4.8. Western Blot Experiments
4.9. Laser Confocal Microscopy Assays
4.10. In Vivo Virulence of E. histolytica Trophozoites
4.11. Immunoprecipitation Assays
4.12. Silencing Assay
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bayer, P.; Arndt, A.; Metzger, S.; Mahajan, R.; Melchior, F.; Jaenicke, R.; Becker, J. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 1998, 280, 275–286. [Google Scholar] [CrossRef]
- Sampson, D.A.; Wang, M.; Matunis, M.J. The Small Ubiquitin-like Modifier-1 (SUMO-1) Consensus Sequence Mediates Ubc9 Binding and Is Essential for SUMO-1 Modification. J. Biol. Chem. 2001, 276, 21664–21669. [Google Scholar] [CrossRef]
- Kamitani, T.; Nguyen, H.P.; Kito, K.; Fukuda-Kamitani, T.; Yeh, E.T. Covalent Modification of PML by the Sentrin Family of Ubiquitin-like Proteins. J. Biol. Chem. 1998, 273, 3117–3120. [Google Scholar] [CrossRef]
- Rangasamy, D.; Woytek, K.; Khan, S.A.; Wilson, V.G. SUMO-1 Modification of Bovine Papillomavirus E1 Protein Is Required for Intranuclear Accumulation. J. Biol. Chem. 2000, 275, 37999–38004. [Google Scholar] [CrossRef] [PubMed]
- Hoege, C.; Pfander, B.; Moldovan, G.-L.; Pyrowolakis, G.; Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nat. Cell Biol. 2002, 419, 135–141. [Google Scholar] [CrossRef]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Schwienhorst, I.; Johnson, E.S.; Dohmen, R.J. SUMO conjugation and deconjugation. Mol. Genet. Genom. 2000, 263, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.L.; Ho, J.C.Y.; Oliver, A.; Watts, F.Z. Cell-cycle-dependent localisation of Ulp1, a Schizosaccharomyces pombe Pmt3 (SUMO)-specific protease. J. Cell Sci. 2002, 115, 1113–1122. [Google Scholar] [CrossRef]
- Giri, R.; Yeh, H.-H.; Wu, C.-H.; Liu, H.-S. SUMO-1 overexpression increases RbAp46 protein stability and suppresses cell growth. Anticancer Res. 2009, 28, 3749–3756. [Google Scholar]
- Neyret-Kahn, H.; Benhamed, M.; Ye, T.; Le Gras, S.; Cossec, J.-C.; Lapaquette, P.; Bischof, O.; Ouspenskaia, M.; Dasso, M.; Seeler, J.; et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013, 23, 1563–1579. [Google Scholar] [CrossRef]
- Di Genova, B.M.; Da Silva, R.C.; Da Cunha, J.P.C.; Gargantini, P.R.; Mortara, R.A.; Tonelli, R.R. Protein SUMOylation is Involved in Cell-cycle Progression and Cell Morphology in Giardia lamblia. J. Eukaryot. Microbiol. 2016, 64, 491–503. [Google Scholar] [CrossRef]
- Liao, S.; Wang, T.; Fan, K.; Tu, X. The small ubiquitin-like modifier (SUMO) is essential in cell cycle regulation in Trypanosoma brucei. Exp. Cell Res. 2010, 316, 704–715. [Google Scholar] [CrossRef]
- Maruthi, M.; Singh, D.; Reddy, S.R.; Kumar, K.A.; Mastan, B.S.; Mishra, S. Modulation of host cell SUMOylation facilitates efficient development of Plasmodium berghei and Toxoplasma gondii. Cell. Microbiol. 2017, 19, e12723. [Google Scholar] [CrossRef] [PubMed]
- Avalos-Padilla, Y.; Betanzos, A.; Javier-Reyna, R.; García-Rivera, G.; Chávez-Munguía, B.; Lagunes-Guillén, A.; Ortega, J.; Orozco, E. EhVps32 Is a Vacuole-Associated Protein Involved in Pinocytosis and Phagocytosis of Entamoeaba histolytica. PLOS Pathog. 2015, 11, e1005079. [Google Scholar] [CrossRef][Green Version]
- Bañuelos, C.; García-Rivera, G.; López-Reyes, I.; Mendoza, L.; González-Robles, A.; Herranz, S.; Vincent, O.; Orozco, E. EhADH112 Is a Bro1 Domain-Containing Protein Involved in the Entamoeba histolytica Multivesicular Bodies Pathway. J. Biomed. Biotechnol. 2012, 2012, 1–15. [Google Scholar] [CrossRef]
- López-Reyes, I.; García-Rivera, G.; Bañuelos, C.; Herranz, S.; Vincent, O.; López-Camarillo, C.; Marchat, L.A.; Orozco, E. Detection of the Endosomal Sorting Complex Required for Transport in Entamoeba histolytica and Characterization of the EhVps4 Protein. J. Biomed. Biotechnol. 2010, 2010, 1–15. [Google Scholar] [CrossRef]
- Avalos-Padilla, Y.; Knorr, R.L.; Javier-Reyna, R.; García-Rivera, G.; Lipowsky, R.; Dimova, R.; Orozco, E. The Conserved ESCRT-III Machinery Participates in the Phagocytosis of Entamoeba histolytica. Front. Cell. Infect. Microbiol. 2018, 8. [Google Scholar] [CrossRef]
- Walsh, J.A. Problems in Recognition and Diagnosis of Amebiasis: Estimation of the Global Magnitude of Morbidity and Mortality. Clin. Infect. Dis. 1986, 8, 228–238. [Google Scholar] [CrossRef]
- Vranych, C.V.; Merino, M.C.; Zamponi, N.; Touz, M.C.; Rópolo, A.S. SUMOylation in Giardia lamblia: A Conserved Post-Translational Modification in One of the Earliest Divergent Eukaryotes. Biomolecules 2012, 2, 312–330. [Google Scholar] [CrossRef] [PubMed]
- Lois, L.M.; Lima, C.D.; Chua, N.-H. Small Ubiquitin-Like Modifier Modulates Abscisic Acid Signaling in Arabidopsis. Plant Cell 2003, 15, 1347–1359. [Google Scholar] [CrossRef] [PubMed]
- Gill, G. SUMO and ubiquitin in the nucleus: Different functions, similar mechanisms? Genes Dev. 2004, 18, 2046–2059. [Google Scholar] [CrossRef] [PubMed]
- Kirkin, V.; Dikic, I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr. Opin. Cell Biol. 2007, 19, 199–205. [Google Scholar] [CrossRef]
- Lehembre, F.; Badenhorst, P.; Müller, S.; Travers, A.; Schweisguth, F.; Dejean, A. Covalent Modification of the Transcriptional Repressor Tramtrack by the Ubiquitin-Related Protein Smt3 in Drosophila Flies. Mol. Cell. Biol. 2000, 20, 1072–1082. [Google Scholar] [CrossRef]
- Huang, H.-W.; Tsoi, S.C.-M.; Sun, Y.H.; Li, S.S.-L. Identification and characterization of the SMT3 cDNA and gene encoding ubiquitin-like protein from Drosophila melanogaster. IUBMB Life 1998, 46, 775–785. [Google Scholar] [CrossRef]
- Jones, D.; Crowe, E.; Stevens, T.A.; Candido, E.P.M. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: Ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 2001, 3, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Walker, J.M.; Smalle, J.; Gosink, M.M.; Davis, S.J.; Durham, T.L.; Sung, D.-Y.; Vierstra, R.D. The Small Ubiquitin-like Modifier (SUMO) Protein Modification System in Arabidopsis. J. Biol. Chem. 2003, 278, 6862–6872. [Google Scholar] [CrossRef] [PubMed]
- Braun, L.; Cannella, D.; Pinheiro, A.M.; Kieffer, S.; Belrhali, H.; Garin, J.; Hakimi, M.-A. The small ubiquitin-like modifier (SUMO)-conjugating system of Toxoplasma gondii. Int. J. Parasitol. 2009, 39, 81–90. [Google Scholar] [CrossRef]
- Dohmen, R.J. SUMO protein modification. Biochim. Biophys. Acta Bioenerg. 2004, 1695, 113–131. [Google Scholar] [CrossRef]
- Zhao, J. Sumoylation regulates diverse biological processes. Cell. Mol. Life Sci. 2007, 64, 3017–3033. [Google Scholar] [CrossRef]
- Sheng, Z.; Wang, X.; Ma, Y.; Zhang, D.; Yang, Y.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. MS-based strategies for identification of protein SUMOylation modification. Electrophoresis 2019, 40, 2877–2887. [Google Scholar] [CrossRef]
- Valdez, M.S.; Orozco, E.; Correa-Basurto, J.; Bello, M.; Chávez-Munguía, B.; Betanzos, A. Heterodimerization of the Entamoeba histolytica EhCPADH virulence complex through molecular dynamics and protein–protein docking. J. Biomol. Struct. Dyn. 2016, 35, 486–503. [Google Scholar] [CrossRef]
- Johnson, E.S. Protein Modification by SUMO. Annu. Rev. Biochem. 2004, 73, 355–382. [Google Scholar] [CrossRef]
- Park-Sarge, O.K.; Sarge, K.D. Detection of sumoylated proteins. Methods Mol. Biol. 2008, 464, 255–265. [Google Scholar] [CrossRef]
- Arroyo, R.; Orozco, E. Localization and identification of Entamoeba histolytica ahesin. Mol. Biochem Parasitol. 1987, 23, 151–158. [Google Scholar] [CrossRef]
- Zhao, Q.; Xie, Y.; Zheng, Y.; Jiang, S.; Liu, W.; Mu, W.; Liu, Z.; Zhao, Y.; Xue, Y.; Ren, J. GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014, 42, W325–W330. [Google Scholar] [CrossRef]
- Eberhart, R.; Kennedy, J. A new Optimizer Using Particle Swarm Theory. In Proceedings of the MHS’95 Proceedings of the Sixth International Symposium on Micro Machine and Human Science, Nagoya, Japan, 4–6 October 1995; pp. 39–43. [Google Scholar] [CrossRef]
- Perry, J.J.P.; Tainer, J.A.; Boddy, M.N. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 2008, 33, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Javier-Reyna, R.; Montaño, S.; García-Rivera, G.; Rodríguez, M.A.; González-Robles, A.; Orozco, E. EhRabB mobilises the EhCPADH complex through the actin cytoskeleton during phagocytosis of Entamoeba histolytica. Cell. Microbiol. 2019, 21, e13071. [Google Scholar] [CrossRef] [PubMed]
- Solís, C.F.; Santi-Rocca, J.; Perdomo, R.; Weber, C.; Guillén, N. Use of Bacterially Expressed dsRNA to Downregulate Entamoeba histolytica Gene Expression. PLoS ONE 2009, 4, e8424. [Google Scholar] [CrossRef]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef]
- García-Rivera, G.; Rodríguez, M.A.; Ocádiz, R.; Martínez-López, M.C.; Arroyo, R.; González-Robles, A.; Orozco, E. Entamoeba histolytica: A novel cysteine protease and an adhesin form the 112 kDa surface protein. Mol. Microbiol. 1999, 33, 556–568. [Google Scholar] [CrossRef]
- Orozco, E.; Marchat, L.A.; Gómez, C.; López-Camarillo, C.; Pérez, D.G. Drug Resistance Mechanisms in Entamoeba histolytica, Giardia lamblia, Trichomonas vaginalis, and Opportunistic Anaerobic Protozoa. Antimicrob. Drug Resist. 2009, 549–559. [Google Scholar] [CrossRef]
- Van Der Veen, A.G.; Ploegh, H.L. Ubiquitin-Like Proteins. Annu. Rev. Biochem. 2012, 81, 323–357. [Google Scholar] [CrossRef]
- Karpiyevich, M.; Artavanis-Tsakonas, K. Ubiquitin-Like Modifiers: Emerging Regulators of Protozoan Parasites. Biomolecules 2020, 10, 1403. [Google Scholar] [CrossRef] [PubMed]
- Sylvestersen, K.B.; Young, C.; Nielsen, M.L. Advances in characterizing ubiquitylation sites by mass spectrometry. Curr. Opin. Chem. Biol. 2013, 17, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Sundaramoorthy, V.; Green, D.; Locke, K.; O’Brien, C.M.; Dearnley, M.; Bingham, J. Novel role of SARM1 mediated axonal degeneration in the pathogenesis of rabies. PLoS Pathog. 2020, 16, e1008343. [Google Scholar] [CrossRef]
- Gill, G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005, 15, 536–541. [Google Scholar] [CrossRef]
- Verger, A.; Perdomo, J.; Crossley, M. Modification with SUMO. EMBO Rep. 2003, 4, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, W.-G.; Xu, X. Ubiquitin-like modifications in the DNA damage response. Mutat. Res. Mol. Mech. Mutagen. 2017, 803–805, 56–75. [Google Scholar] [CrossRef]
- Enserink, J.M. Regulation of Cellular Processes by SUMO: Understudied Topics. In SUMO Regulation of Cellular Processes (Advances in Experimental Medicine and Biology, 963), 2nd ed.; Wilson, V., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 89–97. [Google Scholar] [CrossRef]
- Rothenbusch, U.; Sawatzki, M.; Chang, Y.; Caesar, S.; Schlenstedt, G. Sumoylation regulates Kap114-mediated nuclear transport. EMBO J. 2012, 31, 2461–2472. [Google Scholar] [CrossRef]
- Arya, S.; Sharma, G.; Gupta, P.; Tiwari, S. In silico analysis of ubiquitin/ubiquitin-like modifiers and their conjugating enzymes in Entamoeba species. Parasitol. Res. 2012, 111, 37–51. [Google Scholar] [CrossRef]
- Kumari, R.; Gupta, P.; Tiwari, S. Ubc7/Ube2g2 ortholog in Entamoeba histolytica: Connection with the plasma membrane and phagocytosis. Parasitol. Res. 2018, 117, 1599–1611. [Google Scholar] [CrossRef]
- Meulmeester, E.; Kunze, M.; Hsiao, H.H.; Urlaub, H.; Melchior, F. Mechanism and Consequences for Paralog-Specific Sumoylation of Ubiquitin-Specific Protease 25. Mol. Cell 2008, 30, 610–619. [Google Scholar] [CrossRef]
- Shang, Q.; Xu, C.; Zhang, J.; Zhang, X.; Tu, X. Solution structure of SUMO from Trypanosoma brucei and its interaction with Ubc9. Proteins Struct. Funct. Bioinform. 2009, 76, 266–269. [Google Scholar] [CrossRef]
- Kerscher, O.; Felberbaum, R.; Hochstrasser, M. Modification of Proteins by Ubiquitin and Ubiquitin-Like Proteins. Annu. Rev. Cell Dev. Biol. 2006, 22, 159–180. [Google Scholar] [CrossRef]
- Bolaños, J.; Betanzos, A.; Javier-Reyna, R.; Rivera, G.G.; Huerta, M.; Pais-Morales, J.; Gonzalez-Robles, A.; Rodríguez, M.A.; Schnoor, M.; Orozco, E. EhNPC1 and EhNPC2 Proteins Participate in Trafficking of Exogenous Cholesterol in Entamoeba histolytica Trophozoites: Relevance for Phagocytosis. PLoS Pathog. 2016, 12, e1006089. [Google Scholar] [CrossRef]
- McCullough, J.; Fisher, R.D.; Whitby, F.G.; Sundquist, W.I.; Hill, C.P. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 7687–7691. [Google Scholar] [CrossRef]
- Orozco, E.; Suárez, M.E.; Sánchez, T. Differences in adhesion, phagocytosis and virulence of clones from Entamoeba histolytica, strain HM1: IMSS. Int. J. Parasitol. 1985, 15, 655–660. [Google Scholar] [CrossRef]
- Diamond, L.S.; Harlow, D.R.; Cunnick, C.C. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 1978, 72, 431–432. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. William Humphrey, “Visual Molecular Dynamics.Pdf”. J. Mol. Graph. 1996, 4, 33–38. [Google Scholar] [CrossRef]
- Kale, L.V.; Bhatele, A. Scalable Molecular Dynamics with NAMD. Parallel Sci. Eng. Appl. 2016, 26, 91–107. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins†. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Batcho, P.F.; Case, D.A.; Schlick, T. Optimized particle-mesh Ewald/multiple-time step integration for molecular dynamics simulations. J. Chem. Phys. 2001, 115, 4003–4018. [Google Scholar] [CrossRef]
- Koukos, P.I.; Glykos, N.M. Grcarma: A fully automated task-oriented interface for the analysis of molecular dynamics trajectories. J. Comput. Chem. 2013, 34, 2310–2312. [Google Scholar] [CrossRef]
- Comeau, S.R.; Gatchell, D.W.; Vajda, S.; Camacho, C.J. ClusPro: An automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 2004, 20, 45–50. [Google Scholar] [CrossRef]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How Good is Automated Protein Docking? Proteins Struct. Funct. Bioinform. 2013, 81, 2159–2166. [Google Scholar] [CrossRef] [PubMed]
- Luis, B.G.; Pastor-Palacios, G.; Rodriguez-Rocha, R.; Azuara-Liceaga, E. Cloning and initial characterization of a family A DNA polymerase from Entamoeba histolytica: A putative mitochondrial DNA Polymerase. FASEB J. 2007, 21, A1039. [Google Scholar]
- Huerta, M.; Reyes, L.; García-Rivera, G.; Bañuelos, C.; Betanzos, A.; Díaz-Hernández, M.; Galindo, A.; Bolaños, J.; Cárdenas, H.; Azuara-Liceaga, E.; et al. A noncanonical GATA transcription factor of Entamoeba histolytica modulates genes involved in phagocytosis. Mol. Microbiol. 2020, 114, 1019–1037. [Google Scholar] [CrossRef] [PubMed]
- Trasviña-Arenas, C.H.; Cardona-Felix, C.S.; Azuara-Liceaga, E.; Díaz-Quezada, C.; Brieba, L.G. Proliferating cell nuclear antigen restores the enzymatic activity of a DNA ligase I deficient in DNA binding. FEBS Open Biol. 2017, 7, 659–674. [Google Scholar] [CrossRef] [PubMed]
- Novikoff, A.B.; Novikoff, P.M.; Davis, C.; Quintana, N. Studies on microperoxisomes II. A cytochemical method for light and electron microscopy. J. Histochem. Cytochem. 1972, 20, 1006–1023. [Google Scholar] [CrossRef]
- Takiff, H.E.; Chen, S.-M.; Court, D.L. Genetic analysis of the rnc operon of Escherichia coli. J. Bacteriol. 1989, 171, 2581–2590. [Google Scholar] [CrossRef]



| Putative Protein | Access Number |
|---|---|
| Ran GTPase-activating protein | EHI_185290 |
| SP-RING zinc finger domain-containing protein | EHI_069470 |
| SP-RING zinc finger domain-containing protein | EHI_152530 |
| Ubiquitin-conjugating enzyme family protein ubiquitin-conjugating enzyme E2 | EHI_147470 |
| Ubiquitin-conjugating enzyme family protein | EHI_178500 |
| Ulp1 protease family, C-terminal catalytic domain containing | EHI_067510 |
| Ubiquitin-activating enzyme ubiquitin-like 1-activating enzyme E1 B | EHI_035540 |
| Ulp1 protease family, c-terminal catalytic domain containing | EHI_097940 |
| Proliferating cell nuclear antigen (PCNA) | EHI_128450 |
| Protein | E. histolytica | S. cerevisiae | Identity | H. sapiens | Identity | G. lamblia | Identity | Protein Type |
|---|---|---|---|---|---|---|---|---|
| SUMO | EHI_170060 | YDR510W | 55% | NP_008868.3 | 48.35% | GL50803_7760 | 33% | SUMO protein ubiquitin-activating enzyme, putative |
| E1 | EHI_035540 | YDR390C | 35% | NP_005490.1 | 36% | GL50803_6288 | 25% | SUMO-conjugating enzyme UBC9, putative |
| E2a | EHI_178500 | YDL064W | 51% | NP_003336.1 | 49.67% | GL50803_24068 | 46% | SUMO-conjugating enzyme UBC9, putative |
| E2b | EHI_147470 | YDL064W | 56% | NP_003336.1 | 53.55% | GL50803_24068 | 49% | sumo ligase, putative |
| E3a | EHI_098320 | YOR156C | 29% | NP_775298.1 | 27.88% | GL50803_11930 | 38% | sumo ligase, putative |
| E3b | EHI_069470 | YOR156C | 31% | XP_011538282.1 | 32.22% | GL50803_11930 | 40% | sumo ligase, putative |
| * Upl1a | EHI_067510 | YPL020C | 27% | XP_006719425.1 | 27.5% | GL50803_16438 | 25% | sentrin/sumo-specific protease, putative |
| * Upl1b | EHI_097940 | YIL031W | 24% | NP_001070671.1 | 23.6% | GL50803_16438 | 32% | Ulp1 protease family, C-terminal catalytic domain-containing protein |
| ESCRT Complex | Protein | Position | Peptide/Sequence | Score | Site | Protein Size (aa) |
|---|---|---|---|---|---|---|
| ESCRT-III | EhVps2 | 117 | RKVNEATKLPAMQKV | 24,671 | SUMOylation | 246 |
| EhVps20 | 32–36 | VTDLDQKIVDLDRQIRQNI | 65,754 | SIM | 206 | |
| EhVps24 | 176 | EGGIEAVKNEVIAES | 43,439 | SUMOylation | 205 | |
| EhVps32 | 133–137 | NNEKSHEIGDLLGEDLQDI | 64,005 | SIM | 204 | |
| ESCRT Accessory Proteins | EhADH | 154 | QAAGAFQKAADCAQL | 25,680 | SUMOylation | 687 |
| - | 366–370 | EYNSKAQ VILND SKKCES | 58,036 | SIM | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Hernández, M.; Javier-Reyna, R.; Sotto-Ortega, I.; García-Rivera, G.; Montaño, S.; Betanzos, A.; Zanatta, D.; Orozco, E. Protein Sumoylation Is Crucial for Phagocytosis in Entamoeba histolytica Trophozoites. Int. J. Mol. Sci. 2021, 22, 5709. https://doi.org/10.3390/ijms22115709
Díaz-Hernández M, Javier-Reyna R, Sotto-Ortega I, García-Rivera G, Montaño S, Betanzos A, Zanatta D, Orozco E. Protein Sumoylation Is Crucial for Phagocytosis in Entamoeba histolytica Trophozoites. International Journal of Molecular Sciences. 2021; 22(11):5709. https://doi.org/10.3390/ijms22115709
Chicago/Turabian StyleDíaz-Hernández, Mitzi, Rosario Javier-Reyna, Izaid Sotto-Ortega, Guillermina García-Rivera, Sarita Montaño, Abigail Betanzos, Dxinegueela Zanatta, and Esther Orozco. 2021. "Protein Sumoylation Is Crucial for Phagocytosis in Entamoeba histolytica Trophozoites" International Journal of Molecular Sciences 22, no. 11: 5709. https://doi.org/10.3390/ijms22115709
APA StyleDíaz-Hernández, M., Javier-Reyna, R., Sotto-Ortega, I., García-Rivera, G., Montaño, S., Betanzos, A., Zanatta, D., & Orozco, E. (2021). Protein Sumoylation Is Crucial for Phagocytosis in Entamoeba histolytica Trophozoites. International Journal of Molecular Sciences, 22(11), 5709. https://doi.org/10.3390/ijms22115709






