Kawasaki Disease Patient Stratification and Pathway Analysis Based on Host Transcriptomic and Proteomic Profiles
Abstract
1. Introduction
2. Results
2.1. Description of Datasets
| Dataset Name | GEO Accession(s) | Platform(s) Used for Generation | KD | DB | DV | HC | Citation(s) |
|---|---|---|---|---|---|---|---|
| Transcriptomic discovery | GSE73461 | Microarray: HumanHT-12 version 4.0 | 77 | 31 | 92 | 62 | [18] |
| Transcriptomic validation | GSE73462 GSE73463 | Microarrays: 1 × HumanHT-12 version 3.0 1 × HumanHT-12 version 4.0 | 101 | 23 | 28 | 16 * | [19,22] |
| Proteomic discovery | NA | LC–MS/MS | 26 | 73 | 75 | 25 | unpublished |
| Proteomic validation | NA | SomaScan [20] | 26 | 48 | 31 | 25 | unpublished |
2.2. Comparison of Kawasaki Disease to Bacterial and Viral Infection
2.2.1. Differential Abundance Analysis
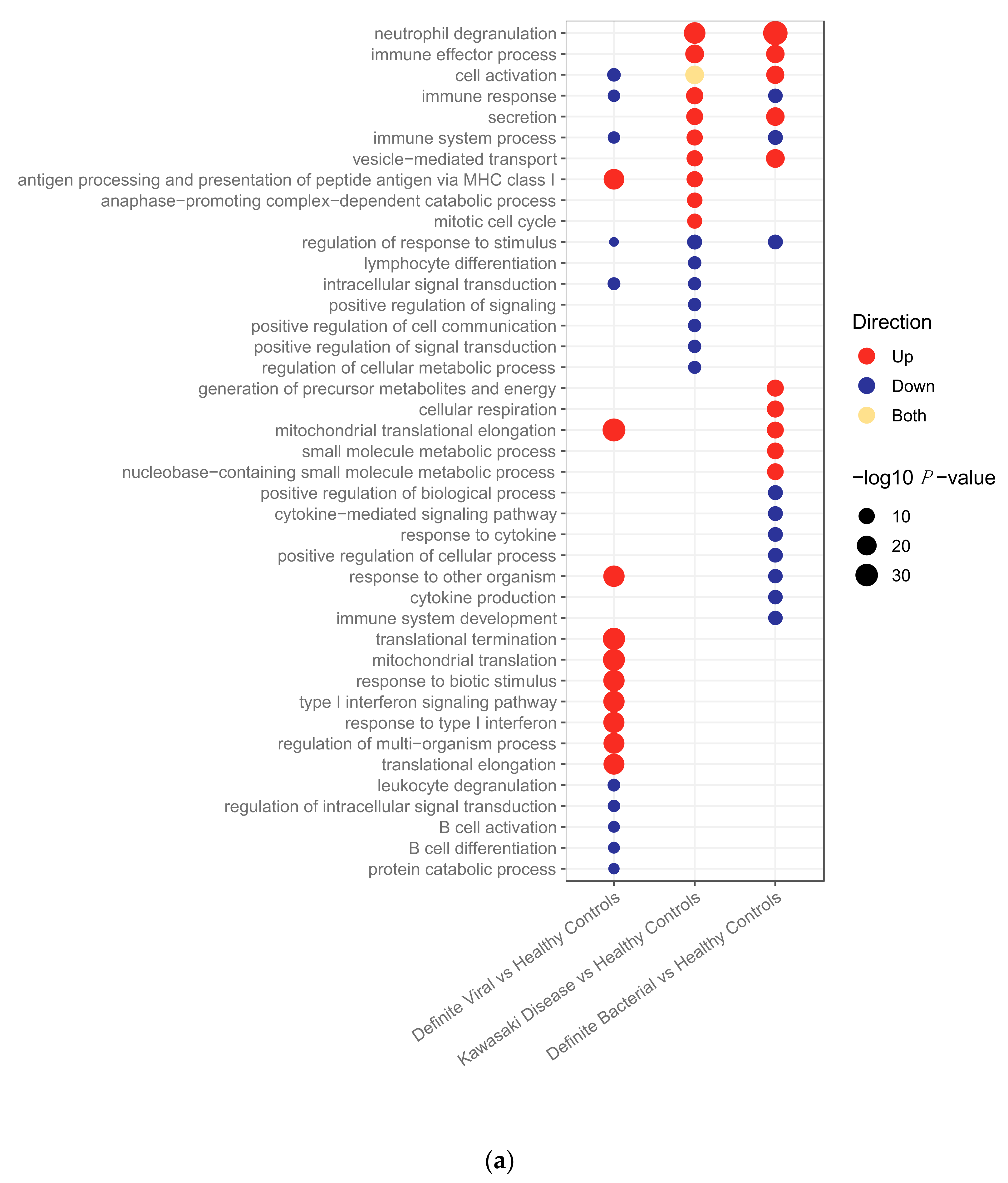
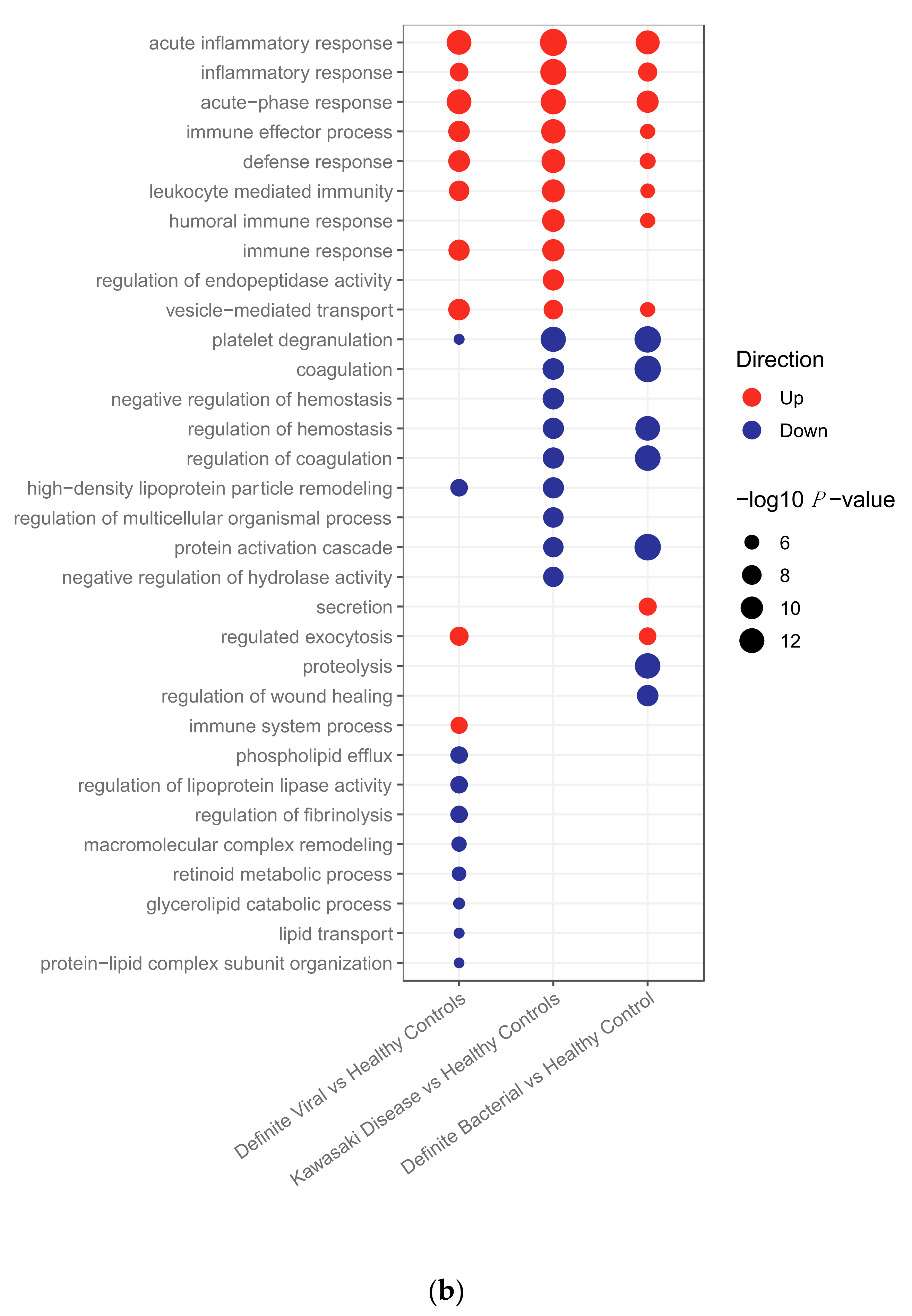
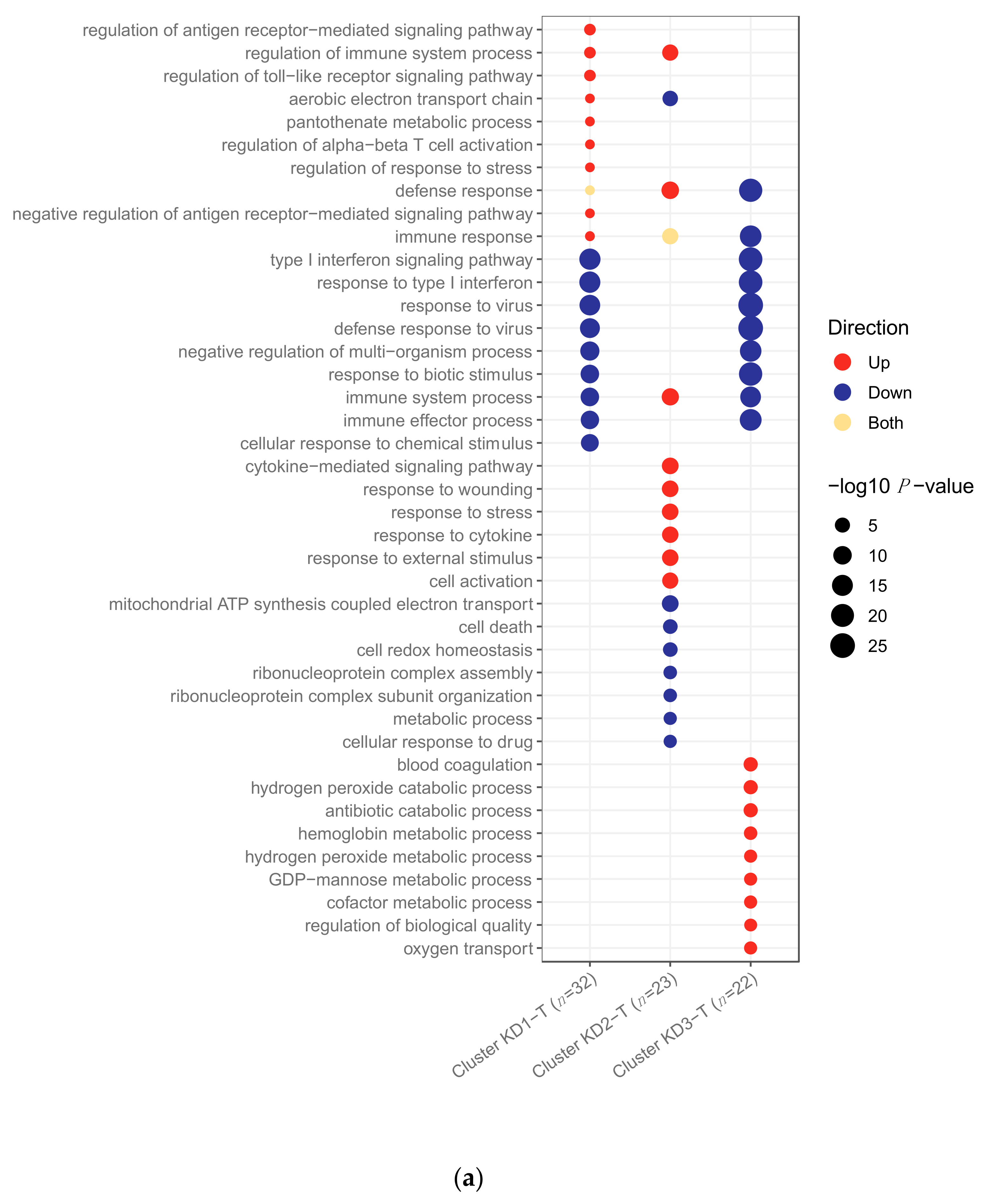
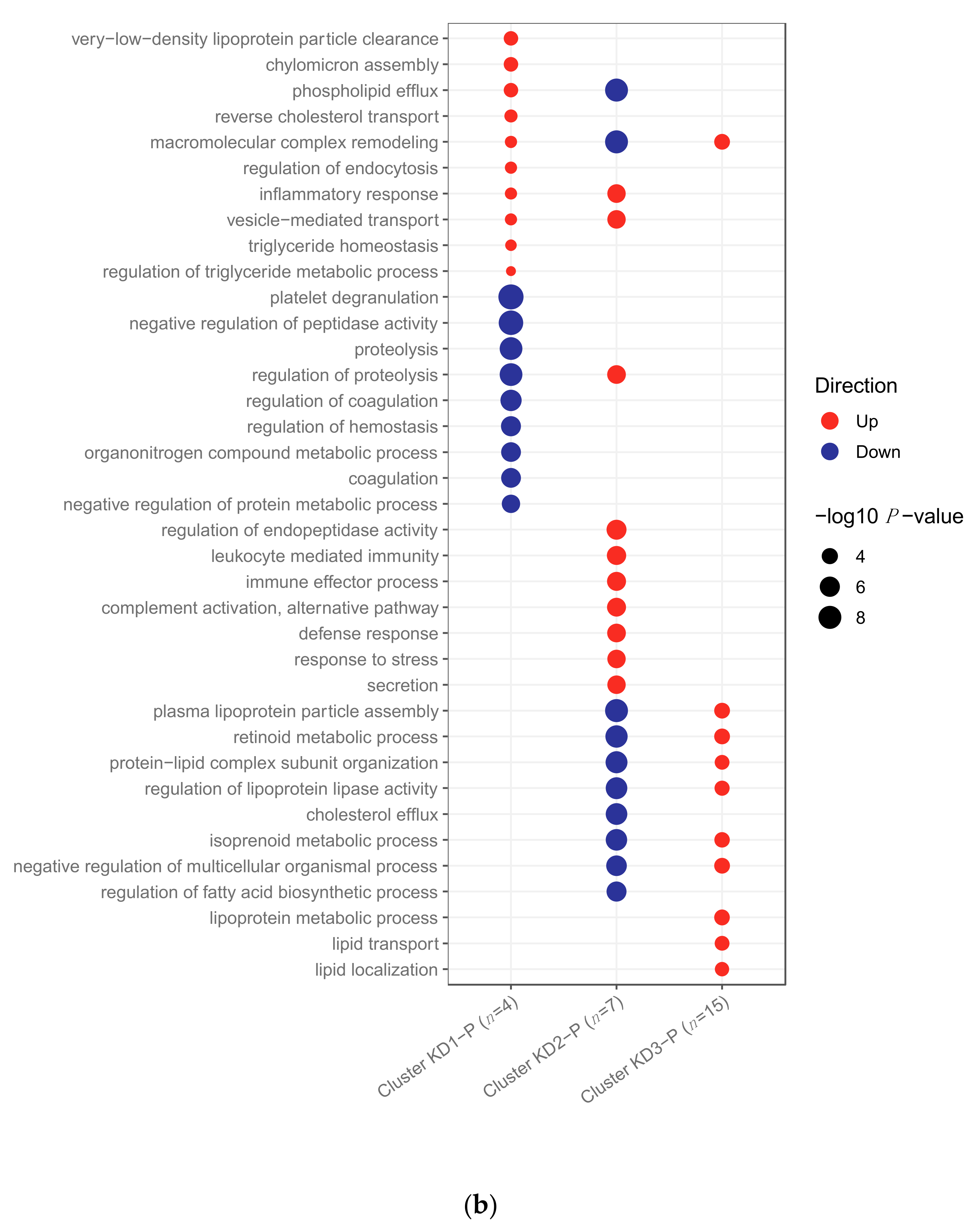
2.2.2. Pathway Analysis
2.2.3. Clustering
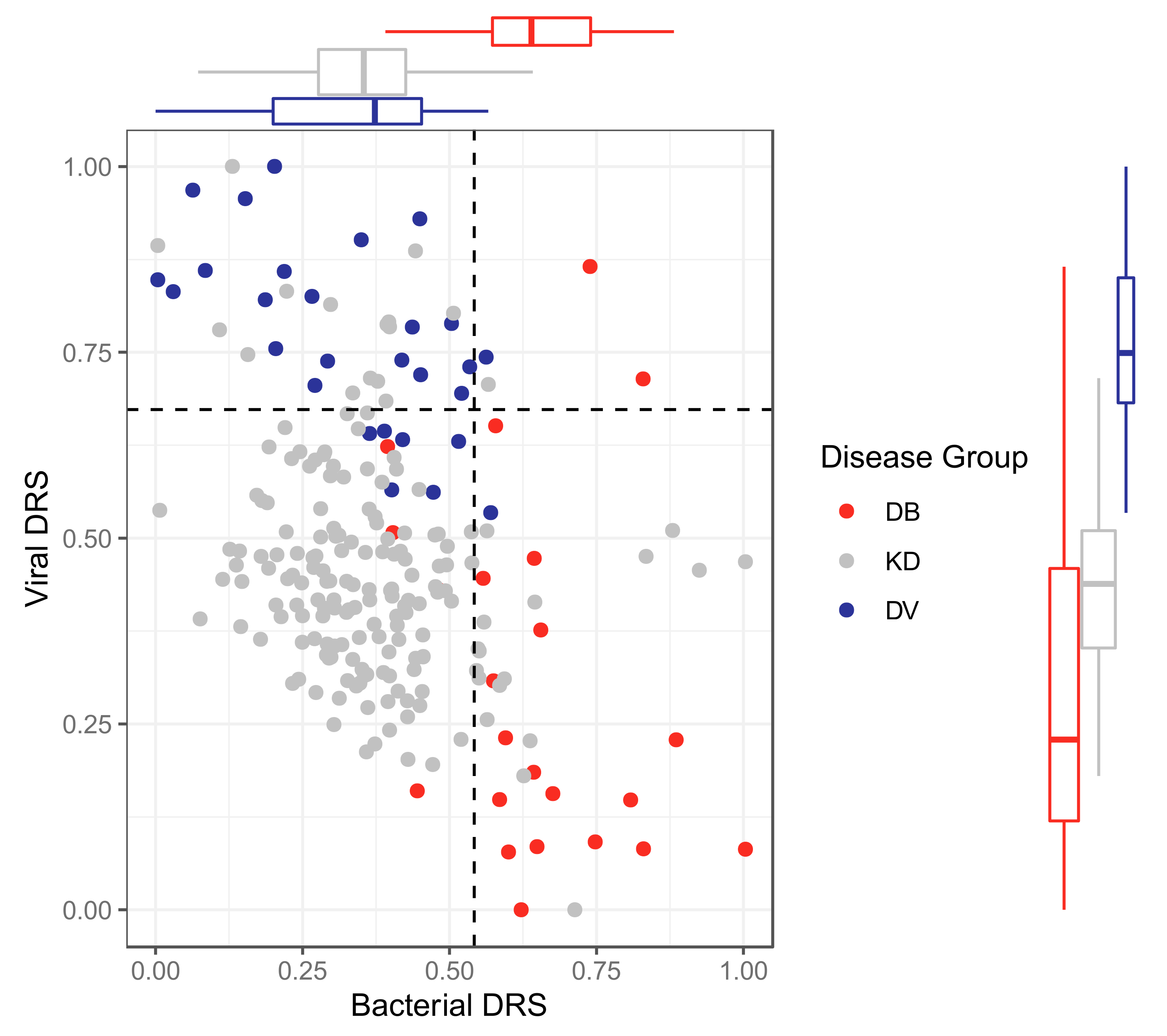
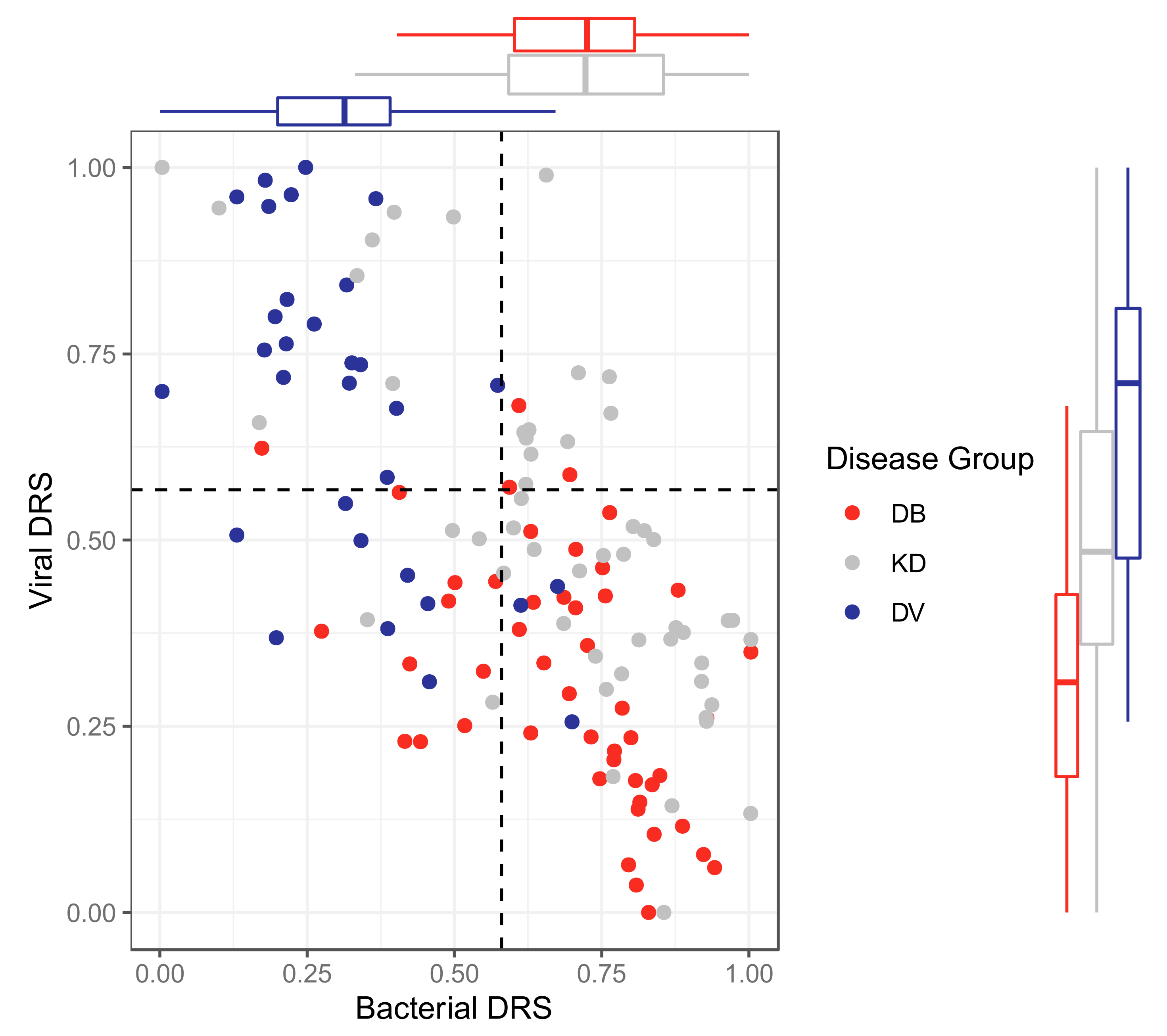
2.2.4. Classification Using Disease Risk Scores
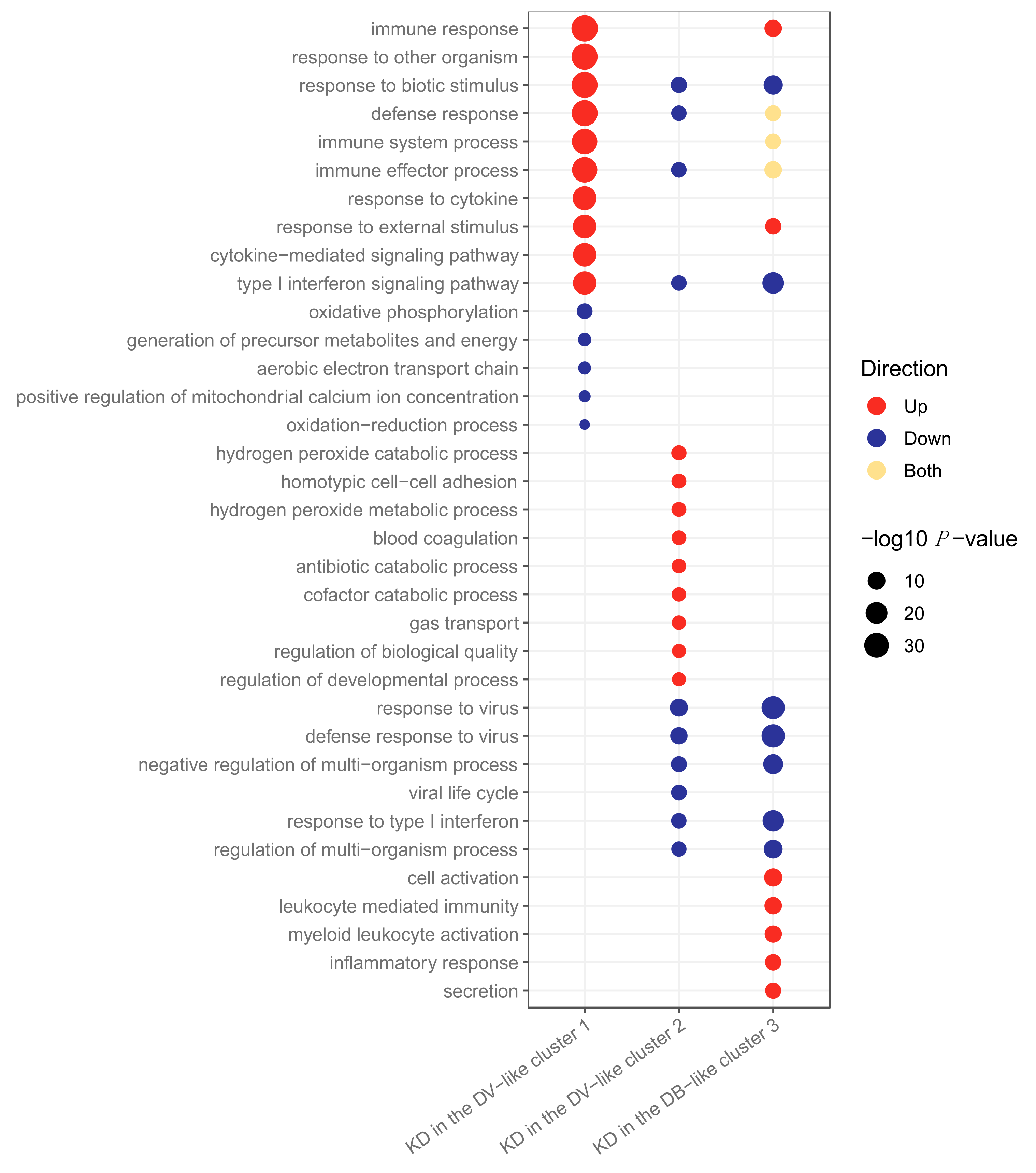
2.3. Clustering of Kawasaki Disease Patients Alone
3. Discussion
4. Materials and Methods
4.1. Patient Recruitment
4.2. Data Generation
4.2.1. Transcriptomic Datasets
4.2.2. Proteomic Datasets
4.3. Statistical Methods
4.3.1. Pre-Processing of Gene Expression Data
4.3.2. Pre-Processing of Protein Abundance Data
4.3.3. Comparison of Kawasaki Disease to Bacterial and Viral Infections
Differential Abundance Analysis
Pathway Analysis
Clustering Analysis
Classification
4.3.4. Exploration of Kawasaki Disease Samples Alone
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CAA | Coronary artery aneurysm |
| DB | Definite bacterial |
| DRS | Disease risk score |
| DV | Definite viral |
| FDR | False discovery rate |
| HC | Healthy control |
| KD | Kawasaki disease |
| LFC | Log-fold change |
| SDA | Significantly differentially abundant |
References
- Kawasaki, T.; Kosaki, F.; Okawa, S.; Shigematsu, I.; Yanagawa, H. A New Infantile Acute Febrile Mucocutaneous Lymph Node Syndrome (MLNS) Prevailing in Japan. Pediatrics 1974, 54. [Google Scholar]
- Ramphul, K.; Mejias, S.G. Kawasaki disease: A comprehensive review. Arch. Med. Sci. Atheroscler. Dis. 2018, 3, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Ogata, S.; Shimizu, C.; Franco, A.; Touma, R.; Kanegaye, J.T.; Choudhury, B.P.; Naidu, N.N.; Kanda, Y.; Hoang, L.T.; Hibberd, M.L.; et al. Treatment Response in Kawasaki Disease Is Associated with Sialylation Levels of Endogenous but Not Therapeutic Intravenous Immunoglobulin G. PLoS ONE 2013, 8, e81448. [Google Scholar] [CrossRef] [PubMed]
- Skochko, S.M.; Jain, S.; Sun, X.; Sivilay, N.; Kanegaye, J.T.; Pancheri, J.; Shimizu, C.; Sheets, R.; Tremoulet, A.H.; Burns, J.C. Kawasaki Disease Outcomes and Response to Therapy in a Multiethnic Community: A 10-Year Experience. J. Pediatr. 2018, 203, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Brogan, P.; Burns, J.C.; Cornish, J.; Diwakar, V.; Eleftheriou, D.; Gordon, J.B.; Gray, H.H.; Johnson, T.W.; Levin, M.; Malik, I.; et al. Lifetime cardiovascular management of patients with previous Kawasaki disease. Heart 2020, 106, 411–420. [Google Scholar] [CrossRef]
- Singh, S.; Vignesh, P.; Burgner, D. The epidemiology of Kawasaki disease: A global update. Arch. Dis. Child. 2015, 100, 1084–1088. [Google Scholar] [CrossRef]
- Nagata, S. Causes of Kawasaki Disease—From Past to Present. Front. Pediatr. 2019, 7, 18. [Google Scholar] [CrossRef]
- Dietz, S.M.; van Stijn, D.; Burgner, D.; Levin, M.; Kuipers, I.M.; Hutten, B.A.; Kuijpers, T.W. Dissecting Kawasaki disease: A state-of-the-art review. Eur. J. Pediatr. 2017, 176, 995–1009. [Google Scholar] [CrossRef]
- Nakamura, A.; Ikeda, K.; Hamaoka, K. Aetiological significance of infectious stimuli in Kawasaki disease. Front. Pediatr. 2019, 7, 244. [Google Scholar] [CrossRef]
- Rodó, X.; Ballester, J.; Cayan, D.; Melish, M.E.; Nakamura, Y.; Uehara, R.; Burns, J.C. Association of Kawasaki disease with tropospheric wind patterns. Sci. Rep. 2011, 1, 150. [Google Scholar] [CrossRef]
- Rypdal, M.; Rypdal, V.; Burney, J.A.; Cayan, D.; Bainto, E.; Skochko, S.; Tremoulet, A.H.; Creamean, J.; Shimizu, C.; Kim, J.; et al. Clustering and climate associations of Kawasaki Disease in San Diego County suggest environmental triggers. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Levin, M. Childhood Multisystem Inflammatory Syndrome—A New Challenge in the Pandemic. N. Engl. J. Med. 2020, 383, 393–395. [Google Scholar] [CrossRef]
- Whittaker, E.; Bamford, A.; Kenny, J.; Kaforou, M.; Jones, C.E.; Shah, P.; Ramnarayan, P.; Fraisse, A.; Miller, O.; Davies, P.; et al. Clinical Characteristics of 58 Children with a Pediatric Inflammatory Multisystem Syndrome Temporally Associated with SARS-CoV-2. JAMA J. Am. Med. Assoc. 2020, 324, 259–269. [Google Scholar] [CrossRef]
- Dufort, E.M.; Koumans, E.H.; Chow, E.J.; Rosenthal, E.M.; Muse, A.; Rowlands, J.; Barranco, M.A.; Maxted, A.M.; Rosenberg, E.S.; Easton, D.; et al. Multisystem Inflammatory Syndrome in Children in New York State. N. Engl. J. Med. 2020, 383, 347–358. [Google Scholar] [CrossRef]
- McCrindle, B.W.; Manlhiot, C. SARS-CoV-2-Related Inflammatory Multisystem Syndrome in Children: Different or Shared Etiology and Pathophysiology as Kawasaki Disease? JAMA J. Am. Med. Assoc. 2020, 324, 246–248. [Google Scholar] [CrossRef]
- Kaforou, M.; Wright, V.J.; Oni, T.; French, N.; Anderson, S.T.; Bangani, N.; Banwell, C.M.; Brent, A.J.; Crampin, A.C.; Dockrell, H.M.; et al. Detection of Tuberculosis in HIV-Infected and -Uninfected African Adults Using Whole Blood RNA Expression Signatures: A Case-Control Study. PLoS Med. 2013, 10, e1001538. [Google Scholar] [CrossRef]
- Kaforou, M.; Herberg, J.A.; Wright, V.J.; Coin, L.J.M.; Levin, M. Diagnosis of Bacterial Infection Using a 2-Transcript Host RNA Signature in Febrile Infants 60 Days or Younger. JAMA 2017, 317, 1577–1578. [Google Scholar] [CrossRef]
- Wright, V.J.; Herberg, J.A.; Kaforou, M.; Shimizu, C.; Eleftherohorinou, H.; Shailes, H.; Barendregt, A.M.; Menikou, S.; Gormley, S.; Berk, M.; et al. Diagnosis of Kawasaki Disease Using a Minimal Whole-Blood Gene Expression Signature. JAMA Pediatr. 2018, 172, e182293. [Google Scholar] [CrossRef]
- Hoang, L.T.; Shimizu, C.; Ling, L.; Naim, A.N.M.; Khor, C.C.; Tremoulet, A.H.; Wright, V.; Levin, M.; Hibberd, M.L.; Burns, J.C. Global gene expression profiling identifies new therapeutic targets in acute Kawasaki disease. Genome Med. 2014. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010. [Google Scholar] [CrossRef]
- McCrindle, B.W.; Rowley, A.H.; Newburger, J.W.; Burns, J.C.; Bolger, A.F.; Gewitz, M.; Baker, A.L.; Jackson, M.A.; Takahashi, M.; Shah, P.B.; et al. Diagnosis, treatment, and long-term management of Kawasaki disease: A scientific statement for health professionals from the American Heart Association. Circulation 2017, 135, e927–e999. [Google Scholar] [CrossRef]
- Herberg, J.A.; Kaforou, M.; Gormley, S.; Sumner, E.R.; Patel, S.; Jones, K.D.J.; Paulus, S.; Fink, C.; Martinon-Torres, F.; Montana, G.; et al. Transcriptomic profiling in childhood H1N1/09 influenza reveals reduced expression of protein synthesis genes. J. Infect. Dis. 2013, 208, 1664–1668. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Charrad, M.; Ghazzali, N.; Boiteau, V.; Niknafs, A. Nbclust: An R package for determining the relevant number of clusters in a data set. J. Stat. Softw. 2014. [Google Scholar] [CrossRef]
- Kimura, Y.; Yanagimachi, M.; Ino, Y.; Aketagawa, M.; Matsuo, M.; Okayama, A.; Shimizu, H.; Oba, K.; Morioka, I.; Imagawa, T.; et al. Identification of candidate diagnostic serum biomarkers for Kawasaki disease using proteomic analysis. Sci. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Whitin, J.C.; Yu, T.T.S.; Ling, X.B.; Kanegaye, J.T.; Burns, J.C.; Cohen, H.J. A novel truncated form of serum amyloid a in kawasaki disease. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Danese, S. Nonimmune cells in inflammatory bowel disease: From victim to villain. Trends Immunol. 2008, 29, 555–564. [Google Scholar] [CrossRef]
- Tanaka, K.; Yoshioka, T.; Bieberich, C.; Jay, G. Role of the Major Histocompatibility Complex Class I Antigens in Tumor Growth and Metastasis. Annu. Rev. Immunol. 1988, 6, 359–380. [Google Scholar] [CrossRef]
- Tremoulet, A.H.; Jain, S.; Chandrasekar, D.; Sun, X.; Sato, Y.; Burns, J.C. Evolution of laboratory values in patients with Kawasaki disease. Pediatr. Infect. Dis. J. 2011, 30, 1022–1026. [Google Scholar] [CrossRef]
- Biezeveld, M.H.; van Mierlo, G.; Lutter, R.; Kuipers, I.M.; Dekker, T.; Hack, C.E.; Newburger, J.W.; Kuijpers, T.W. Sustained activation of neutrophils in the course of Kawasaki disease: An association with matrix metalloproteinases. Clin. Exp. Immunol. 2005, 141, 183–188. [Google Scholar] [CrossRef]
- Asano, T.; Ogawa, S. Expression of IL-8 in Kawasaki disease. Clin. Exp. Immunol. 2000, 122, 514–519. [Google Scholar] [CrossRef]
- Zandstra, J.; van de Geer, A.; Tanck, M.W.T.; van Stijn-Bringas Dimitriades, D.; Aarts, C.E.M.; Dietz, S.M.; van Bruggen, R.; Schweintzger, N.A.; Zenz, W.; Emonts, M.; et al. Biomarkers for the Discrimination of Acute Kawasaki Disease From Infections in Childhood. Front. Pediatr. 2020, 8, 355. [Google Scholar] [CrossRef]
- Manlhiot, C.; Mueller, B.; O’Shea, S.; Majeed, H.; Bernknopf, B.; Labelle, M.; Westcott, K.V.; Bai, H.; Chahal, N.; Birken, C.S.; et al. Environmental epidemiology of Kawasaki disease: Linking disease etiology, pathogenesis and global distribution. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Rodó, X.; Curcoll, R.; Robinson, M.; Ballester, J.; Burns, J.C.; Cayan, D.R.; Lipkin, W.I.; Williams, B.L.; Couto-Rodriguez, M.; Nakamura, Y.; et al. Tropospheric winds from northeastern China carry the etiologic agent of Kawasaki disease from its source to Japan. Proc. Natl. Acad. Sci. USA 2014, 111, 7952–7957. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, L.; Du, H.; Zhang, J.; Li, Y.Y.; Qu, J.; Zhang, W.; Wang, Y.; Bao, S.; Li, Y.; et al. SARS-CoV-2 Infection in Children. N. Engl. J. Med. 2020, 382, 1663–1665. [Google Scholar] [CrossRef]
- Götzinger, F.; Santiago-García, B.; Noguera-Julián, A.; Lanaspa, M.; Lancella, L.; Calò Carducci, F.I.; Gabrovska, N.; Velizarova, S.; Prunk, P.; Osterman, V.; et al. COVID-19 in children and adolescents in Europe: A multinational, multicentre cohort study. Lancet Child Adolesc. Heal. 2020, 0. [Google Scholar] [CrossRef]
- Davies, P.; Evans, C.; Kanthimathinathan, H.K.; Lillie, J.; Brierley, J.; Waters, G.; Johnson, M.; Griffiths, B.; du Pré, P.; Mohammad, Z.; et al. Intensive care admissions of children with paediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2 (PIMS-TS) in the UK: a multicentre observational study. Lancet Child Adolesc. Health 2020. [Google Scholar] [CrossRef]
- Burgner, D.; Davila, S.; Breunis, W.B.; Ng, S.B.; Li, Y.; Bonnard, C.; Ling, L.; Wright, V.J.; Thalamuthu, A.; Odam, M.; et al. A genome-wide association study identifies novel and functionally related susceptibility Loci for Kawasaki disease. PLoS Genet. 2009. [Google Scholar] [CrossRef] [PubMed]
- Curtis, N.; Zheng, R.; Lamb, J.R.; Levin, M. Evidence for a superantigen mediated process in Kawasaki disease. Arch. Dis. Child. 1995, 72, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Han, S.B.; Lee, S.Y. Antibiotic use in children with Kawasaki disease. World J. Pediatr. 2018, 14, 621–622. [Google Scholar] [CrossRef] [PubMed]
- Diz, A.P.; Martínez-Fernández, M.; Rolán-Alvarez, E. Proteomics in evolutionary ecology: Linking the genotype with the phenotype. Mol. Ecol. 2012, 21, 1060–1080. [Google Scholar] [CrossRef]
- Benseler, S.M.; McCrindle, B.W.; Silverman, E.D.; Tyrrell, P.N.; Wong, J.; Yeung, R.S.M. Infections and Kawasaki disease: Implications for coronary artery outcome. Pediatrics 2005, 116, e760–e766. [Google Scholar] [CrossRef]
- Martinón-Torres, F.; Salas, A.; Rivero-Calle, I.; Cebey-López, M.; Pardo-Seco, J.; Herberg, J.A.; Boeddha, N.P.; Klobassa, D.S.; Secka, F.; Paulus, S.; et al. Life-threatening infections in children in Europe (the EUCLIDS Project): a prospective cohort study. Lancet Child Adolesc. Health 2018, 2, 404–414. [Google Scholar] [CrossRef]
- R Foundation for Statistical Computing R: A language and environment for statistical computing. R A Lang. Environ. Stat. Comput. 3.3.1 2016.
- Du, P.; Kibbe, W.A.; Lin, S.M. Lumi: A pipeline for processing Illumina microarray. Bioinformatics 2008. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Sweeney, T.E.; COCONUT: COmbat CO-Normalization Using conTrols (COCONUT). R Package Version 1.0.2. 2017. Available online: https://rdrr.io/cran/COCONUT/ (accessed on 25 May 2021).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A K-Means Clustering Algorithm. Appl. Stat. 1979, 28, 100. [Google Scholar] [CrossRef]
- Krzanowski, W.J.; Lai, Y.T. A Criterion for Determining the Number of Groups in a Data Set Using Sum-of-Squares Clustering. Biometrics 1988, 44, 23. [Google Scholar] [CrossRef]
- Caliñski, T.; Harabasz, J. A Dendrite Method For Cluster Analysis. Commun. Stat. 1974, 3, 1–27. [Google Scholar] [CrossRef]
- Gordon, A.D.; Hartigan, J.A. Clustering Algorithms. J. Am. Stat. Assoc. 1976. [Google Scholar] [CrossRef]
- McClain, J.O.; Rao, V.R. CLUSTISZ: A Program to Test for the Quality of Clustering of a Set of Objects. J. Mark. Res. 1975, 12, 456–460. [Google Scholar]
- Dunn, J.C. Well-separated clusters and optimal fuzzy partitions. J. Cybern. 1974, 4, 95–104. [Google Scholar] [CrossRef]
- Halkidi, M.; Vazirgiannis, M.; Balislakis, V. Quality scheme assessment in the clustering process. In Principles of Data Mining and Knowledge Discovery; Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heisenberg, Germany, 2000; Volume 1910, pp. 265–276. [Google Scholar]
- Halkidi, M.; Vazirgiannis, M. Clustering validity assessment: Finding the optimal partitioning of a data set. In Proceedings of the IEEE International Conference on Data Mining, ICDM, San Jose, CA, USA, 29 November–2 December 2001; pp. 187–194. [Google Scholar] [CrossRef]
- Hubert, L.J.; Levin, J.R. A general statistical framework for assessing categorical clustering in free recall. Psychol. Bull. 1976, 83, 1072–1080. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Ball, G.H.; Hall, D.J. Isodata, A Novel Method of Data Analysis and Pattern Classification; Stanford Research Institute: Menlo Park, CA, USA, 1965. [Google Scholar]
- Milligan, G.W. An examination of the effect of six types of error perturbation on fifteen clustering algorithms. Psychometrika 1980, 45, 325–342. [Google Scholar] [CrossRef]
- Milligan, G.W. A monte carlo study of thirty internal criterion measures for cluster analysis. Psychometrika 1981, 46, 187–199. [Google Scholar] [CrossRef]
- Ratkowsky, D.; Lance, G. A Criterion for Determining the Number of Groups in a Classification. Aust. Comput. J. 1978, 10, 115–117. [Google Scholar]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jackson, H.; Menikou, S.; Hamilton, S.; McArdle, A.; Shimizu, C.; Galassini, R.; Huang, H.; Kim, J.; Tremoulet, A.; Thorne, A.; et al. Kawasaki Disease Patient Stratification and Pathway Analysis Based on Host Transcriptomic and Proteomic Profiles. Int. J. Mol. Sci. 2021, 22, 5655. https://doi.org/10.3390/ijms22115655
Jackson H, Menikou S, Hamilton S, McArdle A, Shimizu C, Galassini R, Huang H, Kim J, Tremoulet A, Thorne A, et al. Kawasaki Disease Patient Stratification and Pathway Analysis Based on Host Transcriptomic and Proteomic Profiles. International Journal of Molecular Sciences. 2021; 22(11):5655. https://doi.org/10.3390/ijms22115655
Chicago/Turabian StyleJackson, Heather, Stephanie Menikou, Shea Hamilton, Andrew McArdle, Chisato Shimizu, Rachel Galassini, Honglei Huang, Jihoon Kim, Adriana Tremoulet, Adam Thorne, and et al. 2021. "Kawasaki Disease Patient Stratification and Pathway Analysis Based on Host Transcriptomic and Proteomic Profiles" International Journal of Molecular Sciences 22, no. 11: 5655. https://doi.org/10.3390/ijms22115655
APA StyleJackson, H., Menikou, S., Hamilton, S., McArdle, A., Shimizu, C., Galassini, R., Huang, H., Kim, J., Tremoulet, A., Thorne, A., Fischer, R., de Jonge, M. I., Kuijpers, T., Wright, V., Burns, J. C., Casals-Pascual, C., Herberg, J., Levin, M., Kaforou, M., & on behalf of the PERFORM Consortium. (2021). Kawasaki Disease Patient Stratification and Pathway Analysis Based on Host Transcriptomic and Proteomic Profiles. International Journal of Molecular Sciences, 22(11), 5655. https://doi.org/10.3390/ijms22115655







