Genome-Wide Analysis of nsLTP Gene Family and Identification of SiLTPs Contributing to High Oil Accumulation in Sesame (Sesamum indicum L.)
Abstract
:1. Introduction
2. Results
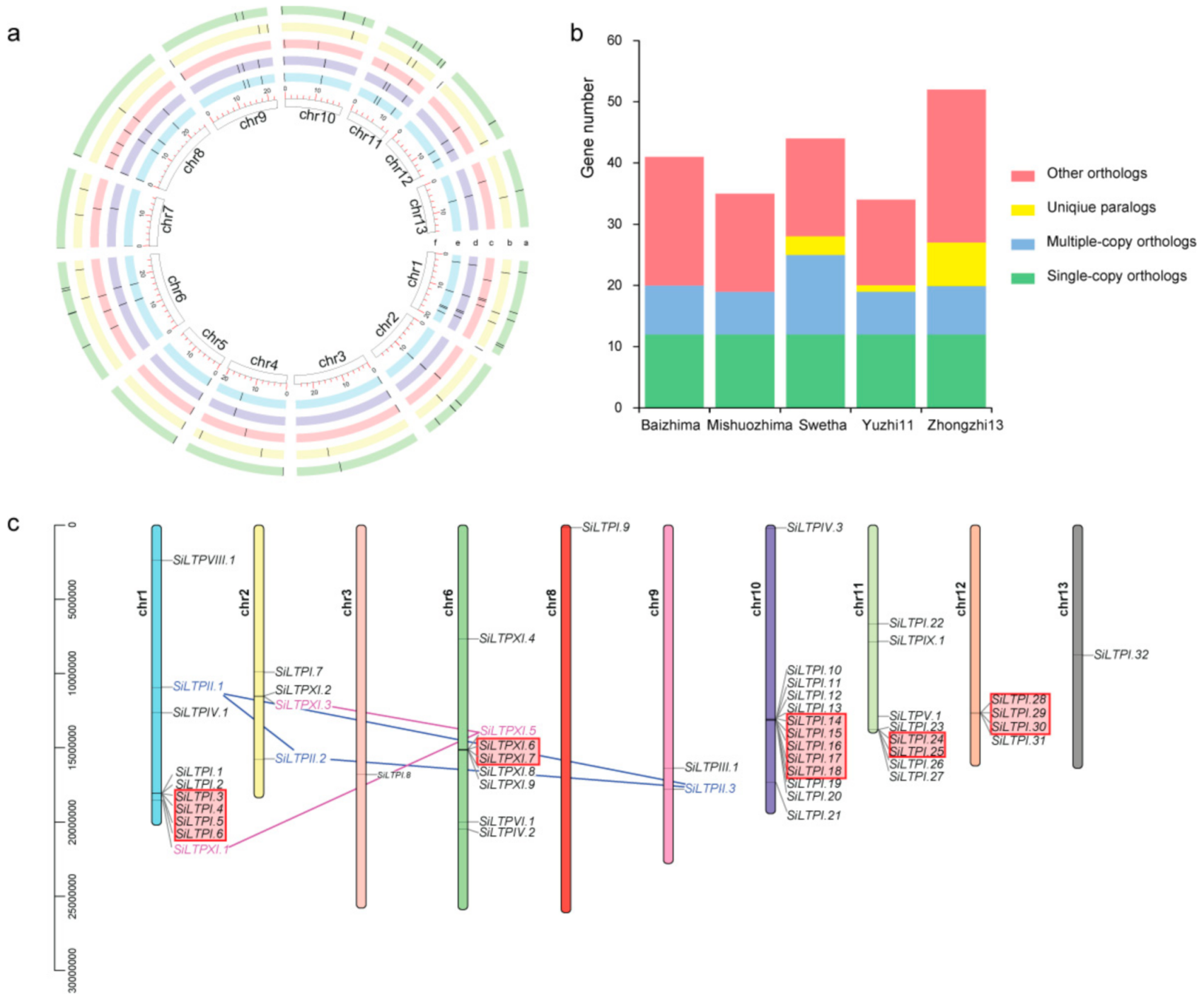
2.1. Diversity, Classification and Distribution of Sesamum indicum LTPs (SiLTPs)
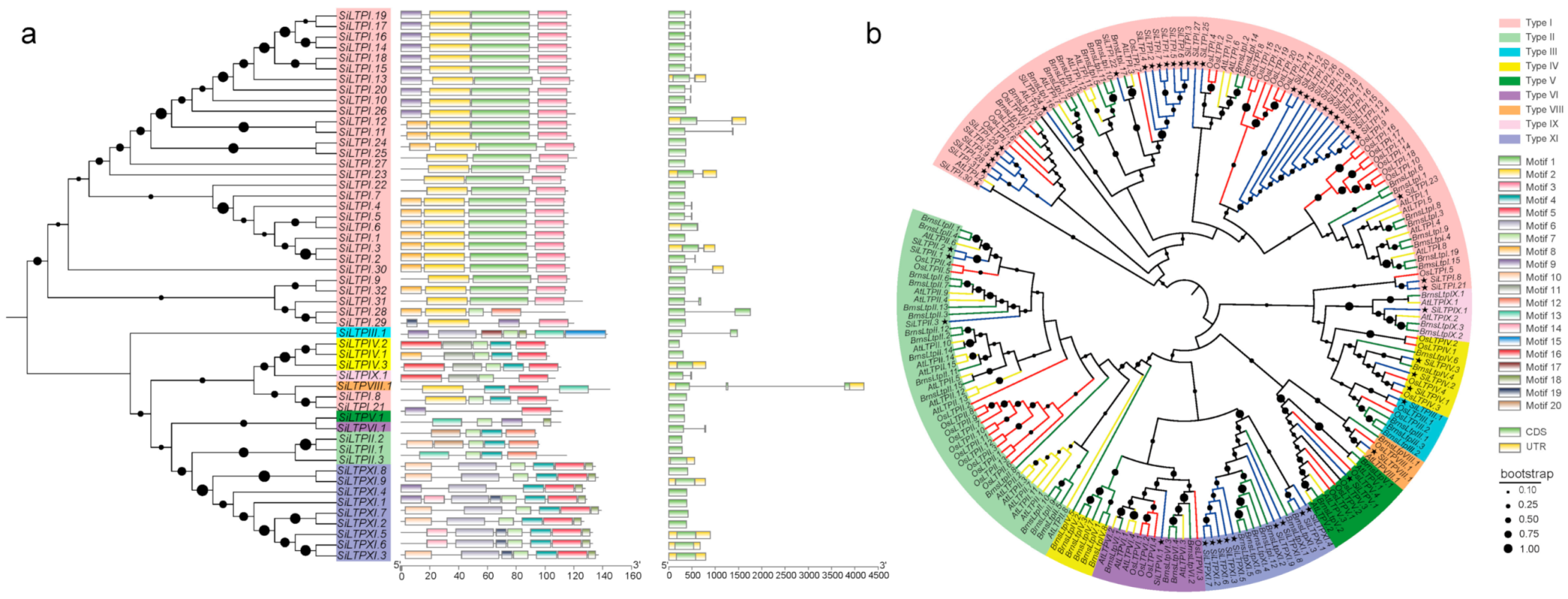
2.2. Characterization and Phylogenetic Analysis of the Conserved Motifs of Sesame SiLTP Genes
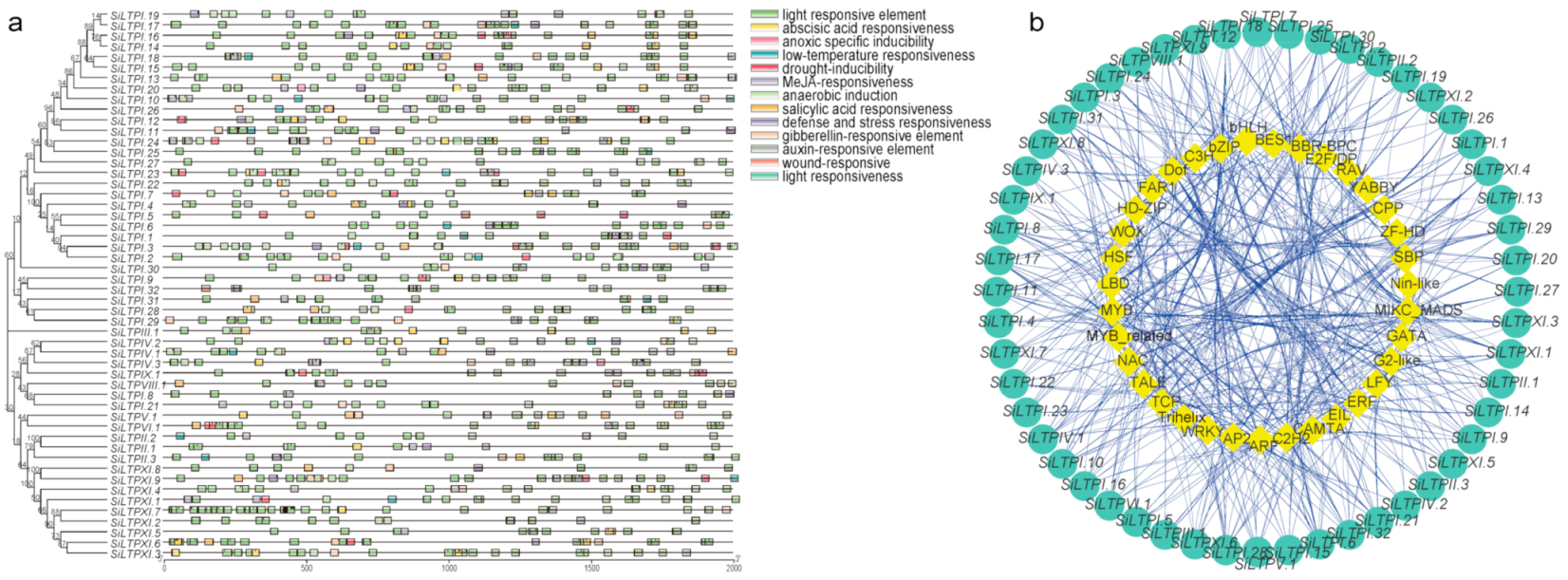
2.3. Cis-Acting Elements and Interacting Transcription Factors among the SiLTPs
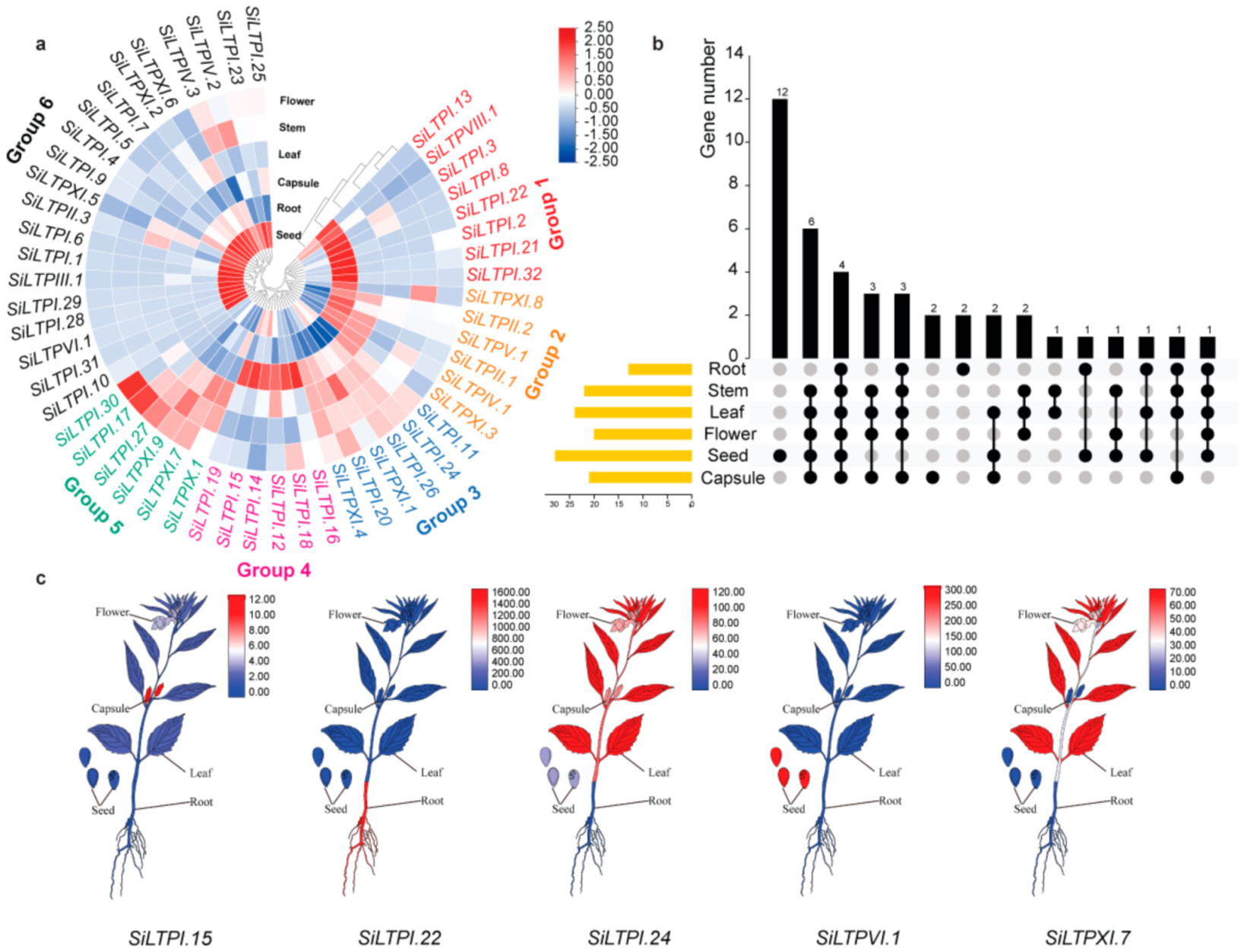
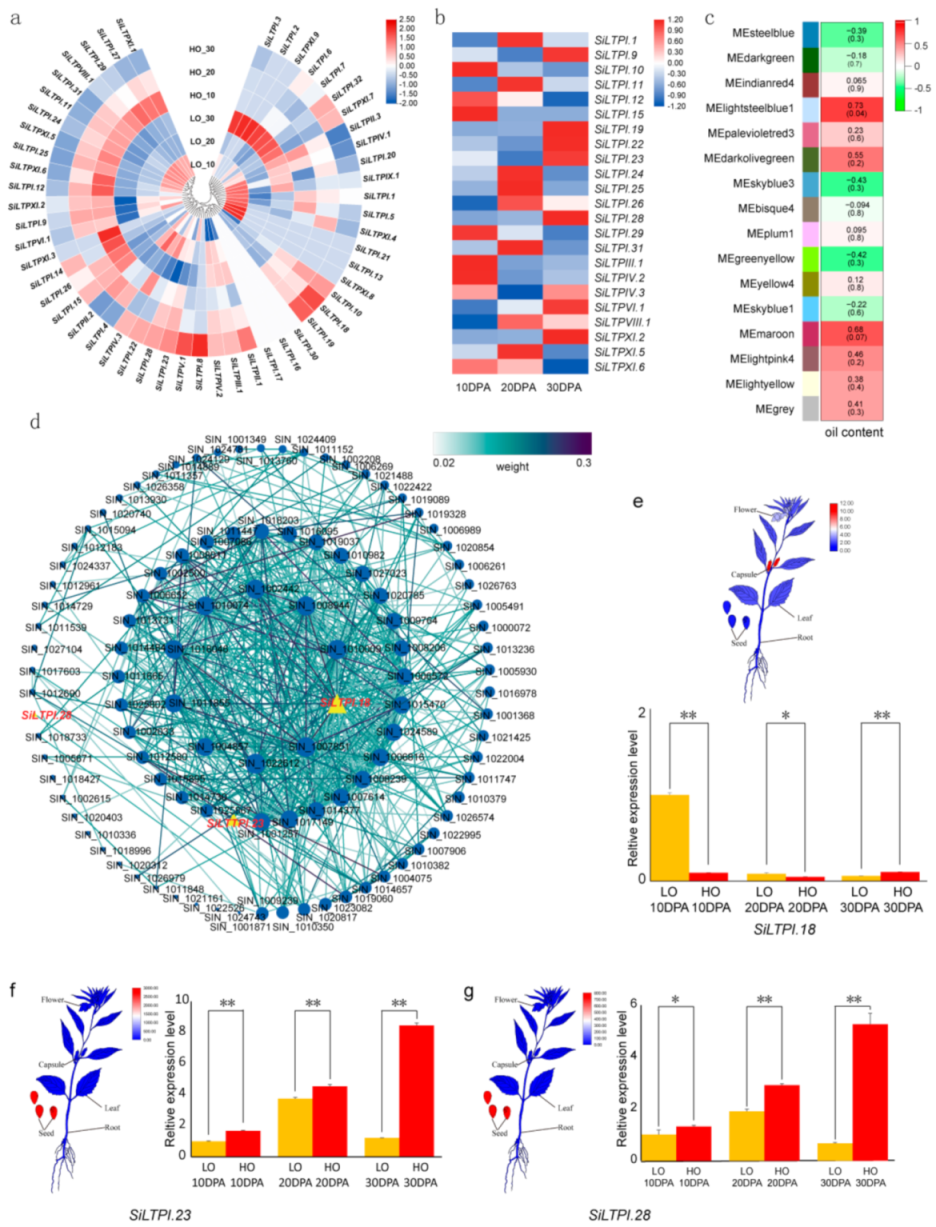
2.4. Expression Profiles of SiLTP Genes in Different Tissues of Sesame
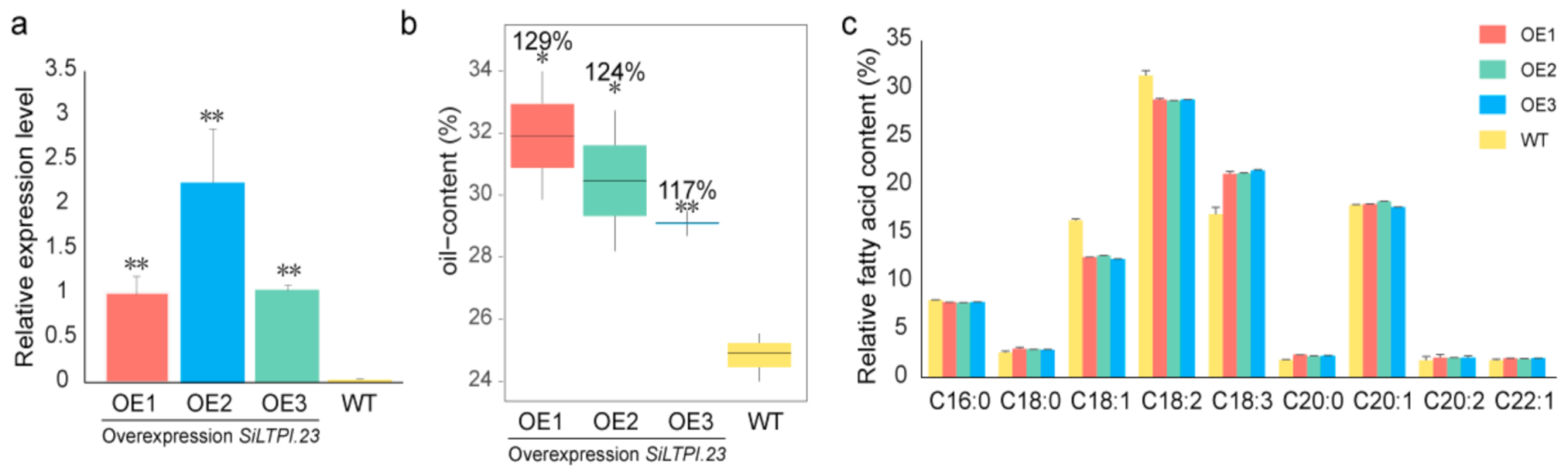
2.5. The SiLTPs Genes Involved in High Oil Content in Sesame Seeds
3. Discussion
4. Materials and Methods
4.1. Identification of the nsLTP Genes in Sesame Genome
4.2. Sequence Analysis, Phylogenetic Analysis and WGCNA Analysis
4.3. Plant Growth, RNA Extraction and SiLTPs Expression Analysis
4.4. Arabidopsis Transgenics Experiment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnson, L.A.; Suleiman, T.M.; Lusas, E.W. Sesame protein: A review and prospectus. J. Am. Oil Chem. Soc. 1979, 56, 463–468. [Google Scholar] [CrossRef]
- Wang, L.H.; Yu, S.; Tong, C.B.; Zhao, Y.Z.; Liu, Y.; Song, C.; Zhang, Y.X.; Zhang, X.D.; Wang, Y.; Hua, W.; et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014, 15, R39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Liu, K.; Zhang, Y.; Feng, Q.; Wang, L.; Zhao, Y.; Li, D.; Zhao, Q.; Zhu, X.; Zhu, X.; et al. Genetic discovery for oil production and quality in sesame. Nat. Commun. 2015, 6, 8609. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, Y.; Li, D.; Dossa, K.; Wang, M.L.; Zhou, R.; Yu, J.; Zhang, X. Gene expression profiles that shape high and low oil content sesames. BMC Genet. 2019, 20, 45. [Google Scholar] [CrossRef] [PubMed]
- Santi, C.; Molesini, B.; Guzzo, F.; Pii, Y.; Vitulo, N.; Pandolfini, T. Genome-wide transcriptional changes and lipid profile modifications induced by Medicago truncatula n5 overexpression at an early stage of the symbiotic interaction with Sinorhizobium meliloti. Genes 2017, 8, 396. [Google Scholar] [CrossRef] [Green Version]
- Wong, L.H.; Copic, A.; Levine, T.P. Advances on the transfer of lipids by Lipid Transfer Proteins. Trends Biochem. Sci. 2017, 42, 516–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutrot, F.; Chantret, N.; Gautier, M.-F. Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genom. 2008, 9, 86. [Google Scholar] [CrossRef] [Green Version]
- Salminen, T.A.; Blomqvist, K.; Edqvist, J. Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 2016, 244, 971–997. [Google Scholar] [CrossRef] [Green Version]
- Wong, L.H.; Levine, T.P. Lipid transfer proteins do their thing anchored at membrane contact sites… but what is their thing? Biochem. Soc. Trans. 2016, 44, 517–527. [Google Scholar] [CrossRef]
- Kader, J.C.; Julienne, M.; Vergnolle, C. Purification and characterization of a spinach-leaf protein capable of transferring phospholipids from liposomes to mitochondria or chloroplasts. Eur. J. Biochem. 1984, 139, 411–416. [Google Scholar] [CrossRef]
- Carvalho Ade, O.; Gomes, V.M. Role of plant lipid transfer proteins in plant cell physiology-a concise review. Peptides 2007, 28, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Edqvist, J.; Blomqvist, K.; Nieuwland, J.; Salminen, T.A. Plant lipid transfer proteins: Are we finally closing in on the roles of these enigmatic proteins? J. Lipid Res. 2018, 59, 1374–1382. [Google Scholar] [CrossRef] [Green Version]
- Edstam, M.M.; Laurila, M.; Hoglund, A.; Raman, A.; Dahlstrom, K.M.; Salminen, T.A.; Edqvist, J.; Blomqvist, K. Characterization of the GPI-anchored lipid transfer proteins in the moss Physcomitrella patens. Plant Physiol. Biochem. 2014, 75, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, X.; Lu, C.; Zeng, X.; Li, Y.; Fu, D.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Huang, D.; Liu, K.; Hu, S.; Yu, J.; Gao, G.; Song, S. Discovery, identification and comparative analysis of non-specific lipid transfer protein (nsLtp) family in Solanaceae. Genom. Proteom. Bioinf. 2010, 8, 229–237. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Gao, G.; Xu, K.; Chen, B.; Yan, G.; Li, F.; Qiao, J.; Zhang, T.; Wu, X. Genome-wide survey and expression analysis of the putative non-specific lipid transfer proteins in Brassica rapa L. PLoS ONE 2014, 9, e84556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, J.; Lv, H.; Yang, L.; Fang, Z.; Zhuang, M.; Zhang, Y.; Liu, Y.; Li, Z. Genome-wide identification and characterization of non-specific lipid transfer proteins in cabbage. PeerJ 2018, 6, e5379. [Google Scholar] [CrossRef]
- Kouidri, A.; Whitford, R.; Suchecki, R.; Kalashyan, E.; Baumann, U. Genome-wide identification and analysis of non-specific Lipid Transfer Proteins in hexaploid wheat. Sci. Rep. 2018, 8, 17087. [Google Scholar] [CrossRef]
- Meng, C.; Yan, Y.; Liu, Z.; Chen, L.; Zhang, Y.; Li, X.; Wu, L.; Zhang, G.; Wang, X.; Ma, Z. Systematic analysis of cotton non-specific lipid transfer protein family revealed a special group that is involved in fiber elongation. Front. Plant Sci. 2018, 9, 1285. [Google Scholar] [CrossRef]
- D’Agostino, N.; Buonanno, M.; Ayoub, J.; Barone, A.; Monti, S.M.; Rigano, M.M. Identification of non-specific Lipid Transfer Protein gene family members in Solanum lycopersicum and insights into the features of Solal3 protein. Sci. Rep. 2019, 9, 1607. [Google Scholar] [CrossRef]
- Zhang, M.; Kim, Y.; Zong, J.; Lin, H.; Dievart, A.; Li, H.; Zhang, D.; Liang, W. Genome-wide analysis of the barley non-specific lipid transfer protein gene family. Crop J. 2019, 7, 65–76. [Google Scholar] [CrossRef]
- Debono, A.; Yeats, T.H.; Rose, J.K.C.; Bird, D.; Jetter, R.; Kunst, L.; Samuels, L. Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 2009, 21, 1230–1238. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhou, W.; Lu, Z.; Ouyang, Y.; Su, O.C.; Yao, J. A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice. Plant Sci. 2015, 239, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Pagnussat, L.A.; Oyarburo, N.; Cimmino, C.; Pinedo, M.L.; de la Canal, L. On the role of a lipid-transfer protein Arabidopsis Ltp3 mutant is compromised in germination and seedling growth. Plant Signal. Behav. 2015, 10, e1105417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.Y.; Jauh, G.Y.; Mollet, G.-C.; Eckard, K.J.; Nothnagel, E.A.; Walling, L.L.; Lord, E.M. A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell 2000, 12, 151–164. [Google Scholar] [PubMed]
- Deeken, R.; Saupe, S.; Klinkenberg, J.; Riedel, M.; Leide, J.; Hedrich, R.; Mueller, T.D. The nonspecific lipid transfer protein AtLtpI-4 is involved in suberin formation of Arabidopsis thaliana crown galls. Plant Physiol. 2016, 172, 1911–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiapparino, A.; Maeda, K.; Turei, D.; Saez-Rodriguez, J.; Gavin, A.C. The orchestra of lipid-transfer proteins at the crossroads between metabolism and signaling. Prog. Lipid Res. 2016, 61, 30–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkina, E.I.; Melnikova, D.N.; Bogdanov, I.V.; Ovchinnikova, T.V. Lipid transfer proteins as components of the plant innate immune system: Structure, functions, and applications. Acta Nat. 2016, 8, 47–61. [Google Scholar] [CrossRef]
- Hairat, S.; Baranwal, V.K.; Khurana, P. Identification of Triticum aestivum nsLTPs and functional validation of two members in development and stress mitigation roles. Plant Physiol. Biochem. 2018, 130, 418–430. [Google Scholar] [CrossRef]
- Pan, Y.; Li, J.; Jiao, L.; Li, C.; Zhu, D.; Yu, J. A non-specific setaria italica lipid transfer protein gene plays a critical role under abiotic stress. Front. Plant Sci. 2016, 7, 1752. [Google Scholar] [CrossRef] [Green Version]
- Zachowski, A.; Guerbette, F.; Grosbois, M.; Jolliot-Croquin, A.; Kader, J.C. Characterisation of acyl binding by a plant lipid-transfer protein. Eur. J. Biochem. 1998, 257, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, P.O.; Raguideau, S.; Quince, C.; Holden, J.; Zhang, L.; Williams, T.A.; Gubry-Rangin, C. Gene duplication drives genome expansion in a major lineage of Thaumarchaeota. Nat. Commun. 2020, 11, 5494. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajima, D.; Kaneko, A.; Sakamoto, M.; Ito, Y.; Hue, N.T.; Miyazaki, M.; Ishibashi, Y.; Yuasa, T.; Iwayainoue, M. Wrinkled 1 (WRI1) Homologs, AP2-Type transcription factors involving master regulation of seed storage oil synthesis in castor bean (Ricinus communis L.). Amer. J. Plant Sci. 2013, 4, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Pré, M.; Atallah, M.; Champion, A.; Vos, M.D.; Pieterse, C.M.J.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [Green Version]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Wang, H.W.; Zhang, B.; Hao, Y.J.; Huang, J.; Tian, A.G.; Liao, Y.; Zhang, J.S.; Chen, S.Y. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 2007, 52, 716–729. [Google Scholar] [PubMed]
- Ibáez-Salazar, A.; Rosales-Mendoza, S.; Rocha-Uribe, A.; Ramírez-Alonso, J.I.; Lara-Hernández, I.; Hernández-Torres, A.; Paz-Maldonado, L.M.T.; Silva-Ramírez, A.S.; Bauelos-Hernández, B.; Martínez-Salgado, J.L.; et al. Over-expression of Dof-type transcription factor increases lipid production in Chlamydomonas reinhardtii. J. Biotechnol. 2014, 184, 27–38. [Google Scholar] [CrossRef]
- Bedigian, D.; Harlan, J.R. Evidence for cultivation of sesame in the ancient world. Econ. Bot. 1986, 40, 137–154. [Google Scholar] [CrossRef]
- Murata, J.; Ono, E.; Yoroizuka, S.; Toyonaga, H.; Shiraishi, A.; Mori, S.; Tera, M.; Azuma, T.; Nagano, A.J.; Nakayasu, M.; et al. Oxidative rearrangement of (+)-sesamin by CYP92B14 co-generates twin dietary lignans in sesame. Nat. Commun. 2017, 8, 2155. [Google Scholar] [CrossRef] [PubMed]
- Edstam, M.M.; Viitanen, L.; Salminen, T.A.; Edqvist, J. Evolutionary history of the non-specific lipid transfer proteins. Mol. Plant 2011, 4, 947–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, J.W.; Donoghue, P.C.J. Whole-genome duplication and plant macroevolution. Trends Plant Sci. 2018, 23, 933–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Zhu, Y.X. Plant polyploidy and evolution. J. Integr. Plant Biol. 2019, 61, 4–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Li, X.; Chen, W.; Peng, X.; Xiao, C.; Zhu, S.; Cheng, B. Whole-genome survey and characterization of MADS-box gene family in maize and sorghum. Plant Cell Tissue Organ Cult. 2011, 105, 159–173. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef] [Green Version]
- Gincel, E.; Simorre, J.P.; Caille, A.; Marion, D.; Ptak, M.; Vovelle, F. Three-dimensional structure in solution of a wheat lipid-transfer protein from multidimensional 1H-NMR data. A new folding for lipid carriers. Eur. J. Biochem. 1994, 226, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Shoeva, O.Y.; Mock, H.P.; Kukoeva, T.V.; Borner, A.; Khlestkina, E.K. Regulation of the flavonoid biosynthesis pathway genes in purple and black grains of Hordeum vulgare. PLoS ONE 2016, 11, e0163782. [Google Scholar] [CrossRef]
- Gong, L.; Zhang, H.; Liu, X.; Gan, X.; Nie, F.; Yang, W.; Zhang, L.; Chen, Y.; Song, Y.; Zhang, H. Ectopic expression of HaNAC1, an ATAF transcription factor from Haloxylon ammodendron, improves growth and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 151, 535–544. [Google Scholar] [CrossRef]
- Arslan, Ç.; Uzun, B.; Ülger, S.; Çağırgan, M.İ. Determination of oil content and fatty acid composition of sesame mutants suited for intensive management conditions. J. Am. Oil Chem. Soc. 2007, 84, 917–920. [Google Scholar] [CrossRef]
- Unver, T.; Wu, Z.; Sterck, L.; Turktas, M.; Lohaus, R.; Li, Z.; Yang, M.; He, L.; Deng, T.; Escalante, F.J.; et al. Genome of wild olive and the evolution of oil biosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, E9413–E9422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gacek, K.; Bartkowiak-Broda, I.; Batley, J. Genetic and molecular regulation of seed storage proteins (SSPs) to improve protein nutritional value of oilseed rape (Brassica napus L.) seeds. Front. Plant Sci. 2018, 9, 890. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.M.; Lee, S.B.; Cho, S.H.; Hwang, I.; Hur, C.G.; Suh, M.C. Isolation and characterization of multiple abundant lipid transfer protein isoforms in developing sesame (Sesamum indicum L.) seeds. Plant Physiol. Biochem. 2008, 46, 127–139. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Sudhir, K.; Glen, S.; Li, M.; Christina, K.; Koichiro, T. Evolution, MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar]
- Ivica, L.; Peer, B. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, J.; Wang, Y.; Zhang, Y.; Dossa, K.; Li, D.; Zhou, R.; Wang, L.; Zhang, X. Genome-wide identification and expression analyses of genes involved in raffinose accumulation in sesame. Sci. Rep. 2018, 8, 4331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Zhou, R.; Li, D.; Liu, A.; Qin, L.; Mmadi, M.A.; Su, R.; Zhang, Y.; Wang, J.; Gao, Y.; et al. A novel motif in the 5′-UTR of an orphan gene ‘Big Root Biomass’ modulates root biomass in sesame. Plant Biotechnol. J. 2020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, S.; You, J.; Shi, L.; Sheng, C.; Zhou, W.; Dossou, S.S.K.; Dossa, K.; Wang, L.; Zhang, X. Genome-Wide Analysis of nsLTP Gene Family and Identification of SiLTPs Contributing to High Oil Accumulation in Sesame (Sesamum indicum L.). Int. J. Mol. Sci. 2021, 22, 5291. https://doi.org/10.3390/ijms22105291
Song S, You J, Shi L, Sheng C, Zhou W, Dossou SSK, Dossa K, Wang L, Zhang X. Genome-Wide Analysis of nsLTP Gene Family and Identification of SiLTPs Contributing to High Oil Accumulation in Sesame (Sesamum indicum L.). International Journal of Molecular Sciences. 2021; 22(10):5291. https://doi.org/10.3390/ijms22105291
Chicago/Turabian StyleSong, Shengnan, Jun You, Lisong Shi, Chen Sheng, Wangyi Zhou, Senouwa Segla Koffi Dossou, Komivi Dossa, Linhai Wang, and Xiurong Zhang. 2021. "Genome-Wide Analysis of nsLTP Gene Family and Identification of SiLTPs Contributing to High Oil Accumulation in Sesame (Sesamum indicum L.)" International Journal of Molecular Sciences 22, no. 10: 5291. https://doi.org/10.3390/ijms22105291








