Biochemical and Initial Structural Characterization of the Monocot Chimeric Jacalin OsJAC1
Abstract
1. Introduction
2. Results
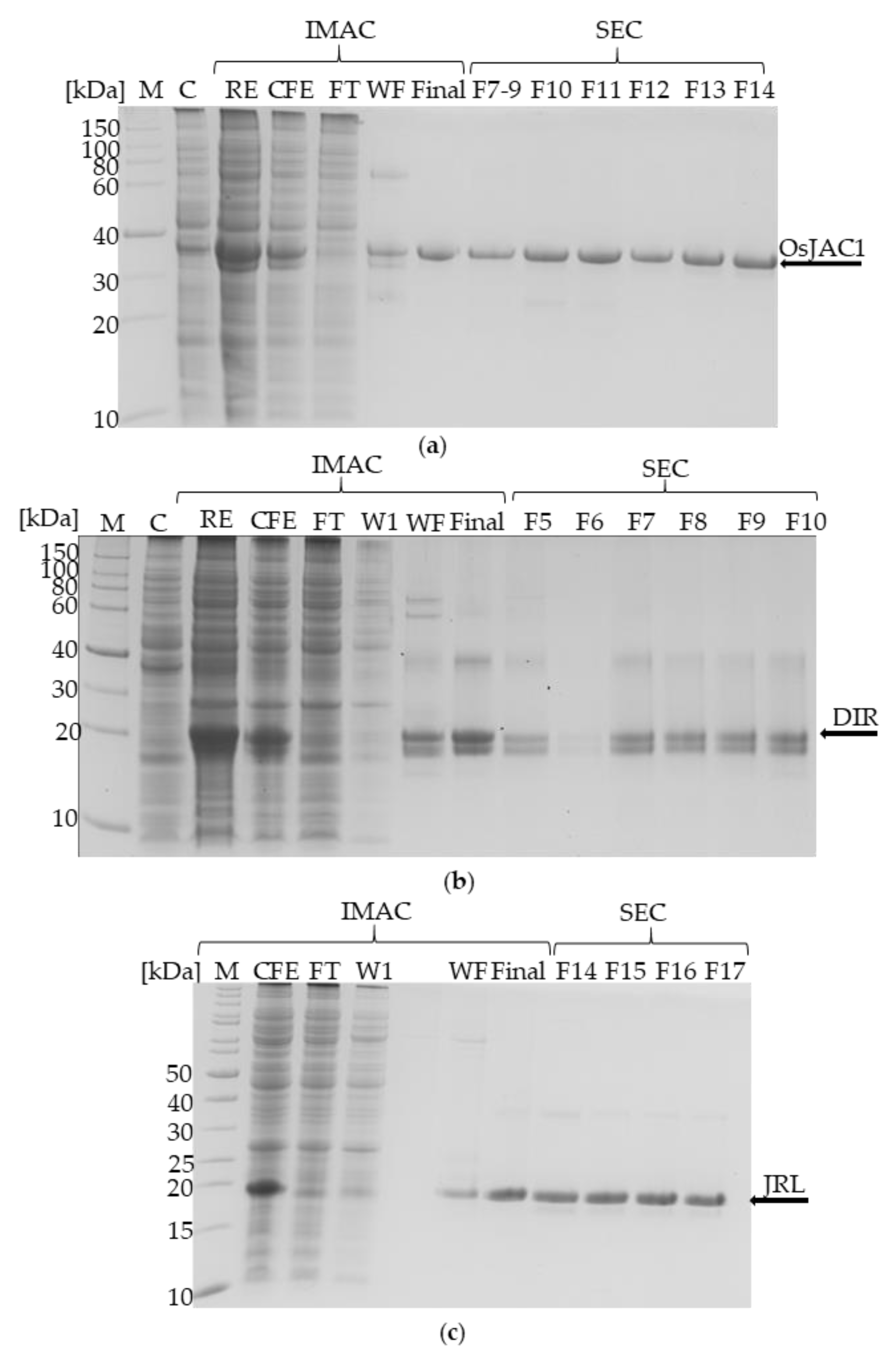
2.1. Heterologous Expression and Isolation of the OsJAC1 Protein from Oryza sativa and Its JRL and DIR Domains
2.2. Properties of the Chimeric Lectin OsJAC1—Initial Structural Characterization
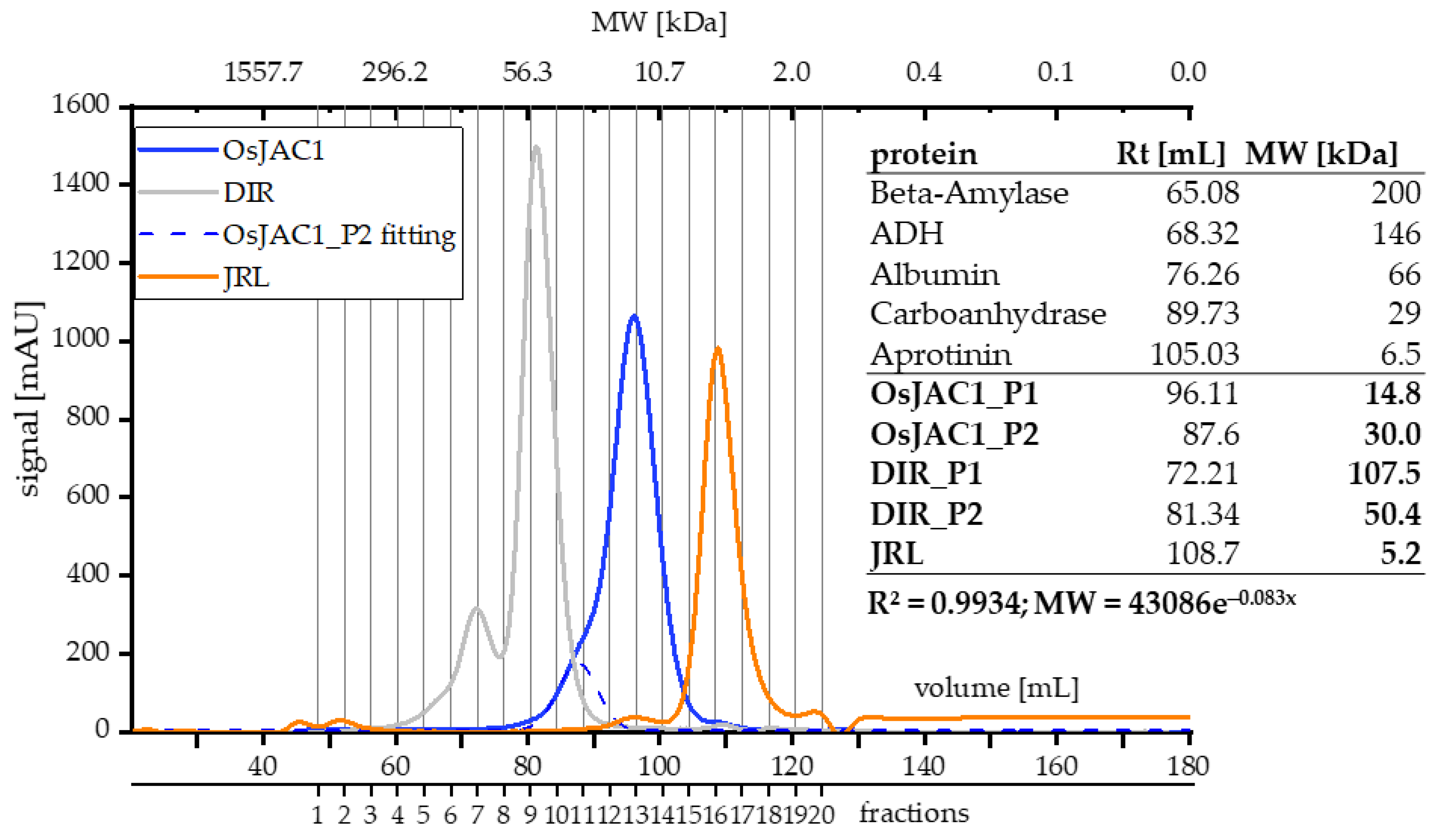
2.2.1. Investigation of Oligomericity
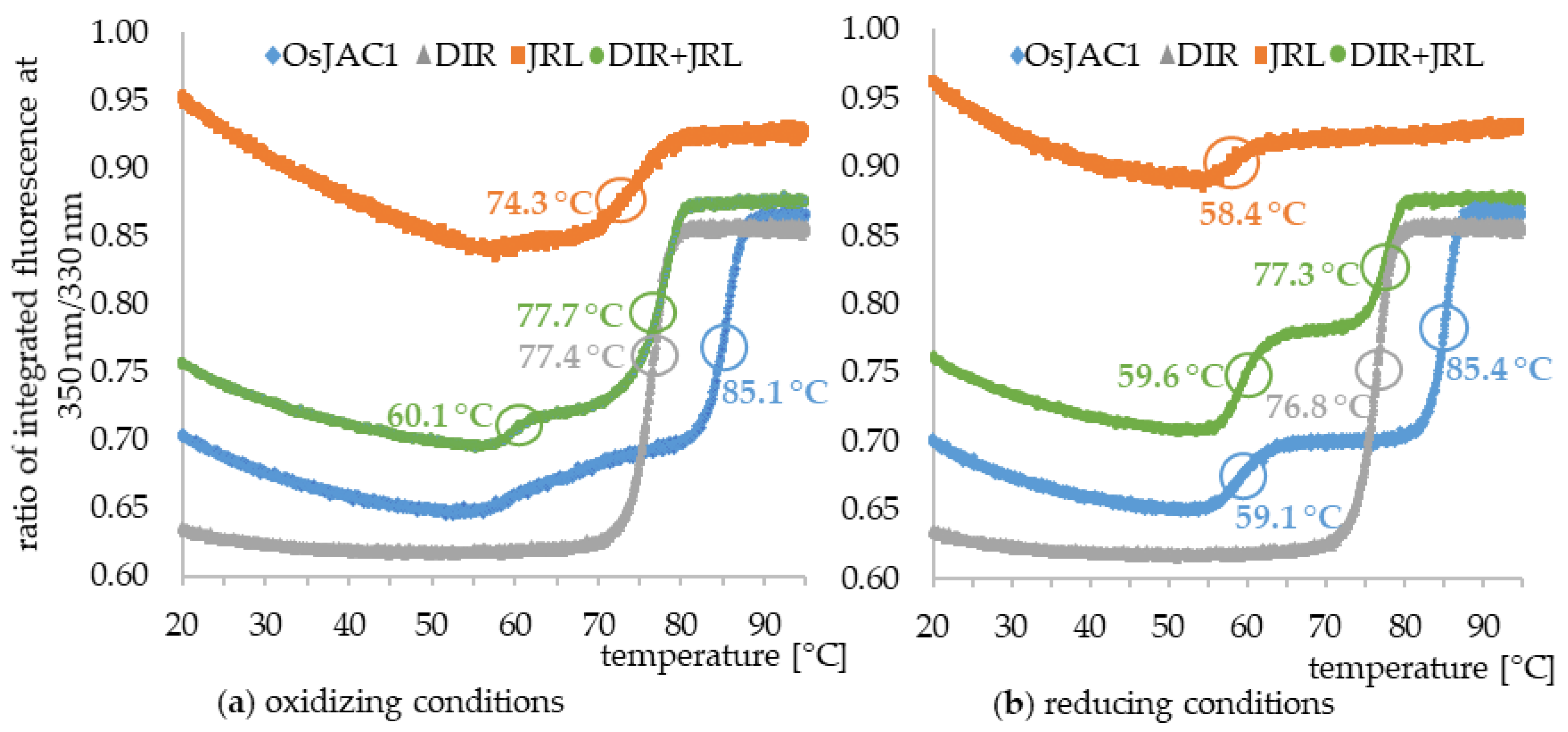
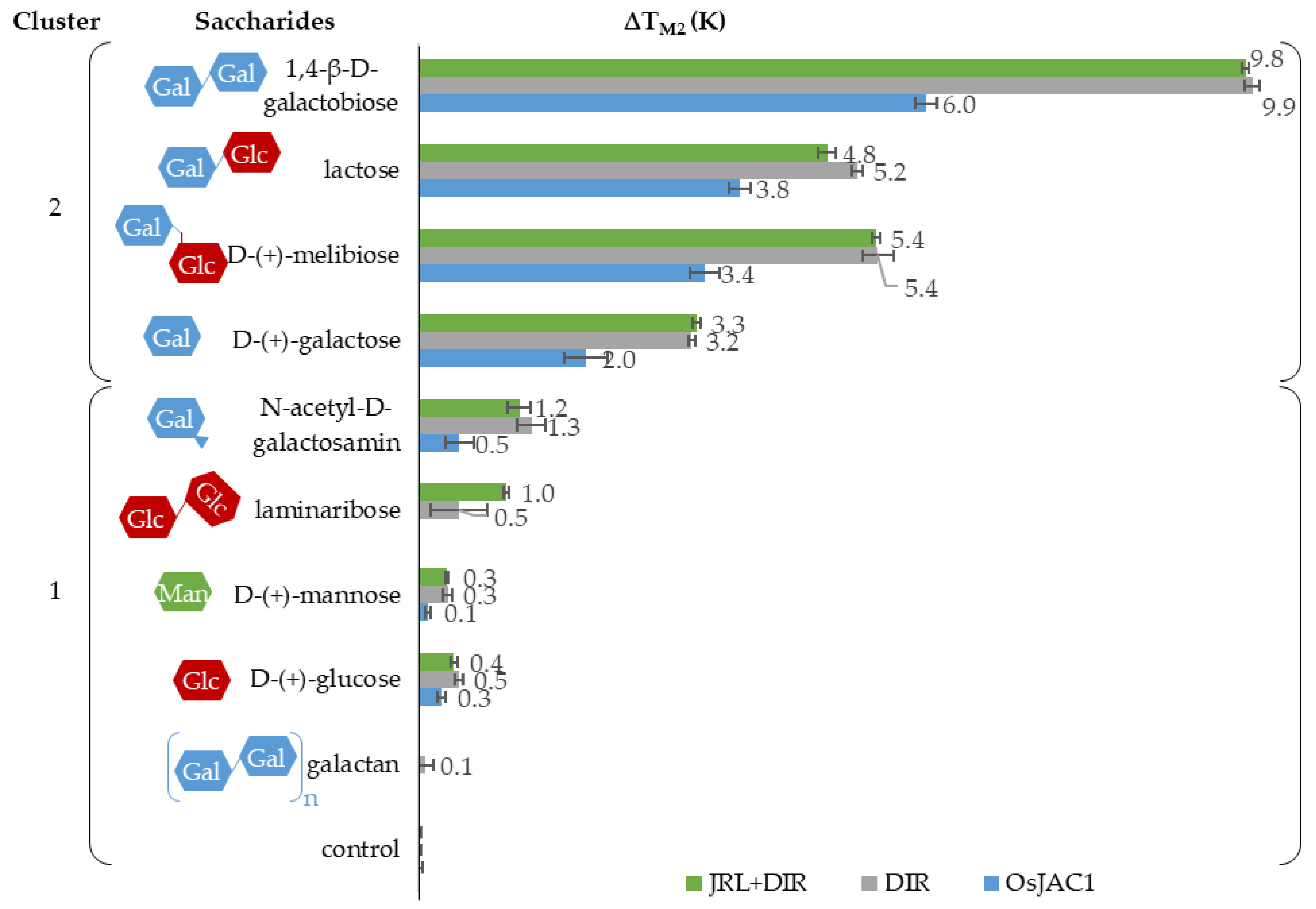
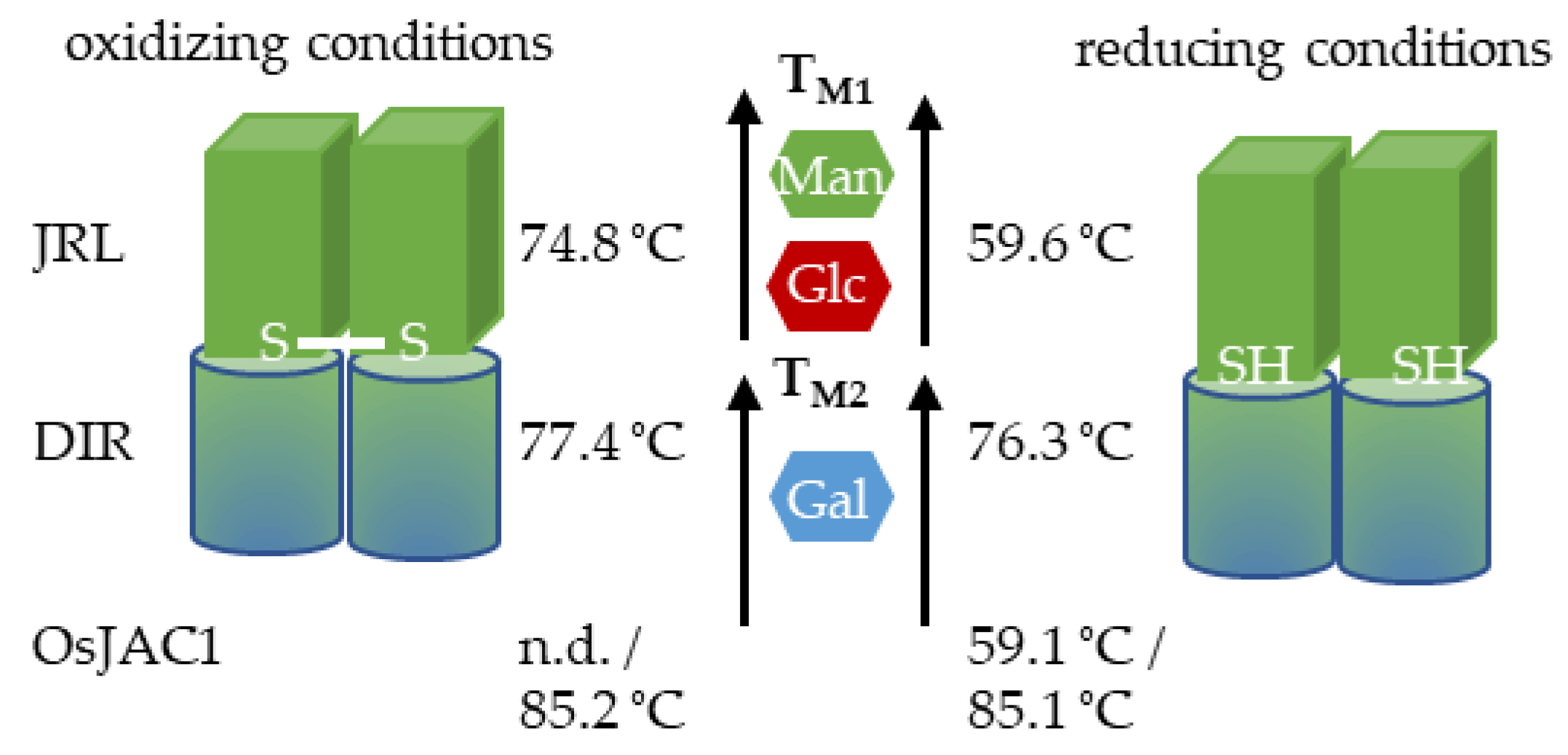
2.2.2. Melting Point (Tm)
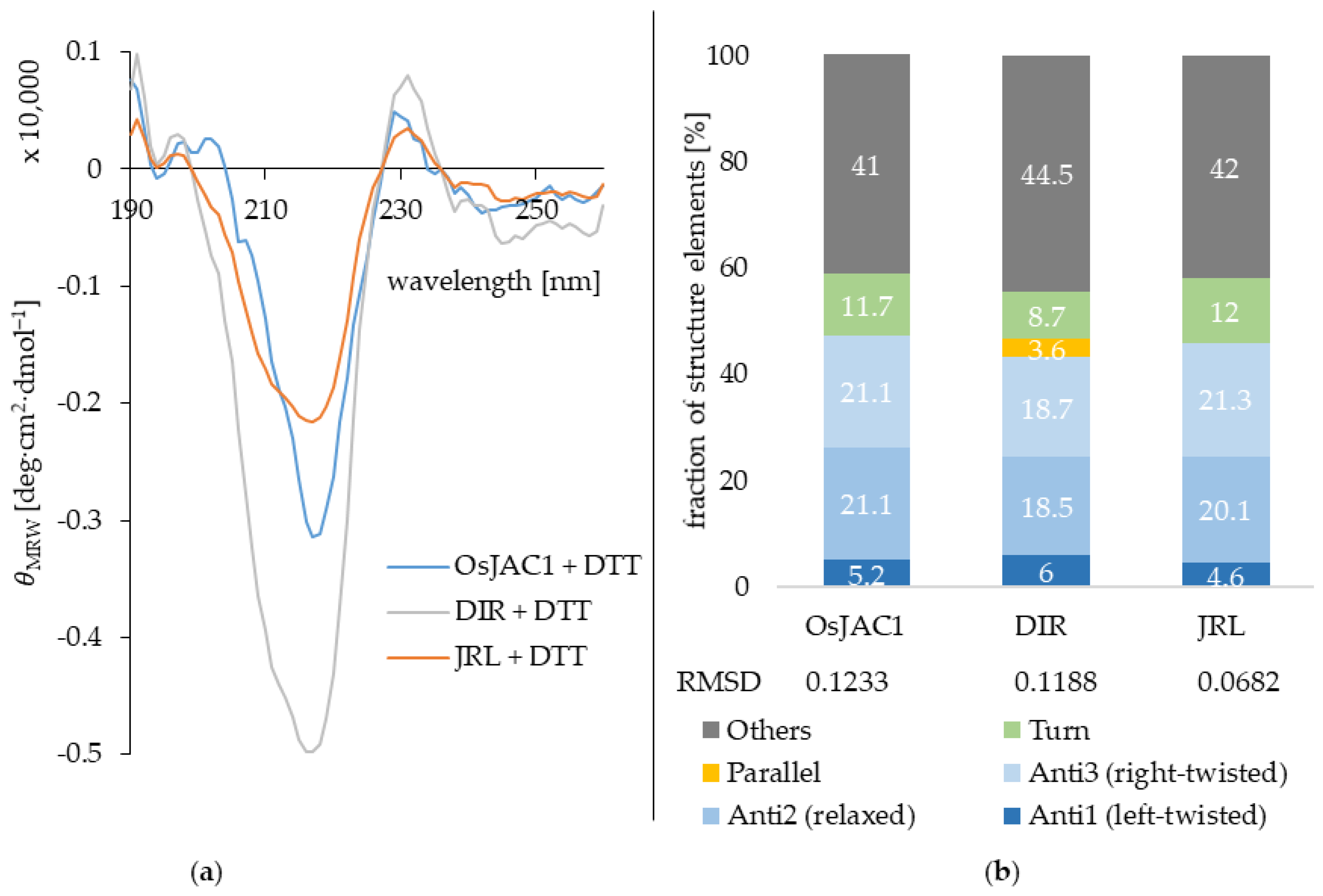
2.2.3. Circular Dichroism (CD) Spectroscopy
2.3. Interaction Partners of the Lectin Domain
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Western Blot Using Transgenic Plants Tissue
4.3. Construct and Cloning
4.4. Heterologous Expression of OsJAC1 and Its DIR and JRL Domains
4.5. Isolation of OsJAC1 and the Single Domains (DIR and JRL)
4.6. Protein Concentration Determination
4.7. MALDI-TOF-MS Analysis
4.8. DLS
4.9. Differential Scanning Fluorimetry
4.10. CD Spectroscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| C | control |
| CD | circular dichroism |
| CFE | cell free extract |
| DIR | dirigent protein |
| DLS | dynamic light scattering |
| DSF | differential scanning fluorimetry |
| DTT | dithiothreitol |
| F# | SEC fraction |
| FT | flow through |
| GFP | green fluorescent protein |
| GST | glutathione S-transferase-tag |
| IMAC | immobilized metal-chelate affinity chromatography |
| JRL | jacalin-related lectin |
| JRL + DIR | stoichiometric mixture of JRL and DIR domain |
| gJRL | galactose-specific jacalin-related lectins |
| mJRL | mannose-specific jacalin-related lectins |
| M | marker |
| OsJAC1_P1 | main OsJAC1 peak of SEC |
| OsJAC1_P2 | secondary OsJAC1 peak of SEC |
| PMSF | phenylmethylsulfonyl fluoride |
| RE | raw extract/disrupted cell suspension |
| RT | room temperature |
| SEC | size-exclusion chromatography |
| Tm | melting point |
| W1 | wash fraction with 100% buffer A (20 mm imidazol) |
| WF | wash fraction with 30% buffer B (250 mm imidazol) |
References
- Vijayan, M.; Chandra, N. Lectins. Curr. Opin. Struct. Biol. 1999, 9, 707–714. [Google Scholar] [CrossRef]
- Van Holle, S.; Van Damme, E.J. Messages from the past: New insights in plant lectin evolution. Front. Recent Dev. Plant Sci. 2019, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.; Peumans, W.J.; Barre, A.; Rougé, P. Plant lectins: A composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. Crit. Rev. Plant Sci. 1998, 17, 575–692. [Google Scholar] [CrossRef]
- Van Holle, S.; Van Damme, E.J. Signaling through plant lectins: Modulation of plant immunity and beyond. Biochem. Soc. Trans. 2018, 46, 217–233. [Google Scholar] [CrossRef]
- Song, M.; Xu, W.; Xiang, Y.; Jia, H.; Zhang, L.; Ma, Z. Association of jacalin-related lectins with wheat responses to stresses revealed by transcriptional profiling. Plant Mol. Biol. 2014, 84, 95–110. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Li, L.; Xu, H.; Xi, J.; Cao, X.; Xu, H.; Rong, S.; Dong, Y.; Wang, C.; Chen, R. A rice jacalin-related mannose-binding lectin gene, OsJRL, enhances Escherichia coli viability under high salinity stress and improves salinity tolerance of rice. Plant Biol. 2017, 19, 257–267. [Google Scholar] [CrossRef] [PubMed]
- De Schutter, K.; Van Damme, E.J. Protein-carbohydrate interactions as part of plant defense and animal immunity. Molecules 2015, 20, 9029–9053. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.-H. Monocot chimeric jacalins: A novel subfamily of plant lectins. Crit. Rev. Biotechnol. 2014, 34, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Esch, L.; Schaffrath, U. An update on jacalin-like lectins and their role in plant defense. Int. J. Mol. Sci. 2017, 18, 1592. [Google Scholar] [CrossRef]
- Jiang, J.-F.; Han, Y.; Xing, L.-J.; Xu, Y.-Y.; Xu, Z.-H.; Chong, K. Cloning and expression of a novel cDNA encoding a mannose-specific jacalin-related lectin from Oryza sativa. Toxicon 2006, 47, 133–139. [Google Scholar] [CrossRef]
- Weidenbach, D.; Esch, L.; Möller, C.; Hensel, G.; Kumlehn, J.; Höfle, C.; Hückelhoven, R.; Schaffrath, U. Polarized defense against fungal pathogens is mediated by the Jacalin-related lectin domain of modular Poaceae-specific proteins. Mol. Plant 2016, 9, 514–527. [Google Scholar] [CrossRef]
- Jiang, J.F.; Xu, Y.Y.; Chong, K. Overexpression of OsJAC1, a lectin gene, suppresses the coleoptile and stem elongation in rice. J. Integr. Plant Biol. 2007, 49, 230–237. [Google Scholar] [CrossRef]
- Jung, I.J.; Ahn, J.-W.; Jung, S.; Hwang, J.E.; Hong, M.J.; Choi, H.-I.; Kim, J.-B. Overexpression of rice jacalin-related mannose-binding lectin (OsJAC1) enhances resistance to ionizing radiation in Arabidopsis. BMC Plant Biol. 2019, 19, 561. [Google Scholar] [CrossRef] [PubMed]
- Maschberger, M.; Breitsprecher, D. Rapid Quantification of Unfolded Proteins for Quality Control and Optimization of Storage Conditions. Available online: https://resources.nanotempertech.com/application-notes/rapid-quantification-of-unfolded-proteins-for-quality-control-and-optimization-of-storage-conditions (accessed on 3 July 2020).
- Meagher, J.L.; Winter, H.C.; Ezell, P.; Goldstein, I.J.; Stuckey, J.A. Crystal structure of banana lectin reveals a novel second sugar binding site. Glycobiology 2005, 15, 1033–1042. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef] [PubMed]
- Datta, D.; Pohlentz, G.; Schulte, M.; Kaiser, M.; Goycoolea, F.M.; Müthing, J.; Mormann, M.; Swamy, M.J. Physico-chemical characteristics and primary structure of an affinity-purified α-d-galactose-specific, jacalin-related lectin from the latex of mulberry (Morus indica). Arch. Biochem. Biophys. 2016, 609, 59–68. [Google Scholar] [CrossRef]
- Swanson, M.D.; Boudreaux, D.M.; Salmon, L.; Chugh, J.; Winter, H.C.; Meagher, J.L.; André, S.; Murphy, P.V.; Oscarson, S.; Roy, R. Engineering a therapeutic lectin by uncoupling mitogenicity from antiviral activity. Cell 2015, 163, 746–758. [Google Scholar] [CrossRef]
- Meng, Q.; Moinuddin, S.G.; Kim, S.-J.; Bedgar, D.L.; Costa, M.A.; Thomas, D.G.; Young, R.P.; Smith, C.A.; Cort, J.R.; Davin, L.B. Pterocarpan synthase (PTS) structures suggest a common quinone methide—Stabilizing function in dirigent proteins and proteins with dirigent-like domains. J. Biol. Chem. 2020, 295, 11584–11601. [Google Scholar] [CrossRef]
- Gasper, R.; Effenberger, I.; Kolesinski, P.; Terlecka, B.; Hofmann, E.; Schaller, A. Dirigent protein mode of action revealed by the crystal structure of AtDIR6. Plant Physiol. 2016, 172, 2165–2175. [Google Scholar] [CrossRef]
- Solís, D.; Maté, M.J.; Lohr, M.; Ribeiro, J.P.; López-Merino, L.; André, S.; Buzamet, E.; Cañada, F.J.; Kaltner, H.; Lensch, M. N-domain of human adhesion/growth-regulatory galectin-9: Preference for distinct conformers and non-sialylated N-glycans and detection of ligand-induced structural changes in crystal and solution. Int. J. Biochem. Cell Biol. 2010, 42, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, R.; Sekar, K.; Banerjee, R.; Sharma, V.; Surolia, A.; Vijayan, M. A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a β-prism fold. Nat. Struct. Biol. 1996, 3, 596–603. [Google Scholar] [CrossRef]
- Barre, A.; Bourne, Y.; Van Damme, E.J.; Peumans, W.J.; Rougé, P. Mannose-binding plant lectins: Different structural scaffolds for a common sugar-recognition process. Biochimie 2001, 83, 645–651. [Google Scholar] [CrossRef]
- Kim, K.-W.; Smith, C.A.; Daily, M.D.; Cort, J.R.; Davin, L.B.; Lewis, N.G. Trimeric structure of (+)-pinoresinol-forming dirigent protein at 1.95 Å resolution with three isolated active sites. J. Biol. Chem. 2015, 290, 1308–1318. [Google Scholar] [CrossRef]
- Meyer, A.J.; Brach, T.; Marty, L.; Kreye, S.; Rouhier, N.; Jacquot, J.P.; Hell, R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007, 52, 973–986. [Google Scholar] [CrossRef] [PubMed]
- Nakae, S.; Shionyu, M.; Ogawa, T.; Shirai, T. Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization. Proteins Struct. Funct. Bioinform. 2018, 86, 644–653. [Google Scholar] [CrossRef]
- Nagata, Y.; Yamashita, M.; Honda, H.; Akabane, J.; Uehara, K.; Saito, A.; Sumisa, F.; Nishibori, K.; Oodaira, Y. Characterization, occurrence, and molecular cloning of a lectin from Grifola frondosa: Jacalin-related lectin of fungal origin. Biosci. Biotechnol. Biochem. 2005, 69, 2374–2380. [Google Scholar] [CrossRef] [PubMed]
- Gabrielsen, M.; Abdul-Rahman, P.S.; Othman, S.; Hashim, O.H.; Cogdell, R.J. Structures and binding specificity of galactose-and mannose-binding lectins from champedak: Differences from jackfruit lectins. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 709–716. [Google Scholar] [CrossRef]
- Neto, A.E.V.; de Sousa, F.D.; Pereira, H.D.M.; Moreno, F.B.M.B.; Lourenzoni, M.R.; Grangeiro, T.B.; Moreira, A.C.d.O.M.; Moreira, R.d.A. New structural insights into anomeric carbohydrate recognition by frutalin: An α-d-galactose-binding lectin from breadfruit seeds. Biochem. J. 2019, 476, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Kittur, F.S.; Yu, H.Y.; Bevan, D.R.; Esen, A. Deletion of the N-terminal dirigent domain in maize β-glucosidase aggregating factor and its homolog sorghum lectin dramatically alters the sugar-specificities of their lectin domains. Plant Physiol. Biochem. 2010, 48, 731–734. [Google Scholar] [CrossRef]
- Gabius, H.-J.; André, S.; Jiménez-Barbero, J.; Romero, A.; Solís, D. From lectin structure to functional glycomics: Principles of the sugar code. Trends Biochem. Sci. 2011, 36, 298–313. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Hause, B.; Van Damme, E.J. The galactose-binding and mannose-binding jacalin-related lectins are located in different sub-cellular compartments. FEBS Lett. 2000, 477, 186–192. [Google Scholar] [CrossRef]
- Rougé, P.; Peumans, W.J.; Barre, A.; Van Damme, E.J. A structural basis for the difference in specificity between the two jacalin-related lectins from mulberry (Morus nigra) bark. Biochem. Biophys. Res. Commun. 2003, 304, 91–97. [Google Scholar] [CrossRef]
- Kittur, F.S.; Yu, H.Y.; Bevan, D.R.; Esen, A. Homolog of the maize β-glucosidase aggregating factor from sorghum is a jacalin-related GalNAc-specific lectin but lacks protein aggregating activity. Glycobiology 2009, 19, 277–287. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kittur, F.S.; Lalgondar, M.; Yu, H.Y.; Bevan, D.R.; Esen, A. Maize β-glucosidase-aggregating factor is a polyspecific jacalin-related chimeric lectin, and its lectin domain is responsible for β-glucosidase aggregation. J. Biol. Chem. 2007, 282, 7299–7311. [Google Scholar] [CrossRef]
- Ma, Q.-H.; Han, J.-Q. Identification of monocot chimeric jacalin family reveals functional diversity in wheat. Planta 2021, 253, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Halls, S.C.; Davin, L.B.; Kramer, D.M.; Lewis, N.G. Kinetic study of coniferyl alcohol radical binding to the (+)-pinoresinol forming dirigent protein. Biochemistry 2004, 43, 2587–2595. [Google Scholar] [CrossRef]
- Ma, Q.-H.; Liu, Y.-C. TaDIR13, a dirigent protein from wheat, promotes lignan biosynthesis and enhances pathogen resistance. Plant Mol. Biol. Rep. 2015, 33, 143–152. [Google Scholar] [CrossRef]


| OsJAC1 | DIR | JRL | |
|---|---|---|---|
| residue range | 1–329 | 28–182 | 183–329 |
| molecular mass [kDa] | 35.57 | 19.3 | 18.39 |
| plasmid | pColdIV | pET15b(+) | pET15b(+) |
| expression condition | 10 °C for 3.5 d | 10 °C for 3.5 d | 10 °C for 3.5 d |
| pellet size [g] | 7 | 5.8 | 8.6 |
| purity [%] a | >95 | >95 | >95 |
| conc. after IMAC [mg mL−1] | 10.7 | 9.7 | 8.1 |
| VFinal [mL] | 3.5 | 3.5 | 3.5 |
| appearance | light yellow color | light yellow color | colorless |
| Buffer Condition | Min. MW/kDa | Max. MW/MDa | No. of Distinct Species |
|---|---|---|---|
| 15 mm TRIS-HCl pH 7.4 | 308 | 26.95 | two |
| +150 mm CaCl2 | 268.8 | 3317 | many |
| +50 mm lactose | 187.4 | - | one |
| +150 mm MgSO4 | 183.7 | - | one |
| +4 mm DTT | 137.5 | - | one |
| DTT | Sample | Initial ratio a | ∆ ratio b | TM1 (°C) (±) | TM2 (°C) (±) |
|---|---|---|---|---|---|
| − | JRL | 0.95 | −0.03 | n.d. | 74.3 ± 0.04 |
| − | DIR | 0.63 | 0.23 | n.a. | 77.4 ± 0.01 |
| − | OsJAC1 | 0.70 | 0.17 | n.d. | 85.1 ± 0.03 |
| − | JRL + DIR | 0.76 | 0.12 | 60.1 ± 0.02 | 77.7 ± 0.02 |
| + | JRL | 0.96 | −0.03 | 58.4 ± 0.13 | n.d. |
| + | DIR | 0.56 | 0.28 | n.a. | 76.8 ± 0.03 |
| + | OsJAC1 | 0.70 | 0.16 | 59.1 ± 0.01 | 85.4 ± 0.01 |
| + | JRL + DIR | 0.77 | 0.12 | 59.6 ± 0.51 | 77.3 ± 0.05 |
| Protein | α-Helix | β-Sheet | β-Turns | Other | Reference |
|---|---|---|---|---|---|
| OsJAC1 JRL domain | 0% | 46% | 12% | 42% | - |
| gJRL [18] | 2% | 34% | 23% | 39% | [18] |
| mJRL | 0% | 73% 1 | - | 27% | pdb:4PIF [19] |
| OsJAC1 DIR domain | 1% | 47% | 9% | 44% | - |
| Dirigent protein GePTS1 | 4% | 56% 1 | - | 40% | pdb:6OOC [20] |
| Dirigent protein AtDIR6 | 0% | 56% 1 | - | 44% | pdb:5LAL [21] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huwa, N.; Weiergräber, O.H.; Kirsch, C.; Schaffrath, U.; Classen, T. Biochemical and Initial Structural Characterization of the Monocot Chimeric Jacalin OsJAC1. Int. J. Mol. Sci. 2021, 22, 5639. https://doi.org/10.3390/ijms22115639
Huwa N, Weiergräber OH, Kirsch C, Schaffrath U, Classen T. Biochemical and Initial Structural Characterization of the Monocot Chimeric Jacalin OsJAC1. International Journal of Molecular Sciences. 2021; 22(11):5639. https://doi.org/10.3390/ijms22115639
Chicago/Turabian StyleHuwa, Nikolai, Oliver H. Weiergräber, Christian Kirsch, Ulrich Schaffrath, and Thomas Classen. 2021. "Biochemical and Initial Structural Characterization of the Monocot Chimeric Jacalin OsJAC1" International Journal of Molecular Sciences 22, no. 11: 5639. https://doi.org/10.3390/ijms22115639
APA StyleHuwa, N., Weiergräber, O. H., Kirsch, C., Schaffrath, U., & Classen, T. (2021). Biochemical and Initial Structural Characterization of the Monocot Chimeric Jacalin OsJAC1. International Journal of Molecular Sciences, 22(11), 5639. https://doi.org/10.3390/ijms22115639






