Comparison and Characterization of a Cell Wall Invertase Promoter from Cu-Tolerant and Non-Tolerant Populations of Elsholtzia haichowensis
Abstract
1. Introduction
2. Results
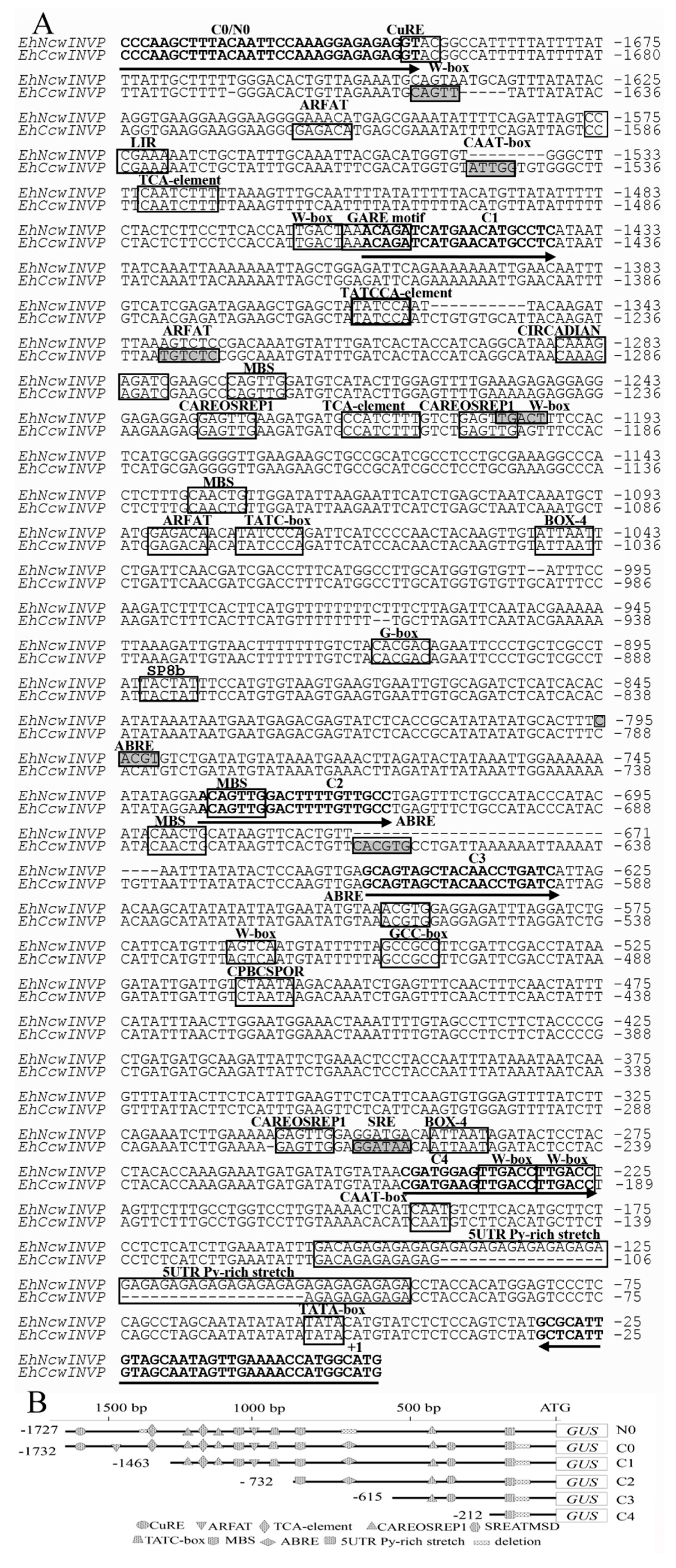
2.1. Comparative Bioinformatics Analysis of Promoters
2.2. Promoter–Reporter Constructs and GUS Expression Analysis in Transgenic Arabidopsis
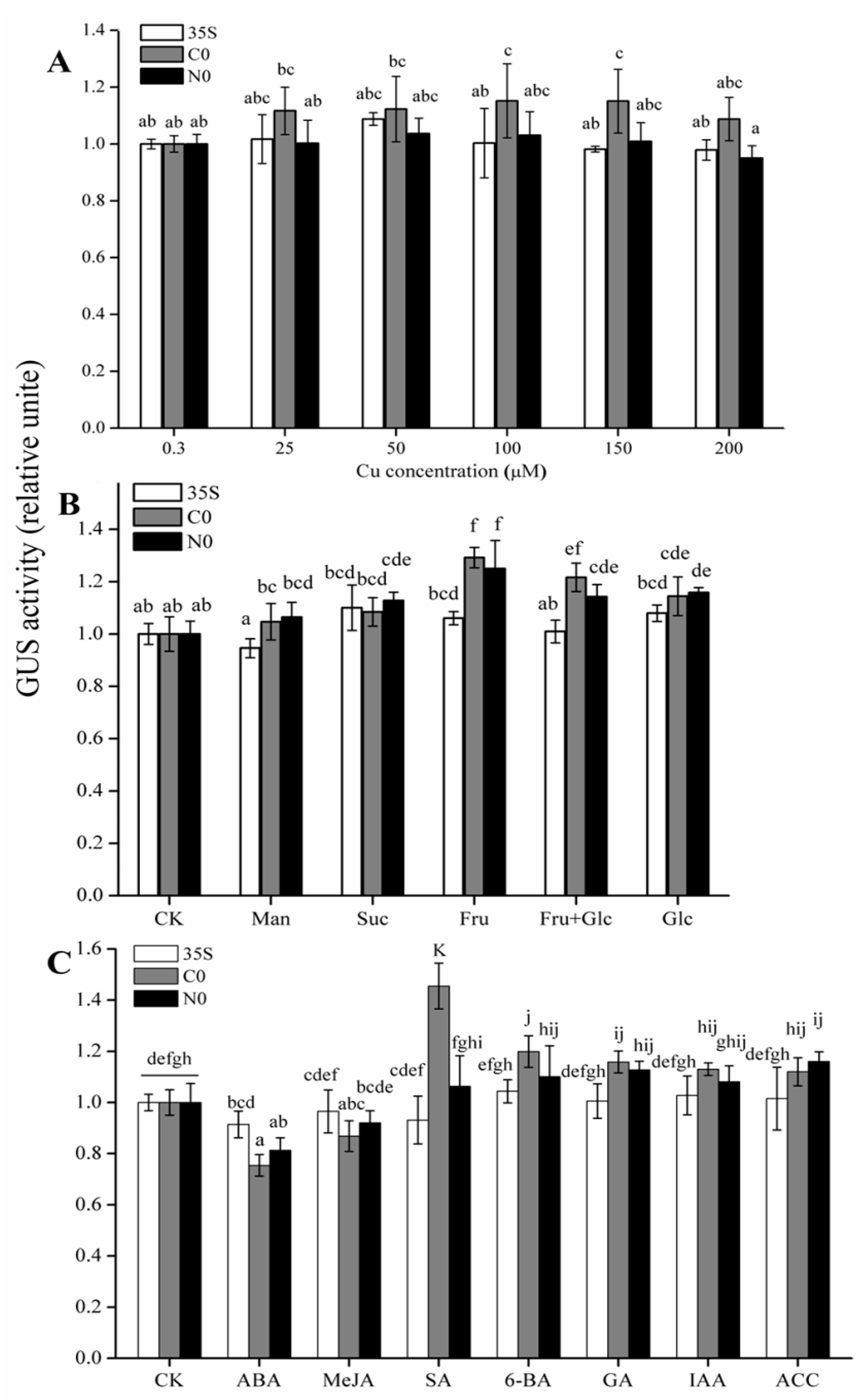
2.3. Quantitative Analysis of GUS Activity Driven by Full-Length Promoters in Response to Cu, Sugar, and Hormone Treatments
2.4. Transcriptional Responses of Full-Length Promoters to SA Treatments
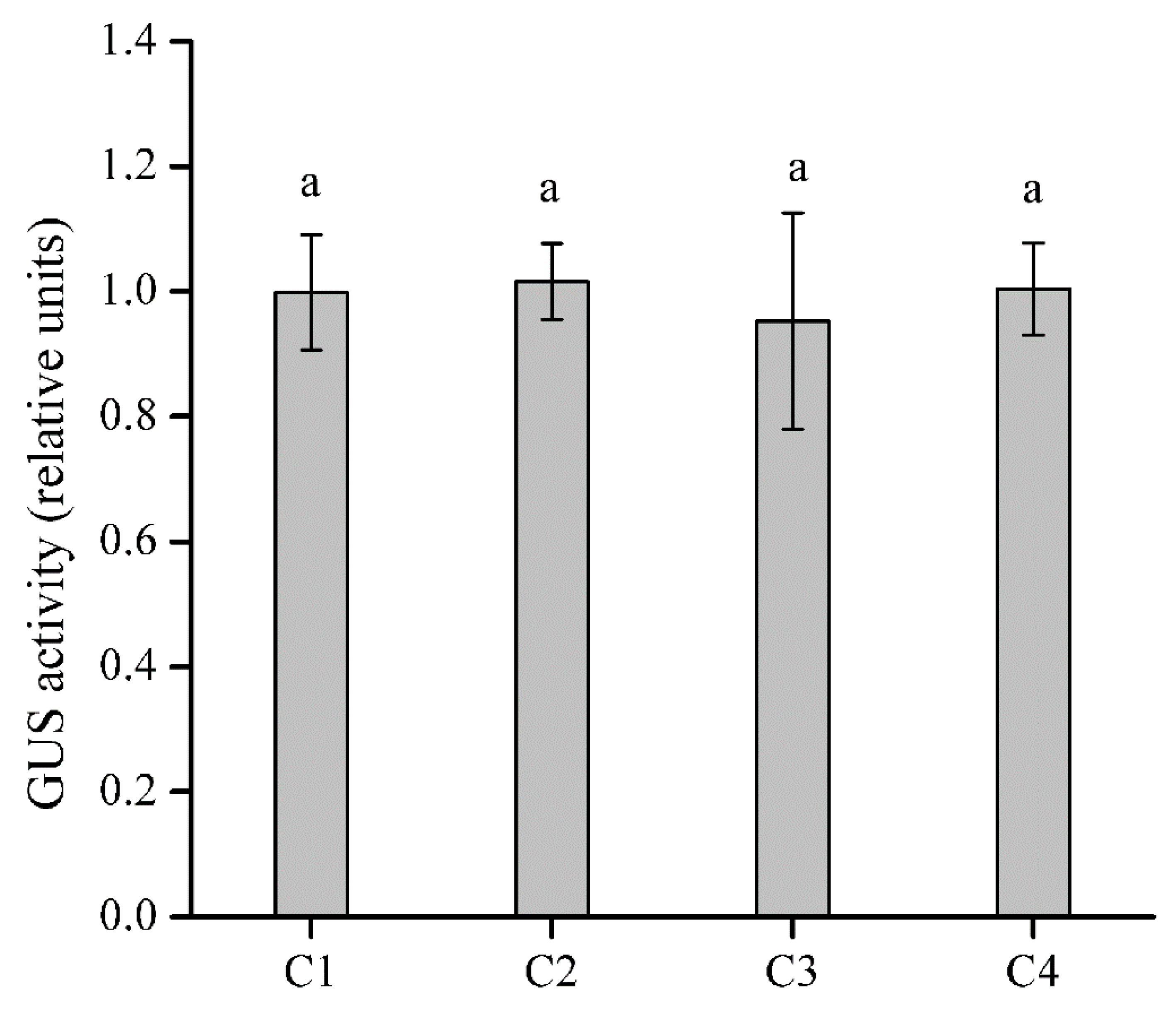
2.5. Identification of the CWIN Promoter Region Affected by SA in EhCcwINVP
3. Discussion
3.1. Copper Does Not Induce Differential Expression of CWIN Promoters from the Cu-Tolerant and Non-Tolerant Populations
3.2. Glucose and Fructose Significantly Induced the Activity of the CWIN Promoter in Both Cu-Tolerant and Non-Tolerant Populations
3.3. SA Induces Differential Expression of CWIN Promoters from Two Populations
3.4. A 270-bp Promoter Fragment Is Required for the SA Response
4. Materials and Methods
4.1. Bioinformatics Analysis of Promoters
4.2. Construction of Plant Expression Vectors and Plant Transformation
4.3. Plant Materials, Stress Treatments, and Sampling
4.4. Histochemical Staining and Fluorometric Quantification of GUS Activity
4.5. RNA Extraction and qRT–PCR Analysis of GUS Gene Expression
4.6. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sturm, A.; Tang, G.-Q. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 1999, 4, 401–407. [Google Scholar] [CrossRef]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and Novel Mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef]
- Ruan, Y.-L.; Jin, Y.; Yang, Y.-J.; Li, G.-J.; Boyer, J.S. Sugar Input, Metabolism, and Signaling Mediated by Invertase: Roles in Development, Yield Potential, and Response to Drought and Heat. Mol. Plant 2010, 3, 942–955. [Google Scholar] [CrossRef]
- Roitsch, T.; González, M.-C. Function and regulation of plant invertases: Sweet sensations. Trends Plant Sci. 2004, 9, 606–613. [Google Scholar] [CrossRef]
- Li, M.J.; Xiong, Z.T.; Liu, H.; Kuo, Y.M.; Tong, L. Copper-induced alteration in sucrose partitioning and its relationship to the root growth of two Elsholtzia haichowensis Sun populations. Int. J. Phytoremediation 2016, 18, 966–976. [Google Scholar] [CrossRef]
- Roitsch, T.; Balibrea, M.E.; Hofmann, M.; Proels, R.; Sinha, A.K. Extracellular invertase: Key metabolic enzyme and PR protein. J. Exp. Bot. 2003, 54, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Sherson, S.M.; Alford, H.L.; Forbes, S.M.; Wallace, G.; Smith, S.M. Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J. Exp. Bot. 2003, 54, 525–531. [Google Scholar] [CrossRef]
- Schweinichen, C.; Büttner, M. Expression of a Plant Cell Wall Invertase in Roots of Arabidopsis Leads to Early Flowering and an Increase in Whole Plant Biomass. Plant Biol. 2005, 7, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Ru, L.; Osorio, S.; Wang, L.; Fernie, A.R.; Patrick, J.W.; Ruan, Y.L. Transcriptomic and metabolomics responses to elevated cell wall invertase activity during tomato fruit set. J. Exp. Bot. 2017, 68, 4263–4279. [Google Scholar] [CrossRef] [PubMed]
- Ru, L.; He, Y.; Zhu, Z.; Patrick, J.W.; Ruan, Y.L. Integrating Sugar Metabolism with Transport: Elevation of Endogenous Cell Wall Invertase Activity Up-Regulates SlHT2 and SlSWEET12c Expression for Early Fruit Development in Tomato. Front. Genet 2020, 11, 592596. [Google Scholar] [CrossRef]
- Li, Z.; Palmer, W.M.; Martin, A.P.; Wang, R.; Rainsford, F.; Jin, Y.; Patrick, J.W.; Yang, Y.; Ruan, Y.L. High invertase activity in tomato reproductive organs correlates with enhanced sucrose import into, and heat tolerance of, young fruit. J. Exp. Bot. 2012, 63, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.N.; Van Dongen, J.T.; Alfred, S.C.; Mamun, E.A.; Zhao, X.; Saini, H.S.; Fernandes, S.F.; Blanchard, C.L.; Sutton, B.G.; Geigenberger, P.; et al. Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ. 2005, 28, 1534–1551. [Google Scholar] [CrossRef]
- Ji, X.; Shiran, B.; Wan, J.; Lewis, D.C.; Jenkins, C.L.; Condon, A.G.; Richards, R.A.; Dolferus, R. Importance of pre-anthesis anther sink strength for maintenance of grain number during reproductive stage water stress in wheat. Plant Cell Environ. 2010, 33, 926–942. [Google Scholar] [CrossRef]
- Antonovics, J.; Bradshaw, A.D.; Turner, R.G. Heavy Metal Tolerance in Plants. Adv. Ecol. Res. 1971, 7, 1–85. [Google Scholar]
- Tang, S.R.; Wilke, B.M.; Huang, C.Y. The uptake of copper by plants dominantly growing on copper mining spoils along the Yangtze River, the People’s Republic of China. Plant Soil 1999, 209, 225–232. [Google Scholar] [CrossRef]
- Liu, J.; Xiong, Z. Differences in Accumulation and Physiological Response to Copper Stress in three Populations of Elsholtzia haichowensis S. Water Air Soil Pollut. 2005, 168, 5–16. [Google Scholar] [CrossRef]
- Cai, S.; Xiong, Z.; Li, L.; Li, M.; Zhang, L.; Liu, C.; Xu, Z. Differential responses of root growth, acid invertase activity and transcript level to copper stress in two contrasting populations of Elsholtzia haichowensis. Ecotoxicology 2014, 23, 76–91. [Google Scholar] [CrossRef]
- Liu, C.; Xu, Z.R.; Cai, S.W.; Zhang, L.; Xiong, Z.T. cDNA cloning, heterologous expression and characterization of a cell wall invertase from copper tolerant population of Elsholtzia haichowensis. Biologia 2015, 70, 1063–1069. [Google Scholar] [CrossRef]
- Huang, W.X.; Cao, Y.; Huang, L.J.; Ren, C.; Xiong, Z.T. Differential expression of acid invertase genes in roots of metallicolous and non-metallicolous populations of Rumex japonicus under copper stress. Chemosphere 2011, 84, 1432–1439. [Google Scholar] [CrossRef]
- Xu, Z.-R.; Cai, S.-W.; Huang, W.-X.; Liu, R.-X.; Xiong, Z.-T. Differential expression of vacuolar and defective cell wall invertase genes in roots and seeds of metalliferous and non-metalliferous populations of Rumex dentatus under copper stress. Ecotoxicol. Environ. Saf. 2018, 147, 17–25. [Google Scholar] [CrossRef]
- Cai, S.W.; Huang, W.X.; Xiong, Z.T.; Ye, F.Y.; Ren, C.; Xu, Z.R.; Liu, C.; Deng, S.Q.; Zhao, J. Comparative study of root growth and sucrose-cleaving enzymes in metallicolous and non-metallicolous populations of Rumex dentatus under copper stress. Ecotoxicol. Environ. Saf. 2013, 98, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Canam, T.; Unda, F.; Mansfield, S.D. Heterologous expression and functional characterization of two hybrid poplar cell-wall invertases. Planta 2008, 228, 1011. [Google Scholar] [CrossRef]
- Liao, S.; Wang, L.; Li, J.; Ruan, Y.-L. Cell Wall Invertase Is Essential for Ovule Development through Sugar Signaling Rather Than Provision of Carbon Nutrients. Plant Physiol. 2020, 183, 1126. [Google Scholar] [CrossRef]
- Shrestha, A.; Khan, A.; Dey, N. cis–trans Engineering: Advances and Perspectives on Customized Transcriptional Regulation in Plants. Mol. Plant 2018, 11, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.R.; Nissim, L.; Stupp, D.; Pery, E.; Binder-Nissim, A.; Weisinger, K.; Enghuus, C.; Palacios, S.R.; Humphrey, M.; Zhang, Z.; et al. A high-throughput screening and computation platform for identifying synthetic promoters with enhanced cell-state specificity (SPECS). Nat. Commun. 2019, 10, 2880. [Google Scholar] [CrossRef]
- Korkmaz, G.; Manber, Z.; Lopes, R.; Prekovic, S.; Schuurman, K.; Kim, Y.; Teunissen, H.; Flach, K.; Wit, E.; Galli, G.G.; et al. A CRISPR-Cas9 screen identifies essential CTCF anchor sites for estrogen receptor-driven breast cancer cell proliferation. Nucleic Acids Res. 2019, 47, 9557–9572. [Google Scholar] [CrossRef] [PubMed]
- Lovell, J.T.; Schwartz, S.; Lowry, D.B.; Shakirov, E.V.; Bonnette, J.E.; Weng, X.; Wang, M.; Johnson, J.; Sreedasyam, A.; Plott, C.; et al. Drought responsive gene expression regulatory divergence between upland and lowland ecotypes of a perennial C4 grass. Genome Res. 2016, 26, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Gould, B.A.; Chen, Y.; Lowry, D.B. Gene regulatory divergence between locally adapted ecotypes in their native habitats. Mol. Ecol. 2018, 27, 4174–4188. [Google Scholar] [CrossRef]
- Miura, K.; Tada, Y. Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 2014, 5, 4. [Google Scholar] [CrossRef]
- Mishra, P.; Dubey, R.S. Excess nickel modulates activities of carbohydrate metabolizing enzymes and induces accumulation of sugars by upregulating acid invertase and sucrose synthase in rice seedlings. BioMetals 2013, 26, 97–111. [Google Scholar] [CrossRef]
- Asthir, B.; Kaur, A.; Basra, A.S. Cultivar variation in heat stability and kinetic properties of soluble invertase in wheat grains. Acta Physiol. Plant. 1998, 20, 339. [Google Scholar] [CrossRef]
- Verma, S.; Dubey, R.S. Effect of Cadmium on Soluble Sugars and Enzymes of their Metabolism in Rice. Biol. Plant. 2001, 44, 117–123. [Google Scholar] [CrossRef]
- Roitsch, T.; Bittner, M.; Godt, D.E. Induction of Apoplastic Invertase of Chenopodium rubrum by D-Glucose and a Glucose Analog and Tissue-Specific Expression Suggest a Role in Sink-Source Regulation. Plant Physiol. 1995, 108, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.; Guivarćh, A.; Hirsche, J.; Bauerfeind, M.A.; González, M.C.; Hyun, T.K.; Eom, S.H.; Chriqui, D.; Engelke, T.; Großkinsky, D.K.; et al. Metabolic Control of Tobacco Pollination by Sugars and Invertases. Plant Physiol. 2017, 173, 984–997. [Google Scholar] [CrossRef]
- Proels, R.K.; Roitsch, T. Extracellular invertase LIN6 of tomato: A pivotal enzyme for integration of metabolic, hormonal, and stress signals is regulated by a diurnal rhythm. J. Exp. Bot. 2009, 60, 1555–1567. [Google Scholar] [CrossRef]
- Haak, D.C.; Fukao, T.; Grene, R.; Hua, Z.H.; Ivanov, R.; Perrella, G.; Li, S. Multilevel Regulation of Abiotic Stress Responses in Plants. Front. Plant Sci. 2017, 8, 1564. [Google Scholar] [CrossRef]
- Lenhard, B.; Sandelin, A.; Carninci, P. Metazoan promoters: Emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012, 13, 233. [Google Scholar] [CrossRef]
- Kropat, J.; Tottey, S.; Birkenbihl, R.P.; Depege, N.; Huijser, P.; Merchant, S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 2005, 102, 18730–18735. [Google Scholar] [CrossRef]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein–Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 2009, 21, 347. [Google Scholar] [CrossRef]
- Nagae, M.; Nakata, M.; Takahashi, Y. Identification of negative cis-acting elements in response to copper in the chloroplastic iron superoxide dismutase gene of the moss Barbula unguiculata. Plant Physiol. 2008, 146, 1687–1696. [Google Scholar] [CrossRef]
- Maas, C.; Laufs, J.; Grant, S.; Korfhage, C.; Werr, W. The combination of a novel stimulatory element in the first exon of the maize Shrunken-1 gene with the following intron 1 enhances reporter gene expression up to 1000-fold. Plant Mol. Biol. 1991, 16, 199–207. [Google Scholar] [CrossRef]
- He, Y.; Xu, Z.; Xiong, Z. DNA methylation patterns of acid invertase gene promoters from cu-tolerant and non-tolerant populations of Elsholtzia haichowensis under copper stress. Plant Sci. J. 2017, 35, 574–582. [Google Scholar]
- Yong-Villalobos, L.; Gonzalez-Morales, S.I.; Wrobel, K.; Gutierrez-Alanis, D.; Cervantes-Perez, S.A.; Hayano-Kanashiro, C.; Oropeza-Aburto, A.; Cruz-Ramirez, A.; Martinez, O.; Herrera-Estrella, L. Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proc. Natl. Acad. Sci. USA 2015, 112, E7293–E7302. [Google Scholar] [CrossRef] [PubMed]
- Tymowska-Lalanne, Z.; Kreis, M. Expression of the Arabidopsis thaliana invertase gene family. Planta 1998, 207, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Tatematsu, K.; Ward, S.; Leyser, O.; Kamiya, Y.; Nambara, E. Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in arabidopsis. Plant Physiol. 2005, 138, 757–766. [Google Scholar] [CrossRef]
- Lu, C.A.; Lim, E.K.; Yu, S.M. Sugar response sequence in the promoter of a rice alpha-amylase gene serves as a transcriptional enhancer. J. Biol. Chem. 1998, 273, 10120–10131. [Google Scholar] [CrossRef]
- Sakr, S.; Wang, M.; Dedaldechamp, F.; Perez-Garcia, M.D.; Oge, L.; Hamama, L.; Atanassova, R. The Sugar-Signaling Hub: Overview of Regulators and Interaction with the Hormonal and Metabolic Network. Int. J. Mol. Sci. 2018, 19, 2506. [Google Scholar] [CrossRef]
- Grierson, C.; Du, J.S.; Zabala, M.D.; Beggs, K.; Smith, C.; Holdsworth, M.; Bevan, M. Separate Cis Sequences and Trans Factors Direct Metabolic and Developmental Regulation of a Potato-Tuber Storage Protein Gene. Plant J. 1994, 5, 815–826. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a Cdna-Encoding a Novel DNA-Binding Protein, Spf1, That Recognizes Sp8 Sequences in the 5’ Upstream Regions of Genes-Coding for Sporamin and Beta-Amylase from Sweet-Potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef]
- Sun, C.X.; Palmqvist, S.; Olsson, H.; Boren, M.; Ahlandsberg, S.; Jansson, C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 2003, 15, 2076–2092. [Google Scholar] [CrossRef]
- Maeo, K.; Tomiya, T.; Hayashi, K.; Akaike, M.; Morikami, A.; Ishiguro, S.; Nakamura, K. Sugar-responsible elements in the promoter of a gene for beta-amylase of sweet potato. Plant Mol. Biol. 2001, 46, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.B.; Song, B.T.; Liu, X.; Xie, C.H.; Li, M.; Lin, Y.; Zhang, H.L.; Liu, J. Promoter regions of potato vacuolar invertase gene in response to sugars and hormones. Plant Physiol. Biochem. 2013, 69, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.J.; Xiao, B.; Wang, L.; Hao, X.Y.; Yue, C.; Cao, H.L.; Wang, Y.C.; Li, N.N.; Yu, Y.B.; Zeng, J.M.; et al. CsINV5, a tea vacuolar invertase gene enhances cold tolerance in transgenic Arabidopsis. BMC Plant Biol. 2018, 18, 228. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.R.; Asgher, M.; Khan, N.A. Alleviation of salt-induced photosynthesis and growth inhibition by salicylic acid involves glycinebetaine and ethylene in mungbean (Vigna radiata L.). Plant Physiol. Biochem. 2014, 80, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Tan, H.; Huang, L.; Cai, C.; Ding, Y.; Bao, H.; Chen, Z.; Zhu, C. Application of exogenous salicylic acid reduces Cd toxicity and Cd accumulation in rice. Ecotoxicol. Environ. Saf. 2021, 207, 111198. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Liu, C.; Liang, Y.; Li, N.; Fu, Q. Salicylic Acid Signals Plant Defence against Cadmium Toxicity. Int. J. Mol. Sci. 2019, 20, 2960. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.-Q.; Lüscher, M.; Sturm, A. Antisense Repression of Vacuolar and Cell Wall Invertase in Transgenic Carrot Alters Early Plant Development and Sucrose Partitioning. Plant Cell 1999, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- LeClere, S.; Schmelz, E.A.; Chourey, P.S. Cell wall invertase-deficient miniature1 kernels have altered phytohormone levels. Phytochemistry 2008, 69, 692–699. [Google Scholar] [CrossRef]
- Odell, J.T.; Nagy, F.; Chua, N.-H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 1985, 313, 810. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hou, J.; Jiang, P.; Qi, S.; Xu, C.; He, Q.; Ding, Z.; Wang, Z.; Zhang, K.; Li, K. Identification of a 467 bp Promoter of Maize Phosphatidylinositol Synthase Gene (ZmPIS) Which Confers High-Level Gene Expression and Salinity or Osmotic Stress Inducibility in Transgenic Tobacco. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef][Green Version]
- Landschulz, W.H.; Johnson, P.F.; Adashi, E.Y.; Graves, B.J.; Mcknight, S.L. Isolation of a Recombinant Copy of the Gene Encoding C/Ebp. Genes Dev. 1988, 2, 786–800. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.N.; Zhang, L.L.; Zeng, B.J.; Zheng, S.C.; Feng, Q.L. Transcription factor CAAT/enhancer-binding protein is involved in regulation of expression of sterol carrier protein x in Spodoptera litura. Insect Mol. Biol. 2015, 24, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, B.; Sardina, J.L.; van Oevelen, C.; Collombet, S.; Kallin, E.M.; Vicent, G.P.; Lu, J.; Thieffry, D.; Beato, M.; Graf, T. C/EBP alpha poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 2014, 506, 235. [Google Scholar] [CrossRef] [PubMed]
- Mora, A.; Sandve, G.K.; Gabrielsen, O.S.; Eskeland, R. In the loop: Promoter-enhancer interactions and bioinformatics. Brief. Bioinform. 2016, 17, 980–995. [Google Scholar] [CrossRef]
- Singer, S.D.; Cox, K.D.; Liu, Z.R. Enhancer-promoter interference and its prevention in transgenic plants. Plant Cell Rep. 2011, 30, 723–731. [Google Scholar] [CrossRef]
- Mohr, T.J.; Mammarella, N.D.; Hoff, T.; Woffenden, B.J.; Jelesko, J.G.; McDowell, J.M. The Arabidopsis Downy Mildew Resistance Gene RPP8 Is Induced by Pathogens and Salicylic Acid and Is Regulated by W Box cis Elements. Mol. Plant-Microbe Interact. 2010, 23, 1303–1315. [Google Scholar] [CrossRef]
- Hussain, R.M.F.; Sheikh, A.H.; Haider, I.; Quareshy, M.; Linthorst, H.J.M. Arabidopsis WRKY50 and TGA Transcription Factors Synergistically Activate Expression of PR1. Front. Plant Sci. 2018, 9, 930. [Google Scholar] [CrossRef]
- Liu, Y.G.; Chen, Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. BioTechniques 2007, 43, 649. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Higo, H. PLACE: A database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998, 26, 358–359. [Google Scholar] [CrossRef]
- Zhao, J.; Cai, S.; Xu, Z.; Xiong, Z. Isolation and activity analysis of cell wall Invertase gene promoter (EhcwINVP) from Elsholtzia haichowensis Sun. Plant Sci. J. 2016, 34, 420–429. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.J.; Zhao, J. Functional analysis of the rice metallothionein gene OsMT2b promoter in transgenic Arabidopsis plants and rice germinated embryos. Plant Sci. 2009, 176, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Van der Does, D.; Leon-Reyes, A.; Koornneef, A.; Van Verk, M.C.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Korbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic Acid Suppresses Jasmonic Acid Signaling Downstream of SCFCOI1-JAZ by Targeting GCC Promoter Motifs via Transcription Factor ORA59. Plant Cell 2013, 25, 744–761. [Google Scholar] [CrossRef]
- Liu, H.; WU, J.; B, H. Synergistic effect of jasmonic acid and other plant growth regulators on laticifer differentiation in Hevea brasiliensis. Chin. J. Trop. Crops 2001, 22, 6–16. [Google Scholar]
- Bhalothia, P.; Sangwan, C.; Alok, A.; Mehrotra, S.; Mehrotra, R. PP2C-like Promoter and Its Deletion Variants Are Induced by ABA but Not by MeJA and SA in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 547. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. Gus Fusions - Beta-Glucuronidase as a Sensitive and Versatile Gene Fusion Marker in Higher-Plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Zhao, J.; Xu, Z.; Xiong, Z. Comparison and Characterization of a Cell Wall Invertase Promoter from Cu-Tolerant and Non-Tolerant Populations of Elsholtzia haichowensis. Int. J. Mol. Sci. 2021, 22, 5299. https://doi.org/10.3390/ijms22105299
Liu R, Zhao J, Xu Z, Xiong Z. Comparison and Characterization of a Cell Wall Invertase Promoter from Cu-Tolerant and Non-Tolerant Populations of Elsholtzia haichowensis. International Journal of Molecular Sciences. 2021; 22(10):5299. https://doi.org/10.3390/ijms22105299
Chicago/Turabian StyleLiu, Rongxiang, Jing Zhao, Zhongrui Xu, and Zhiting Xiong. 2021. "Comparison and Characterization of a Cell Wall Invertase Promoter from Cu-Tolerant and Non-Tolerant Populations of Elsholtzia haichowensis" International Journal of Molecular Sciences 22, no. 10: 5299. https://doi.org/10.3390/ijms22105299
APA StyleLiu, R., Zhao, J., Xu, Z., & Xiong, Z. (2021). Comparison and Characterization of a Cell Wall Invertase Promoter from Cu-Tolerant and Non-Tolerant Populations of Elsholtzia haichowensis. International Journal of Molecular Sciences, 22(10), 5299. https://doi.org/10.3390/ijms22105299




