Abstract
Osteogenesis imperfecta (OI) is a bone fragility disorder that is usually caused by mutations affecting collagen type I. We compared the calvaria bone tissue transcriptome of male 10-week-old heterozygous Jrt (Col1a1 mutation) and homozygous oim mice (Col1a2 mutation) to their respective littermate results. We found that Jrt and oim mice shared 185 differentially expressed genes (upregulated: 106 genes; downregulated: 79 genes). A total of seven genes were upregulated by a factor of two or more in both mouse models (Cyp2e1, Slc13a5, Cgref1, Smpd3, Ifitm5, Cthrc1 and Rerg). One gene (Gypa, coding for a blood group antigen) was downregulated by a factor of two or more in both OI mouse models. Overrepresentation analyses revealed that genes involved in ‘ossification’ were significantly overrepresented among upregulated genes in both Jrt and oim mice, whereas hematopoietic genes were downregulated. Several genes involved in Wnt signaling and transforming growth factor beta signaling were upregulated in oim mice, but less so in Jrt mice. Thus, this study identified a set of genes that are dysregulated across various OI mouse models and are likely to play an important role in the pathophysiology of this disorder.
1. Introduction
Osteogenesis imperfecta (OI) is a heritable connective tissue disorder that is clinically characterized by low bone mass, bone fragility, long bone deformity, scoliosis, discolored sclera and dentinogenesis imperfecta [1]. In the large majority of individuals with OI, the condition is caused by mutations in Col1a1 or Col1a2 that directly affect the bone matrix protein collagen type I [2]. However, pathogenic variants in about 20 other genes can also give rise to an OI phenotype [1].
How mutations in Col1a1 and Col1a2 lead to low bone mass and bone fragility is not entirely clear. The low bone mass of OI is not simply caused by an inability of the osteoblast system to produce bone. Indeed, many children with severe OI have increased bone formation rates, as shown by iliac bone histomorphometry [3,4]. Low bone mass in OI is therefore likely caused by dysregulation of bone cell activity rather than the incapacity of osteoblasts to produce enough organic bone matrix.
Several mouse models with mutations in Col1a1 or Col1a2 are available to study the dysregulation of bone cells. A widely used model is the oim mouse that harbors a frameshift mutation in Col1a2 [5]. Homozygous oim mice are small and have spontaneous fractures, mimicking the manifestations of severe OI in humans. The Jrt mouse is a model of dominant severe OI caused by a splice-site mutation in Col1a1 that leads to an 18-amino acid deletion in the collagen type I alpha 1 chain [6]. Similar to the homozygous oim mouse, the heterozygous Jrt mouse has spontaneous fractures and bone deformities.
Thus, homozygous oim mice and heterozygous Jrt mice harbor quite different mutations in Col1a2 and Col1a1, respectively, but share a similarly severe phenotype. Identifying molecular pathways and genes that are dysregulated in both mouse models might therefore help to delineate common factors that are involved in causing low bone mass and bone fragility in the context of mutations in collagen type I encoding genes. In the present study, we therefore performed RNA sequencing in the calvaria bone tissues of homozygous oim and heterozygous Jrt mice and performed differential gene expression analyses. We identified a set of genes that are dysregulated across various OI mouse models and are likely to play an important role in the pathophysiology of this disorder.
2. Results
Compared to wild type (WT) mice, 231 genes were differentially expressed in Jrt mice, and 2969 genes were differentially expressed in oim mice (Figure 1). We found 185 genes that were differentially expressed in both Jrt and oim mice, which were all concordantly upregulated (106 genes) or downregulated (79 genes) between the two models (Supplementary Tables S1–S6).
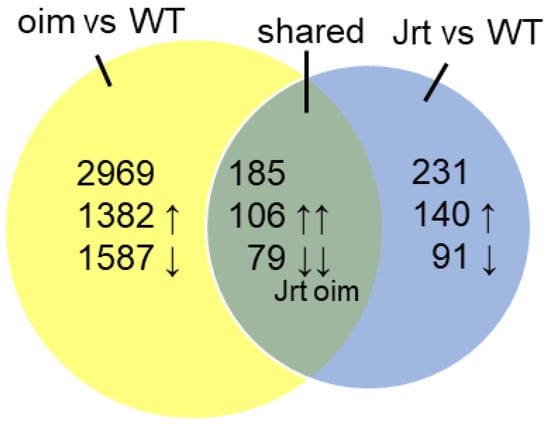
Figure 1.
Results of RNA sequencing in calvaria of 10-week-old mice. Venn diagram of differentially expressed genes in oim and Jrt mice.
The list of the top 20 upregulated genes in Jrt mice contained genes that are known to be important for the osteoblast function (e.g., Bglap—the gene coding for osteocalcin; Ifitm5—coding for BRIL) and the synthesis of extracellular matrix (Smpd3, Col11a1, Col11a2 and Adamts14) (Table 1).

Table 1.
List of the top 20 upregulated genes in Jrt mice compared to WT mice. p values are adjusted by the Benjamini–Hochberg procedure, as determined by deseq2.
In oim mice, the list of the top 20 upregulated genes also contained genes that play a role in the function of osteoblasts (e.g., Edn1, Ddit4, Cdkn1a, Igfbp3 and Stc2) (Table 2). Only one gene (Cyp2e1) was among the top 20 upregulated genes in both Jrt and oim mice. When all upregulated genes were considered, Jrt and oim mice shared several genes that play a role in the pathogenesis of various OI types (Figure 2).

Table 2.
List of the top 20 upregulated genes in oim mice compared to WT mice. p values are adjusted by the Benjamini–Hochberg procedure, as determined by deseq2.
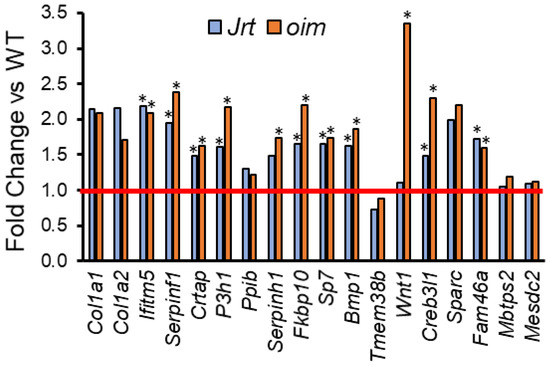
Figure 2.
Fold change in Jrt and oim versus WT mice in genes that, when mutated, give rise to OI. The red line indicates parity to WT. Asterisks (*) indicate significant upregulated genes (adjusted p < 0.05, as determined by deseq2).
To identify a core set of genes that are dysregulated in OI, we searched for genes where expression levels compared to WT mice differed by a factor of at least two in both OI models. A total of eight such genes were identified, seven of which were upregulated (Cyp2e1, Slc13a5, Cgref1, Smpd3, Ifitm5, Cthrc1 and Rerg), and one (Gypa, coding for a blood group antigen) was downregulated (Table 3). The results for these eight genes were validated by RT-PCR (Figure 3). This confirmed the observations from the RNA sequencing analyses, even though the differences in Gypa levels were not significant in Jrt mice. As all previous experiments were conducted in male mice, we also verified differential gene expression in the calvaria of female oim mice using RT-PCR. Except for Rerg and Smpd3, results were similar to those of male oim mice (Supplementary Figure S1).

Table 3.
Genes that are up- or downregulated 2-fold or more in the calvaria of both Jrt and oim mice. p values are adjusted by the Benjamini–Hochberg procedure, as determined by deseq2.
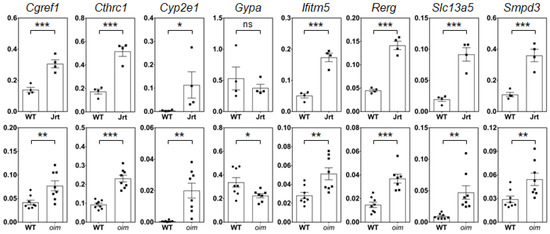
Figure 3.
Real-time PCR results in calvaria RNA of 10-week-old male mice. Results for Jrt are shown in the upper panel, results for oim in the lower panel. Values represent the 2−∆Ct normalized to Rpl27. Significant differences in gene expression compared to WT mice are indicated by asterisks (one-tailed unpaired t-test): ns, non-significant, * p < 0.05, ** p < 0.01, *** p < 0.001. Error bars represent standard errors.
Six of the seven common upregulated genes (Cyp2e1, Cgref1, Slc13a5, Cthrc1, Smpd3 and Ifitm5) were also upregulated in tibia osteocytes of oim mice, as previously reported by Zimmerman et al. [7]. In addition, four of these six genes (Cgref1, Slc13a5, Smpd3 and Ifitm5) were upregulated in tibia osteocytes of CRTAP deficient mice, another mouse model of OI, as reported in the same study.
Overrepresentation analysis in Jrt mice showed that genes involved in ‘ossification’ (GO:0001503) were the most significantly enriched group among upregulated genes (enrichment ratio: 10.2) (Table 4). Other biological processes with significantly enriched upregulated genes included ‘extracellular structure organization’ (GO:0043062), ‘peptidyl-proline modification’ (GO:0018208) and ‘collagen metabolic process’ (GO:0032963). Genes involved in ‘myeloid cell differentiation’ (GO:0030099) were significantly overrepresented among downregulated genes (enrichment ratio: 8.2). Other biological processes significantly enriched for downregulated genes included ‘homeostasis of number of cells’ (GO:0048872) and ‘tetrapyrrole metabolic process’ (GO:0033013).

Table 4.
Overrepresentation analysis of genes that are differentially expressed in Jrt mice. FDR: false discovery rate.
Overrepresentation analysis in oim mice revealed that genes involved in ‘ossification’ (GO:0001503) (enrichment ratio 4.0), ‘response to transforming growth factor beta’ (GO:0071559) (enrichment ratio 3.2) and ‘cell–cell signaling by Wnt’ (GO:0198738) (enrichment ratio 2.7) were significantly overrepresented among upregulated genes (false discovery rate <0.001 in each case) (Table 5). Among downregulated genes, genes involved in ‘myeloid cell differentiation’ (GO:0030099) (enrichment ratio: 2.6) were the most significantly overrepresented. The transcription factor target analysis revealed that there were no significantly enriched target gene sets in Jrt mice. In oim mice, there was a strong enrichment in genes that are responsive to SMAD3, SMAD4 (both of which are involved in TGF-beta signaling [8]) and FOXO1 (involved in Wnt signaling [9]) (false discovery rate <0.001 for each of these transcription factors) (Supplementary Table S7).

Table 5.
Overrepresentation analysis of genes that are differentially expressed in oim mice. FDR: False discovery rate.
In accordance with the biological processes identified by the overrepresentation analyses, several genes involved in Wnt signaling and TGF-beta signaling were upregulated in oim mice, but not, or to a lesser extent, in Jrt mice (Figure 4).
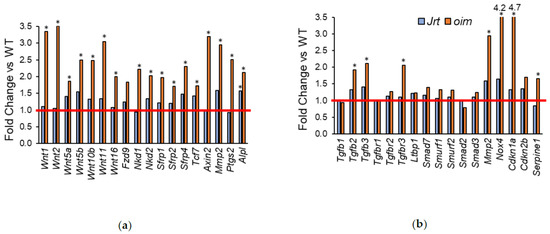
Figure 4.
Fold change in Jrt and oim versus WT mice of genes that are involved in (a) Wnt signaling and (b) TGF-beta signaling. The red line indicates parity to WT. Asterisks (*) indicate significant upregulated genes (adjusted p < 0.05, as determined by deseq2).
As the RNA sequencing analysis indicated that hematopoiesis was downregulated in OI mice, we hypothesized that this decrease in hematopoietic gene expression reflected a decrease in bone marrow space in the calvaria. We therefore performed microCT analysis of the calvaria bone of 10-week-old OI mice and their respective littermates. In Jrt and oim mice, the bone was thinner and contained less bone marrow space (Figure 5).

Figure 5.
Three-dimensional rendering of microCT scans of 10-week-old WT, Jrt and oim mice’s right parietal bones. In Jrt and oim mice, the bone is thinner and contains less bone marrow space (arrows).
3. Discussion
In this study, we assessed differential gene expression in the calvaria bone tissues of two mouse models of severe OI, Jrt and oim. There were almost 13 times more differentially expressed genes in oim mice than in Jrt mice (2969 vs. 231 genes), and the list of the top 20 upregulated genes had little overlap between the two OI models. Genes involved in Wnt signaling and in TGF-beta signaling were upregulated in oim mice, but there was less evidence for dysregulation in these pathways in Jrt mice. Nevertheless, genes involved in ossification and genes that are involved in causing OI were upregulated in both OI models. We finally identified a set of genes that seem to be consistently upregulated across OI mouse models.
Why oim mice have many more upregulated genes than Jrt mice is not clear. Both are models of the severe OI that develop spontaneous fractures [5,6]. The oim model is homozygous for a Col1a2 frameshift mutation, and therefore does not produce any normal collagen type I, whereas the Jrt mouse is heterozygous for a splice-site mutation that leads to the skipping of Col1a1 exon 9 [5,6]. It is possible that the absence of normal collagen type I in the oim mouse triggers a larger number of regulatory cascades than the heterozygous abnormality in the Jrt mouse. It is also possible that the differences in transcriptome dysregulation are influenced by differences in the background strain, as oim mice in the present study were on a B6C3Fe background, whereas the Jrt mice were on an FVB background.
The observation that, compared to oim, Jrt mice seem to have milder or no abnormalities in Wnt and TGF-beta signaling pathways may explain our previous findings that Jrt mice are less responsive to treatments targeting these pathways [10,11]. We have previously noted that treatment with a sclerostin antibody, a therapeutic approach targeting Wnt signaling, appeared to be less effective in Jrt mice than in other mouse models of OI, including oim [10,12]. Similarly, the lower baseline activation of TGF-beta pathways in the Jrt mice may contribute to our observation that antibodies inactivating TGF-beta had less effect on bone mass in the Jrt mouse than in other OI models [11,13].
Among the strongly upregulated genes that are shared between Jrt and oim mice, Cyp2e1, Slc13a5, Cgref1 and Rerg have not previously been implicated in OI pathophysiology to our knowledge. The fold change was highest for Cyp2e1. Cyp2e1 codes for a cytochrome P450 enzyme that metabolizes many substrates and is involved in the generation of reactive oxygen species [14]. The functional importance of Cyp2e1 in the bone is unclear, but prior studies have shown that Cyp2e1 expression in osteoblasts decreases with unloading and increases with differentiation as well as after exposure to tumor necrosis factor alpha [15,16]. Systemically increased tumor necrosis alpha levels have been reported in oim mice [17]. In a separate study, we also observed increased Cyp2e1 expression in the gastrocnemius muscle of both Jrt and oim mice, which supports the hypothesis that Cyp2e1 is upregulated in these mice due to a systemic factor [18].
Slc13a5 codes for a sodium-dependent citrate transporter that mediates the uptake of citrate from blood into many cell types, such as neurons, hepatocytes and osteoblasts [19]. Citrate plays not only a key role in cellular energy metabolism, but also in bone, as it binds to both apatite crystal surfaces and collagen in the extracellular matrix and thereby regulates crystal thickening [20]. Given that OI is characterized by abnormal crystal thickness [21], it seems plausible that citrate and Slc13a5 are involved in the abnormal mineralization seen in the OI bone. Interestingly, the Slc13a5 deficient mouse has some phenotypic overlap with OI, as it shows amelogenesis imperfecta, a tooth enamel defect resembling dentinogenesis imperfecta as well as low bone mineral density [22]. Amelogenesis imperfecta is also present in humans with biallelic loss of function variants in Slc13a5 [23]. It therefore appears plausible that the dysregulation of Slc13a5 contributes to the clinical manifestations associated with OI.
Cgref1 codes for a secretory protein that plays an important role in the regulation of the transcription factor AP-1 [24]. The role of Cgref1 in osteoblasts or bones is not clear. Rerg encodes a transcriptional repressor that is much more highly expressed in calvarial osteoblasts than in femoral osteoblasts and may play a site-specific role in regulating estrogen signaling [25]. It is therefore interesting to note that Rerg was differentially regulated in male but not in female oim mice.
Several of the other strongly upregulated genes have a better characterized role in bones. Ifitm5 codes for BRIL, a protein that is specifically found in osteoblasts [26]. A specific mutation in the 5′ untranslated region of Ifitm5 gives rise to OI type V [27,28]. Although the exact role played by BRIL in regulating osteoblast activity has yet to be determined, Ifitm5 has emerged as a specific and robust marker of osteoblast differentiation [29]. Smpd3 encodes sphingomyelin phosphodiesterase 3, and Smpd3 deficiency is associated with severe bone deformities, fractures and abnormal bone mineralization [30]. Cthrc1 codes for a secreted osteoblast product that decreases bone resorption [31].
Regarding downregulated genes, those involved in ‘myeloid cell differentiation’ were overrepresented in both the Jrt and the oim model. Gypa, the one gene that was strongly downregulated in both Jrt and oim mice, codes for glycophorin A, which is one of the most abundant red cell proteins [32]. Our microCT analyses indicated that Jrt and oim mice had less bone marrow space in their calvaria bones. These observations suggest that the reduced expression of hematopoiesis genes in the OI calvaria reflects a decreased amount of hematopoietic bone marrow.
In conclusion, genes involved in Wnt signaling and in TGF-beta signaling were upregulated in oim mice, but there was less evidence for dysregulation in these pathways in Jrt mice. In both OI models, genes involved in ossification were upregulated, whereas hematopoietic genes were downregulated. This study also identified a set of genes that seem to be consistently upregulated in the bone tissues of several OI mouse models and therefore may play important roles in the pathophysiology of OI.
4. Materials and Methods
All experiments were approved by the Animal Care Committee of McGill University (protocol 2011-5975, approved on 1 April 2018) and conformed to the ethical guidelines of the Canadian Council on Animal Care. The Jrt mice on the FVB background were a gift from Dr. Jane Aubin’s laboratory, University of Toronto, Canada [6]. The breeding colony was maintained at the Animal Care Facility of the Shriners Hospitals for Children—Canada. Male heterozygous Jrt mice and their WT littermates were used for the experiments.
Oim mice on the B6C3Fe background were purchased from Jackson Laboratories (stock 4145670). Breeding pairs were mated to generate WT, heterozygote oim/+ and homozygote oim (hereafter designated oim). Only male WT and oim mice were used for the RNA sequence analyses, whereas real-time PCR tests were performed in both male and female mice. The colony was maintained by breeding oim/+ mice. Genotyping was performed by PCR as described by Zimmerman et al. [33].
4.1. Sample Preparation
Mice were euthanized at the age of 10 weeks (n = 4 for Jrt mice and the corresponding WT mice; n = 8 for oim and the corresponding WT). The calvariae were dissected from mice, immediately immersed in RNAlater (Ambion, Invitrogen, Austin, TX, USA) and stored at −20 °C until processing. While still in RNAlater, the calvariae were carefully cleaned of connective tissues. They were then cut along the sagittal and lambdoid sutures, such that only the parietal and frontal parts were used for the RNA extraction. After mincing with scissors, the bones were blotted to remove excess RNAlater and homogenized in 1 mL of TRIzol™ (Thermo Fisher Scientific, Waltham, MA, USA) for 15 s with a tissue mincer (OMNI International, Kennesaw, GA, USA) equipped with a 5-mm stainless steel generator probe. The total RNA was extracted and further purified on RNeasy MinElute columns (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol.
4.2. Library Preparation and RNA Sequencing
Total RNA was quantified using a NanoDrop Spectrophotometer ND-1000 (NanoDrop Technologies, Inc., Wilmington, DE, USA), and its integrity was assessed by a 2100 Bioanalyzer (Agilent Technologies). The total RNA samples used in the present study had a concentration between 80 and 290 ng/μL, with an RNA integrity number ranging between 8.5 and 9.6. Libraries were generated from 250 ng of the total RNA as follows: mRNA enrichment was performed using the NEBNext Poly(A) Magnetic Isolation Module (New England Biolabs, Ipswich, MA, USA). cDNA synthesis was achieved with the NEBNext RNA First Strand Synthesis and NEBNext Ultra Directional RNA Second Strand Synthesis Modules (New England Biolabs). The remaining steps of the library preparation were done using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs). Adapters and PCR primers were purchased from New England Biolabs. Libraries were quantified using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies, Carlsbad, CA, USA) and the Kapa Illumina GA with Revised Primers—SYBR Fast Universal Kit (Kapa Biosystems, London, UK). Average size fragment was determined using a LabChip GX (PerkinElmer, Waltham, MA, USA) instrument.
The libraries were normalized, pooled and then denatured in 0.05 N NaOH and neutralized using HT1 buffer. The pool was loaded at 225 pM on an NovaSeq S4 l (Illumina, San Diego, CA, USA) ane using Xp protocol as per the manufacturer’s recommendations. The run was performed for 2 × 100 cycles (paired-end mode). A PhiX library was used as a control and mixed with libraries at 1% level. Base calling was performed with RTA v3. Program bcl2fastq2 v2.20 was then used to demultiplex samples and generate fastq reads. Between 36 million and 99 million 100-base paired-end reads were generated per sample.
Data Post-Processing and Statistical Evaluation
Analysis of paired-end sequencing reads was performed using GenPipes, open-source, Python-based standardized analysis pipelines, hosted on the Compute Canada High Performance Computing center (https://www.computecanada.ca). Briefly, paired-end sequencing reads were clipped for an adapter sequence, trimmed for a minimum quality (Q30) in 3’ and filtered for a minimum length of 32 bp using Trimmomatic [34]. Surviving read pairs were aligned to GRCm38 by the RNA-seq aligner STAR using the recommended two-passes approach [35]. Aligned RNA-seq reads were assembled into transcripts, and their relative abundance was estimated using Cufflinks [36] and Cuffdiff [37]. Exploratory analysis was conducted using various functions and packages from R and the Bioconductor project. Differential expression analysis was conducted using DESeq [38]. Expressed short single-nucleotide variants and indel calling were also performed by this pipeline. Significantly differentially expressed genes were defined as those with at least 5 fragments per kilobase of exon per million reads mapped level of expression and an adjusted p value < 0.05 as determined by DESeq. Biologically relevant patterns of differentially expressed genes were identified by Gene Set Enrichment Analysis using Reactome as implemented at the web-based GEne SeT AnaLysis Toolkit (WebGestalt) [39,40,41]. Transcription factor target analysis was also performed with WebGestalt.
4.3. Real-Time PCR Validation
Reverse transcription of 2 µg RNA was performed using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher, Waltham, MA, USA). Real-time PCR was performed in triplicate with 40 ng of cDNA using the QuantStudio™ 7 Flex System (Thermo Fisher), the TaqMan™ Fast Advanced Master Mix (Thermo Fisher) and the following FAM-labeled TaqMan® Gene Expression primers: Cgref1 (Mm01217831_m1), Cthrc1 (Mm01163611_m1), Cyp2e1 (Mm00491127_m1), Gypa (Mm00494848_m1), Ifitm5 (Mm00804741_g1), Rerg (Mm02167721_m1), Slc13a5 (Mm01334459_m1) and Smpd3 (Mm00491359_m1). Results were normalized to Rpl27 (Mm01245874_g1). Gene expression was analyzed according to the delta Ct method and expressed as 2−∆Ct.
4.4. Micro-Computed Tomography (MicroCT)
MicroCT of the calvaria was performed using a SkyScan 1272 device (Bruker, Billerica, MA, USA). The voxel size was 8 μm. Scan parameters included a 0.40-degree increment angle, 3 frames averaged, a 66 kV and 142 mA X-ray source with a 0.5-mm Al filter to reduce beam-hardening artifacts. Cortical analysis was performed in the right parietal bone, considering an ROI of 4-mm length and 2-mm width, between the sagittal and coronal suture. Scans were quantified using the system’s analysis software (SkyScan CT Analyzer, Version 1.16.1.0).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22105290/s1, Figure S1: Real-time PCR results in calvaria RNA of 10-week-old female mice, Table S1: Genes upregulated in Jrt mice, Table S2: Genes downregulated in Jrt mice, Table S3: Genes upregulated in oim mice, Table S4: Genes downregulated in oim mice, Table S5: Genes upregulated in both Jrt and oim mice, Table S6: Genes downregulated in both Jrt and oim mice, Table S7. Results of transcription factor target analysis in male oim mice.
Author Contributions
Conceptualization, P.M. and F.R.; methodology, G.B.; formal analysis, F.R.; investigation, P.M., I.B.-D. and J.M.; writing—original draft preparation, F.R.; writing—review and editing, P.M., J.M. and G.B.; project administration, F.R.; funding acquisition, P.M. and F.R. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Réseau de recherche en santé buccodentaire et osseuse (RSBO, #67070); the Consortium québécois sur la découverte du médicament (CQDM); PreciThera, Inc. and the Shriners of North America.
Institutional Review Board Statement
All experiments were approved by the Animal Care Committee of McGill University (protocol 2011-5975; approved on 1 April 2018) and conformed to the ethical guidelines of the Canadian Council on Animal Care.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
References
- Forlino, A.; Marini, J.C. Osteogenesis imperfecta. Lancet 2016, 387, 1657–1671. [Google Scholar] [CrossRef]
- Bardai, G.; Moffatt, P.; Glorieux, F.H.; Rauch, F. DNA sequence analysis in 598 individuals with a clinical diagnosis of osteogenesis imperfecta: Diagnostic yield and mutation spectrum. Osteoporos. Int. J. Establ. Result Coop. Eur. Found. Osteoporos. Natl. Osteoporos. Found. USA 2016, 27, 3607–3613. [Google Scholar] [CrossRef] [PubMed]
- Rauch, F.; Travers, R.; Parfitt, A.M.; Glorieux, F.H. Static and dynamic bone histomorphometry in children with osteogenesis imperfecta. Bone 2000, 26, 581–589. [Google Scholar] [CrossRef]
- Rauch, F.; Lalic, L.; Roughley, P.; Glorieux, F.H. Relationship between genotype and skeletal phenotype in children and adolescents with osteogenesis imperfecta. J. Bone Miner. Res. Off. J. Am. Soc. Bone Mineral. Res. 2010, 25, 1367–1374. [Google Scholar] [CrossRef] [PubMed]
- Chipman, S.D.; Sweet, H.O.; McBride, D.J.; Davisson, M.T.; Marks, S.C.; Shuldiner, A.R.; Wenstrup, R.J.; Rowe, D.W.; Shapiro, J.R. Defective pro alpha 2(I) collagen synthesis in a recessive mutation in mice: A model of human osteogenesis imperfecta. Proc. Natl. Acad. Sci. USA 1993, 90, 1701–1705. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Guo, R.; Itoh, S.; Moreno, L.; Rosenthal, E.; Zappitelli, T.; Zirngibl, R.A.; Flenniken, A.; Cole, W.; Grynpas, M.; et al. First mouse model for combined osteogenesis imperfecta and Ehlers-Danlos syndrome. J. Bone Miner. Res. Off. J. Am. Soc. Bone Mineral. Res. 2014, 29, 1412–1423. [Google Scholar] [CrossRef]
- Zimmerman, S.M.; Dimori, M.; Heard-Lipsmeyer, M.E.; Morello, R. The osteocyte transcriptome is extensively dysregulated in mouse models of osteogenesis imperfecta. JBMR Plus 2019, 3, e10171. [Google Scholar] [CrossRef]
- Grafe, I.; Alexander, S.; Peterson, J.R.; Snider, T.N.; Levi, B.; Lee, B.; Mishina, Y. TGF-beta family signaling in mesenchymal differentiation. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef]
- Kim, H.N.; Iyer, S.; Ring, R.; Almeida, M. The role of FoxOs in bone health and disease. Curr. Top. Dev. Biol. 2018, 127, 149–163. [Google Scholar] [CrossRef]
- Roschger, A.; Roschger, P.; Keplingter, P.; Klaushofer, K.; Abdullah, S.; Kneissel, M.; Rauch, F. Effect of sclerostin antibody treatment in a mouse model of severe osteogenesis imperfecta. Bone 2014, 66, 182–188. [Google Scholar] [CrossRef]
- Tauer, J.T.; Abdullah, S.; Rauch, F. Effect of anti-TGF-beta treatment in a mouse model of severe osteogenesis imperfecta. J. Bone Miner. Res. Off. J. Am. Soc. Bone Mineral. Res. 2019, 34, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Cardinal, M.; Tys, J.; Roels, T.; Lafont, S.; Ominsky, M.S.; Devogelaer, J.P.; Chappard, D.; Mabilleau, G.; Ammann, P.; Nyssen-Behets, C.; et al. Sclerostin antibody reduces long bone fractures in the oim/oim model of osteogenesis imperfecta. Bone 2019, 124, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Grafe, I.; Yang, T.; Alexander, S.; Homan, E.P.; Lietman, C.; Jiang, M.M.; Bertin, T.; Munivez, E.; Chen, Y.; Dawson, B.; et al. Excessive transforming growth factor-beta signaling is a common mechanism in osteogenesis imperfecta. Nat. Med. 2014, 20, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.K.; Mueller, S. Alcohol and cancer: An overview with special emphasis on the role of acetaldehyde and cytochrome P450 2E1. Adv. Exp. Med. Biol. 2015, 815, 59–70. [Google Scholar] [CrossRef]
- Hong, A.R.; Kim, K.; Lee, J.Y.; Yang, J.Y.; Kim, J.H.; Shin, C.S.; Kim, S.W. Transformation of Mature Osteoblasts into Bone Lining Cells and RNA Sequencing-Based Transcriptome Profiling of Mouse Bone during Mechanical Unloading. Endocrinol. Metab. 2020, 35, 456–469. [Google Scholar] [CrossRef]
- Pathak, J.L.; Liu, L.; Zhu, Y.Q.; Bureik, M. Cytochrome P450 expression patterns in human osteoblasts during osteogenic differentiation with or without TNFalpha treatment. Biopharm. Drug Dispos. 2020, 41, 184–191. [Google Scholar] [CrossRef]
- Matthews, B.G.; Roeder, E.; Wang, X.; Aguila, H.L.; Lee, S.K.; Grcevic, D.; Kalajzic, I. Splenomegaly, myeloid lineage expansion and increased osteoclastogenesis in osteogenesis imperfecta murine. Bone 2017, 103, 1–11. [Google Scholar] [CrossRef]
- Moffatt, P.; Boraschi-Diaz, I.; Bardai, G.; Rauch, F. Muscle transcriptome in mouse models of osteogenesis imperfecta. Bone 2021, 148, 115940. [Google Scholar] [CrossRef]
- Selch, S.; Chafai, A.; Sticht, H.; Birkenfeld, A.L.; Fromm, M.F.; Konig, J. Analysis of naturally occurring mutations in the human uptake transporter NaCT important for bone and brain development and energy metabolism. Sci. Rep. 2018, 8, 11330. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.Y.; Rawal, A.; Schmidt-Rohr, K. Strongly bound citrate stabilizes the apatite nanocrystals in bone. Proc. Natl. Acad. Sci. USA 2010, 107, 22425–22429. [Google Scholar] [CrossRef]
- Fratzl, P.; Paris, O.; Klaushofer, K.; Landis, W.J. Bone mineralization in an osteogenesis imperfecta mouse model studied by small-angle x-ray scattering. J. Clin. Investig. 1996, 97, 396–402. [Google Scholar] [CrossRef][Green Version]
- Irizarry, A.R.; Yan, G.; Zeng, Q.; Lucchesi, J.; Hamang, M.J.; Ma, Y.L.; Rong, J.X. Defective enamel and bone development in sodium-dependent citrate transporter (NaCT) Slc13a5 deficient mice. PLoS ONE 2017, 12, e0175465. [Google Scholar] [CrossRef] [PubMed]
- Schossig, A.; Bloch-Zupan, A.; Lussi, A.; Wolf, N.I.; Raskin, S.; Cohen, M.; Giuliano, F.; Jurgens, J.; Krabichler, B.; Koolen, D.A.; et al. SLC13A5 is the second gene associated with Kohlschutter-Tonz syndrome. J. Med. Genet. 2017, 54, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, L.; Xiong, Y.; Li, J.; Wang, Y.; Shi, T.; Ma, D. The novel secretory protein Cgref1 inhibits the activation of AP-1 transcriptional activity and cell proliferation. Int. J. Biochem. Cell Biol. 2015, 65, 32–39. [Google Scholar] [CrossRef]
- Gharibi, B.; Ghuman, M.S.; Cama, G.; Rawlinson, S.C.F.; Grigoriadis, A.E.; Hughes, F.J. Site-specific differences in osteoblast phenotype, mechanical loading response and estrogen receptor-related gene expression. Mol. Cell. Endocrinol. 2018, 477, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, P.; Gaumond, M.H.; Salois, P.; Sellin, K.; Bessette, M.C.; Godin, E.; de Oliveira, P.T.; Atkins, G.J.; Nanci, A.; Thomas, G. Bril: A novel bone-specific modulator of mineralization. J. Bone Miner. Res. Off. J. Am. Soc. Bone Mineral. Res. 2008, 23, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Cho, T.J.; Lee, K.E.; Lee, S.K.; Song, S.J.; Kim, K.J.; Jeon, D.; Lee, G.; Kim, H.N.; Lee, H.R.; Eom, H.H.; et al. A single recurrent mutation in the 5’-UTR of IFITM5 causes osteogenesis imperfecta type V. Am. J. Hum. Genet. 2012, 91, 343–348. [Google Scholar] [CrossRef]
- Semler, O.; Garbes, L.; Keupp, K.; Swan, D.; Zimmermann, K.; Becker, J.; Iden, S.; Wirth, B.; Eysel, P.; Koerber, F.; et al. A mutation in the 5’-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. Am. J. Hum. Genet. 2012, 91, 349–357. [Google Scholar] [CrossRef]
- Greenblatt, M.B.; Ono, N.; Ayturk, U.M.; Debnath, S.; Lalani, S. The unmixing problem: A guide to applying single-cell RNA sequencing to bone. J. Bone Miner. Res. Off. J. Am. Soc. Bone Mineral. Res. 2019, 34, 1207–1219. [Google Scholar] [CrossRef]
- Aubin, I.; Adams, C.P.; Opsahl, S.; Septier, D.; Bishop, C.E.; Auge, N.; Salvayre, R.; Negre-Salvayre, A.; Goldberg, M.; Guenet, J.L.; et al. A deletion in the gene encoding sphingomyelin phosphodiesterase 3 (Smpd3) results in osteogenesis and dentinogenesis imperfecta in the mouse. Nat. Genet. 2005, 37, 803–805. [Google Scholar] [CrossRef]
- Jin, Y.R.; Stohn, J.P.; Wang, Q.; Nagano, K.; Baron, R.; Bouxsein, M.L.; Rosen, C.J.; Adarichev, V.A.; Lindner, V. Inhibition of osteoclast differentiation and collagen antibody-induced arthritis by CTHRC1. Bone 2017, 97, 153–167. [Google Scholar] [CrossRef]
- Cooling, L. Blood groups in infection and host susceptibility. Clin. Microbiol Rev. 2015, 28, 801–870. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.M.; Heard-Lipsmeyer, M.E.; Dimori, M.; Thostenson, J.D.; Mannen, E.M.; O’Brien, C.A.; Morello, R. Loss of RANKL in osteocytes dramatically increases cancellous bone mass in the osteogenesis imperfecta mouse (oim). Bone Rep. 2018, 9, 61–73. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- DeLuca, D.S.; Levin, J.Z.; Sivachenko, A.; Fennell, T.; Nazaire, M.D.; Williams, C.; Reich, M.; Winckler, W.; Getz, G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 2012, 28, 1530–1532. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Zhang, B.; Kirov, S.; Snoddy, J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33, W741–W748. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).