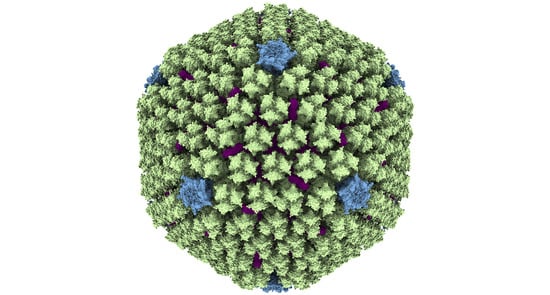
Adenovirus Structure: What Is New?
Abstract
1. Introduction
2. Components and Organization of the Adenovirus Virion
3. Structure of the Capsid Proteins
3.1. Hexon
3.2. Penton Base
3.3. Fibers
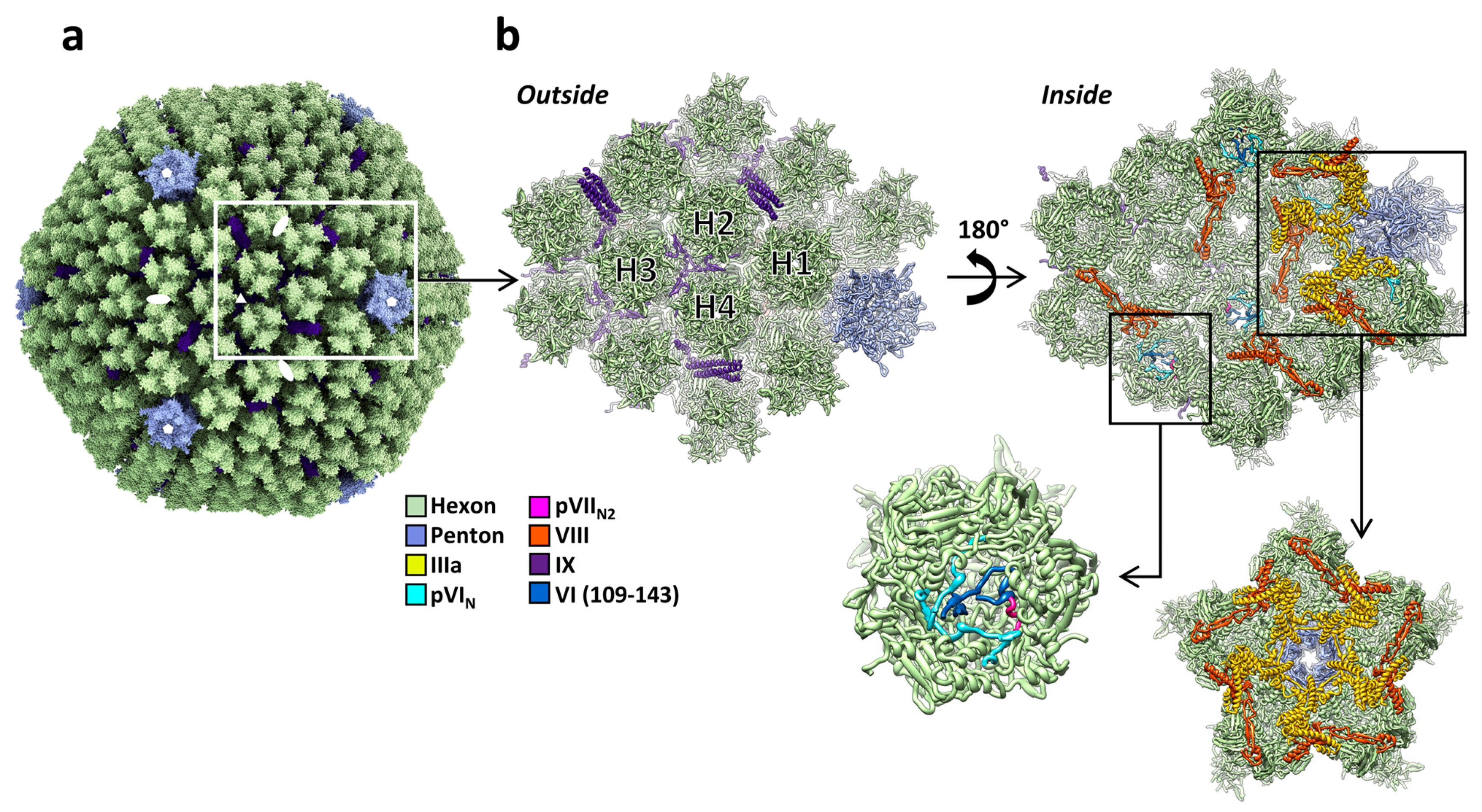
3.4. Protein IIIa
3.5. Protein VI
3.6. Protein VIII
3.7. Protein IX
4. Core Proteins
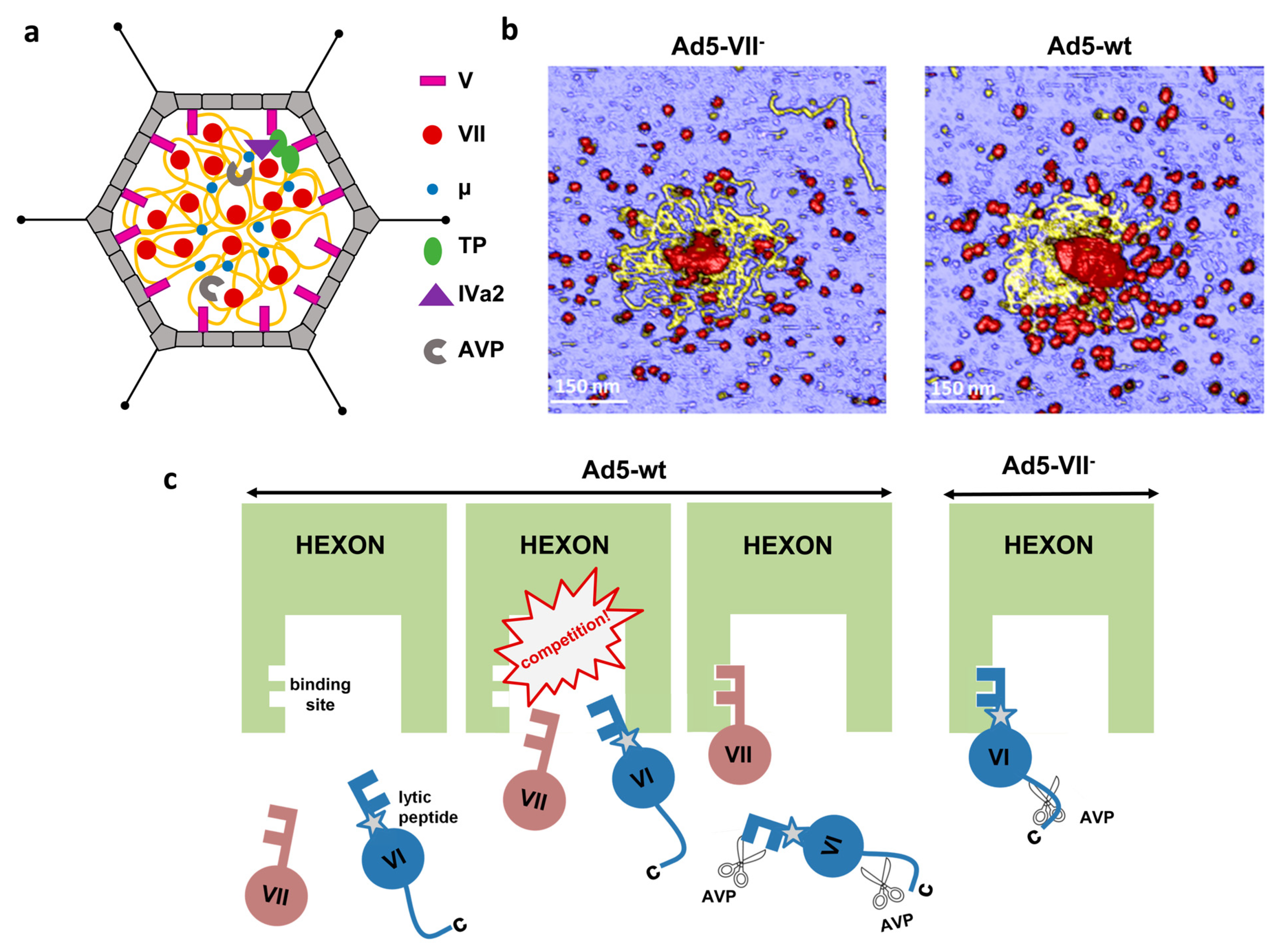
4.1. Protein V
4.2. Proteins VII and μ
4.3. Organization of the Adenovirus Core
4.4. Role of the Core Proteins in Adenovirus Assembly, Maturation and Entry
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rowe, W.P.; Huebner, R.J.; Gilmore, L.K.; Parrott, R.H.; Ward, T.G. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 1953, 84, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Harrach, B.; Tarjan, Z.L.; Benko, M. Adenoviruses across the animal kingdom: A walk in the zoo. FEBS Lett. 2019, 593, 3660–3673. [Google Scholar] [CrossRef] [PubMed]
- Berk, A.J. Adenoviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Wolters Kluwer/Lippincott Williams & Wilkins Health: Philadelphia, PA, USA, 2013; Volume 1, pp. 1704–1731. [Google Scholar]
- Kremer, E.J. Pros and cons of adenovirus-based SARS-CoV-2 vaccines. Mol. Ther. J. Am. Soc. Gene Ther. 2020, 28, 2303–2304. [Google Scholar] [CrossRef] [PubMed]
- Tumban, E. Lead SARS-CoV-2 candidate vaccines: Expectations from phase III trials and recommendations post-vaccine approval. Viruses 2020, 13, 54. [Google Scholar] [CrossRef] [PubMed]
- Hasanpourghadi, M.; Novikov, M.; Ertl, H.C.J. COVID-19 Vaccines Based on Adenovirus Vectors. Trends Biochem Sci 2021, 46, 429–430. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Jin, L.; Koh, S.B.; Atanasov, I.; Schein, S.; Wu, L.; Zhou, Z.H. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 2010, 329, 1038–1043. [Google Scholar] [CrossRef]
- Kundhavai Natchiar, S.; Venkataraman, S.; Mullen, T.M.; Nemerow, G.R.; Reddy, V.S. Revised crystal structure of human adenovirus reveals the limits on protein IX quasi-equivalence and on analyzing large macromolecular complexes. J. Mol. Biol. 2018, 430, 4132–4141. [Google Scholar] [CrossRef]
- San Martín, C. Latest insights on adenovirus structure and assembly. Viruses 2012, 4, 847–877. [Google Scholar] [CrossRef]
- Baker, A.T.; Greenshields-Watson, A.; Coughlan, L.; Davies, J.A.; Uusi-Kerttula, H.; Cole, D.K.; Rizkallah, P.J.; Parker, A.L. Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions. Nat. Commun. 2019, 10, 741. [Google Scholar] [CrossRef]
- Baker, A.T.; Davies, J.A.; Bates, E.A.; Moses, E.; Mundy, R.M.; Marlow, G.; Cole, D.K.; Bliss, C.M.; Rizkallah, P.J.; Parker, A.L. The fiber knob protein of human adenovirus type 49 mediates highly efficient and promiscuous infection of cancer cell lines using a novel cell entry mechanism. J. Virol. 2021, 95, e01849-20. [Google Scholar] [CrossRef]
- Persson, B.D.; John, L.; Rafie, K.; Strebl, M.; Frangsmyr, L.; Ballmann, M.Z.; Mindler, K.; Havenga, M.; Lemckert, A.; Stehle, T.; et al. Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Ballmann, M.Z.; Do, H.T.; Truong, H.N.; Benkő, M.; Harrach, B.; van Raaij, M.J. Crystal structure of raptor adenovirus 1 fibre head and role of the beta-hairpin in siadenovirus fibre head domains. Virol. J. 2016, 13, 106. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Berbís, M.Á.; Ballmann, M.Z.; Kilcoyne, M.; Menéndez, M.; Nguyen, T.H.; Joshi, L.; Cañada, F.J.; Jiménez-Barbero, J.; Benkő, M.; et al. Structure and sialyllactose binding of the carboxy-terminal head domain of the fibre from a siadenovirus, Turkey adenovirus 3. PLoS ONE 2015, 10, e0139339. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Vidovszky, M.Z.; Ballmann, M.Z.; Sanz-Gaitero, M.; Singh, A.K.; Harrach, B.; Benko, M.; Van Raaij, M.J. Crystal structure of the fibre head domain of bovine adenovirus 4, a ruminant atadenovirus. Virol. J. 2015. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wei, Q.; Liu, Y.; Feng, H.; Chen, Y.; Wang, Y.; Bai, Y.; Xing, G.; Deng, R.; Zhang, G. Unravelling the receptor binding property of egg drop syndrome virus (EDSV) from the crystal structure of EDSV fiber head. Int. J. Biol. Macromol. 2019, 139, 587–595. [Google Scholar] [CrossRef]
- Singh, A.K.; Menéndez-Conejero, R.; San Martín, C.; van Raaij, M.J. Crystal structure of the fibre head domain of the atadenovirus snake adenovirus 1. PLoS ONE 2014, 9, e114373. [Google Scholar] [CrossRef] [PubMed]
- Cupelli, K.; Müller, S.; Persson, B.D.; Jost, M.; Arnberg, N.; Stehle, T. Structure of adenovirus type 21 knob in complex with CD46 reveals key differences in receptor contacts among species B adenoviruses. J. Virol. 2010, 84, 3189–3200. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.T.; Mundy, R.M.; Davies, J.A.; Rizkallah, P.J.; Parker, A.L. Human adenovirus type 26 uses sialic acid-bearing glycans as a primary cell entry receptor. Sci. Adv. 2019, 5, eaax3567. [Google Scholar] [CrossRef] [PubMed]
- Vassal-Stermann, E.; Effantin, G.; Zubieta, C.; Burmeister, W.; Iseni, F.; Wang, H.; Lieber, A.; Schoehn, G.; Fender, P. CryoEM structure of adenovirus type 3 fibre with desmoglein 2 shows an unusual mode of receptor engagement. Nat. Commun. 2019, 10, 1181. [Google Scholar] [CrossRef] [PubMed]
- Hograindleur, M.-A.; Effantin, G.; Fenel, D.; Mas, C.; Lieber, A.; Schoehn, G.; Fender, P.; Vassal-Stermann, E. Binding mechanism elucidation of the acute respiratory disease causing agent adenovirus of serotype 7 to desmoglein-2. Viruses 2020, 12, 1075. [Google Scholar] [CrossRef]
- Caraballo, R.; Saleeb, M.; Bauer, J.; Liaci, A.M.; Chandra, N.; Storm, R.J.; Frängsmyr, L.; Qian, W.; Stehle, T.; Arnberg, N.; et al. Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells. Org. Biomol. Chem. 2015, 13, 9194–9205. [Google Scholar] [CrossRef] [PubMed]
- Lenman, A.; Liaci, A.M.; Liu, Y.; Ardahl, C.; Rajan, A.; Nilsson, E.; Bradford, W.; Kaeshammer, L.; Jones, M.S.; Frangsmyr, L.; et al. Human adenovirus 52 uses sialic acid-containing glycoproteins and the coxsackie and adenovirus receptor for binding to target cells. PLoS Pathog. 2015, 11, e1004657. [Google Scholar] [CrossRef]
- Lenman, A.; Liaci, A.M.; Liu, Y.; Frängsmyr, L.; Frank, M.; Blaum, B.S.; Chai, W.; Podgorski, I.I.; Harrach, B.; Benkő, M.; et al. Polysialic acid is a cellular receptor for human adenovirus 52. Proc. Natl. Acad. Sci. USA 2018, 115, E4264–E4273. [Google Scholar] [CrossRef]
- Singh, A.K.; Nguyen, T.H.; Vidovszky, M.Z.; Harrach, B.; Benkő, M.; Kirwan, A.; Joshi, L.; Kilcoyne, M.; Berbis, M.Á.; Cañada, F.J.; et al. Structure and N-acetylglucosamine binding of the distal domain of mouse adenovirus 2 fibre. J. Gen. Virol. 2018, 99, 1494–1508. [Google Scholar] [CrossRef]
- Menéndez-Conejero, R.; Nguyen, T.H.; Singh, A.K.; Condezo, G.N.; Marschang, R.E.; van Raaij, M.J.; San Martín, C. Structure of a reptilian adenovirus reveals a phage tailspike fold stabilizing a vertebrate virus capsid. Structure 2017, 25, 1562–1573.e1565. [Google Scholar] [CrossRef] [PubMed]
- Rafie, K.; Lenman, A.; Fuchs, J.; Rajan, A.; Arnberg, N.; Carlson, L.-A. The structure of enteric human adenovirus 41—A leading cause of diarrhea in children. Sci. Adv. 2021, 7, eabe0974. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Wu, L.; Sun, R.; Zhou, Z.H. Atomic structures of minor proteins VI and VII in human adenovirus. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Veesler, D.; Campbell, M.G.; Barry, M.E.; Asturias, F.J.; Barry, M.A.; Reddy, V.S. Cryo-EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses. Sci. Adv. 2017, 3, e1602670. [Google Scholar] [CrossRef]
- Pérez-Illana, M.; Martinez, M.; Condezo, G.N.; Hernando-Pérez, M.; Mangroo, C.; Brown, M.; Marabini, R.; San Martín, C. Cryo-EM structure of enteric adenovirus HAdV-F41 highlights structural variations among human adenoviruses. Sci. Adv. 2021, 7, eabd9421. [Google Scholar] [CrossRef]
- Marabini, R.; Condezo, G.N.; Krupovic, M.; Menéndez-Conejero, R.; Gómez-Blanco, J.; San Martín, C. Near atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins. Sci. Adv. 2021, 7, eabe6008. [Google Scholar] [CrossRef]
- Cheng, L.; Huang, X.; Li, X.; Xiong, W.; Sun, W.; Yang, C.; Zhang, K.; Wang, Y.; Liu, H.; Huang, X.; et al. Cryo-EM structures of two bovine adenovirus type 3 intermediates. Virology 2014, 450–451, 174–181. [Google Scholar] [CrossRef]
- Davison, A.J.; Wright, K.M.; Harrach, B. DNA sequence of frog adenovirus. J. Gen. Virol. 2000, 81, 2431–2439. [Google Scholar] [CrossRef]
- Doszpoly, A.; Harrach, B.; LaPatra, S.; Benko, M. Unconventional gene arrangement and content revealed by full genome analysis of the white sturgeon adenovirus, the single member of the genus Ichtadenovirus. Infect. Genet. Evol. 2019, 75, 103976. [Google Scholar] [CrossRef]
- Davison, A.J.; Benkő, M.; Harrach, B. Genetic content and evolution of adenoviruses. J. Gen. Virol. 2003, 84, 2895–2908. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Marion, S.; San Martín, C.; Siber, A. Role of condensing particles in polymer confinement: A model for virus-packed “minichromosomes”. Biophys. J. 2017, 113, 1643–1653. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Berná, A.J.; Marion, S.; Chichón, F.J.; Fernández, J.J.; Winkler, D.C.; Carrascosa, J.L.; Steven, A.C.; Šiber, A.; San Martín, C. Distribution of DNA-condensing protein complexes in the adenovirus core. Nucleic Acids Res. 2015, 43, 4274–4283. [Google Scholar] [CrossRef] [PubMed]
- Hoeben, R.C.; Uil, T.G. Adenovirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a013003. [Google Scholar] [CrossRef] [PubMed]
- Mangel, W.F.; San Martín, C. Structure, function and dynamics in adenovirus maturation. Viruses 2014, 6, 4536–4570. [Google Scholar] [CrossRef]
- Ahi, Y.S.; Mittal, S.K. Components of adenovirus genome packaging. Front. Microbiol. 2016, 7, 1503. [Google Scholar] [CrossRef] [PubMed]
- Burnett, R.M.; Grütter, M.G.; White, J.L. The structure of the adenovirus capsid. I. An envelope model of hexon at 6 A resolution. J. Mol. Biol. 1985, 185, 105–123. [Google Scholar] [CrossRef]
- Condezo, G.N.; Martín-González, N.; Pérez-Illana, M.; Hernando-Pérez, M.; Gallardo, J.; San Martín, C. Adenoviruses (Adenoviridae) and their structural relatives. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 329–344. [Google Scholar] [CrossRef]
- Crawford-Miksza, L.; Schnurr, D.P. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 1996, 70, 1836–1844. [Google Scholar] [CrossRef] [PubMed]
- Caspar, D.L.D.; Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol 1962, 27, 1–24. [Google Scholar] [CrossRef]
- Abrishami, V.; Ilca, S.L.; Gomez-Blanco, J.; Rissanen, I.; de la Rosa-Trevin, J.M.; Reddy, V.S.; Carazo, J.M.; Huiskonen, J.T. Localized reconstruction in Scipion expedites the analysis of symmetry mismatches in cryo-EM data. Prog. Biophys. Mol. Biol. 2021, 160, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Ebner, K.; Pinsker, W.; Lion, T. Comparative sequence analysis of the hexon gene in the entire spectrum of human adenovirus serotypes: Phylogenetic, taxonomic, and clinical implications. J. Virol. 2005, 79, 12635–12642. [Google Scholar] [CrossRef]
- Diaz, K.; Hu, C.T.; Sul, Y.; Bromme, B.A.; Myers, N.D.; Skorohodova, K.V.; Gounder, A.P.; Smith, J.G. Defensin-driven viral evolution. PLoS Pathog. 2020, 16, e1009018. [Google Scholar] [CrossRef]
- Alba, R.; Bradshaw, A.C.; Parker, A.L.; Bhella, D.; Waddington, S.N.; Nicklin, S.A.; van Rooijen, N.; Custers, J.; Goudsmit, J.; Barouch, D.H.; et al. Identification of coagulation factor (F)X binding sites on the adenovirus serotype 5 hexon: Effect of mutagenesis on FX interactions and gene transfer. Blood 2009, 114, 965–971. [Google Scholar] [CrossRef] [PubMed]
- Zubieta, C.; Schoehn, G.; Chroboczek, J.; Cusack, S. The structure of the human adenovirus 2 penton. Mol. Cell 2005, 17, 121–135. [Google Scholar] [CrossRef]
- Albinsson, B.; Kidd, A.H. Adenovirus type 41 lacks an RGD alpha(v)-integrin binding motif on the penton base and undergoes delayed uptake in A549 cells. Virus Res. 1999, 64, 125–136. [Google Scholar] [CrossRef]
- Rajan, A.; Persson, B.D.; Frangsmyr, L.; Olofsson, A.; Sandblad, L.; Heino, J.; Takada, Y.; Mould, A.P.; Schnapp, L.M.; Gall, J.; et al. Enteric species F human adenoviruses use laminin-binding integrins as co-receptors for infection of Ht-29 cells. Sci. Rep. 2018, 8, 10019. [Google Scholar] [CrossRef]
- Madisch, I.; Hofmayer, S.; Moritz, C.; Grintzalis, A.; Hainmueller, J.; Pring-Akerblom, P.; Heim, A. Phylogenetic analysis and structural predictions of human adenovirus penton proteins as a basis for tissue-specific adenovirus vector design. J. Virol. 2007, 81, 8270–8281. [Google Scholar] [CrossRef]
- Besson, S.; Vragniau, C.; Vassal-Stermann, E.; Dagher, M.C.; Fender, P. The adenovirus dodecahedron: Beyond the platonic story. Viruses 2020, 12, 718. [Google Scholar] [CrossRef] [PubMed]
- Stasiak, A.C.; Stehle, T. Human adenovirus binding to host cell receptors: A structural view. Med. Microbiol. Immunol. 2020, 209, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Nicklin, S.A.; Wu, E.; Nemerow, G.R.; Baker, A.H. The influence of adenovirus fiber structure and function on vector development for gene therapy. Mol. Ther. J. Am. Soc. Gene Ther. 2005, 12, 384–393. [Google Scholar] [CrossRef]
- Van Raaij, M.J.; Mitraki, A.; Lavigne, G.; Cusack, S. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 1999, 401, 935–938. [Google Scholar] [CrossRef]
- Wu, E.; Pache, L.; Von Seggern, D.J.; Mullen, T.M.; Mikyas, Y.; Stewart, P.L.; Nemerow, G.R. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J Virol 2003, 77, 7225–7235. [Google Scholar] [CrossRef]
- Li, J.; Lad, S.; Yang, G.; Luo, Y.; Iacobelli-Martinez, M.; Primus, F.J.; Reisfeld, R.A.; Li, E. Adenovirus Fiber Shaft Contains a Trimerization Element That Supports Peptide Fusion for Targeted Gene Delivery. Journal of Virology 2006, 80, 12324. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, L.; Zhou, Z.H. Model of the trimeric fiber and its interactions with the pentameric penton base of human adenovirus by cryo-electron microscopy. J. Mol. Biol. 2011, 406, 764–774. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Dong, X.; Wu, X.; Wen, B.; Ji, G.; Cheng, L.; Liu, H. Conserved fiber-penton base interaction revealed by nearly atomic resolution cryo-electron microscopy of the structure of adenovirus provides insight into receptor interaction. J. Virol. 2012, 86, 12322–12329. [Google Scholar] [CrossRef]
- Stass, R.; Ilca, S.L.; Huiskonen, J.T. Beyond structures of highly symmetric purified viral capsids by cryo-EM. Curr. Opin. Struct. Biol. 2018, 52, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Kidd, A.H.; Chroboczek, J.; Cusack, S.; Ruigrok, R.W. Adenovirus type 40 virions contain two distinct fibers. Virology 1993, 192, 73–84. [Google Scholar] [CrossRef]
- Hess, M.; Cuzange, A.; Ruigrok, R.W.H.; Chroboczek, J.; Jacrot, B. The avian adenovirus penton: Two fibres and one base. J. Mol. Biol. 1995, 252, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Pénzes, J.J.; Menéndez-Conejero, R.; Condezo, G.N.; Ball, I.; Papp, T.; Doszpoly, A.; Paradela, A.; Pérez-Berná, A.J.; López-Sanz, M.; Nguyen, T.H.; et al. Molecular characterization of a lizard adenovirus reveals the first atadenovirus with two fiber genes and the first adenovirus with either one short or three long fibers per penton. J. Virol. 2014, 88, 11304–11314. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.L.; Fuller, S.D.; Burnett, R.M. Difference imaging of adenovirus: Bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993, 12, 2589–2599. [Google Scholar] [CrossRef]
- Saban, S.D.; Silvestry, M.; Nemerow, G.R.; Stewart, P.L. Visualization of alpha-helices in a 6-angstrom resolution cryoelectron microscopy structure of adenovirus allows refinement of capsid protein assignments. J. Virol. 2006, 80, 12049–12059. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Nemerow, G.R. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11715–11720. [Google Scholar] [CrossRef]
- Greber, U.F.; Flatt, J.W. Adenovirus entry: From infection to immunity. Annu. Rev. Virol. 2019, 6, 177–197. [Google Scholar] [CrossRef]
- Hernando-Pérez, M.; Martín-González, N.; Pérez-Illana, M.; Suomalainen, M.; Condezo, G.N.; Ostapchuk, P.; Gallardo, J.; Menéndez, M.; Greber, U.F.; Hearing, P.; et al. Dynamic competition for hexon binding between core protein VII and lytic protein VI promotes adenovirus maturation and entry. Proc. Natl. Acad. Sci. USA 2020, 117, 13699–13707. [Google Scholar] [CrossRef]
- Benevento, M.; Di Palma, S.; Snijder, J.; Moyer, C.L.; Reddy, V.S.; Nemerow, G.R.; Heck, A.J. Adenovirus composition, proteolysis, and disassembly studied by in-depth qualitative and quantitative proteomics. J. Biol. Chem. 2014, 289, 11421–11430. [Google Scholar] [CrossRef]
- Reddy, V.S. The role of hexon protein as a molecular mold in patterning the protein IX organization in human adenoviruses. J. Mol. Biol. 2017, 429, 2747–2751. [Google Scholar] [CrossRef] [PubMed]
- Vellinga, J.; van den Wollenberg, D.J.; van der Heijdt, S.; Rabelink, M.J.; Hoeben, R.C. The coiled-coil domain of the adenovirus type 5 protein IX is dispensable for capsid incorporation and thermostability. J. Virol. 2005, 79, 3206–3210. [Google Scholar] [CrossRef]
- Schoehn, G.; El Bakkouri, M.; Fabry, C.M.; Billet, O.; Estrozi, L.F.; Le, L.; Curiel, D.T.; Kajava, A.V.; Ruigrok, R.W.; Kremer, E.J. Three-dimensional structure of canine adenovirus serotype 2 capsid. J. Virol. 2008, 82, 3192–3203. [Google Scholar] [CrossRef] [PubMed]
- Hackenbrack, N.; Rogers, M.B.; Ashley, R.E.; Keel, M.K.; Kubiski, S.V.; Bryan, J.A.; Ghedin, E.; Holmes, E.C.; Hafenstein, S.L.; Allison, A.B. Evolution and cryo-EM capsid structure of a North American bat adenovirus and its relationship to other mastadenoviruses. J. Virol. 2016. [Google Scholar] [CrossRef]
- Matteson, N.L.; Barry, M.A.; Reddy, V.S. Structure-based assessment of protein-protein interactions and accessibility of protein IX in adenoviruses with implications for antigen display. Virology 2018, 516, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Pantelic, R.S.; Lockett, L.J.; Rothnagel, R.; Hankamer, B.; Both, G.W. Cryoelectron microscopy map of Atadenovirus reveals cross-genus structural differences from human adenovirus. J. Virol. 2008, 82, 7346–7356. [Google Scholar] [CrossRef]
- Gorman, J.J.; Wallis, T.P.; Whelan, D.A.; Shaw, J.; Both, G.W. LH3, a “homologue” of the mastadenoviral E1B 55-kDa protein is a structural protein of atadenoviruses. Virology 2005, 342, 159–166. [Google Scholar] [CrossRef][Green Version]
- Van Oostrum, J.; Burnett, R.M. Molecular composition of the adenovirus type 2 virion. J. Virol. 1985, 56, 439–448. [Google Scholar] [CrossRef]
- Chatterjee, P.K.; Vayda, M.E.; Flint, S.J. Interactions among the three adenovirus core proteins. J. Virol. 1985, 55, 379–386. [Google Scholar] [CrossRef]
- Pérez-Vargas, J.; Vaughan, R.C.; Houser, C.; Hastie, K.M.; Kao, C.C.; Nemerow, G.R. Isolation and characterization of the DNA and protein binding activities of adenovirus core protein V. J. Virol. 2014, 88, 9287–9296. [Google Scholar] [CrossRef]
- Puntener, D.; Engelke, M.F.; Ruzsics, Z.; Strunze, S.; Wilhelm, C.; Greber, U.F. Stepwise loss of fluorescent core protein V from human adenovirus during entry into cells. J. Virol. 2011, 85, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Ugai, H.; Borovjagin, A.V.; Le, L.P.; Wang, M.; Curiel, D.T. Thermostability/infectivity defect caused by deletion of the core protein V gene in human adenovirus type 5 is rescued by thermo-selectable mutations in the core protein X precursor. J. Mol. Biol. 2007, 366, 1142–1160. [Google Scholar] [CrossRef]
- Hosokawa, K.; Sung, M.T. Isolation and characterization of an extremely basic protein from adenovirus type 5. J. Virol. 1976, 17, 924–934. [Google Scholar] [CrossRef]
- Johnson, J.S.; Osheim, Y.N.; Xue, Y.; Emanuel, M.R.; Lewis, P.W.; Bankovich, A.; Beyer, A.L.; Engel, D.A. Adenovirus protein VII condenses DNA, represses transcription, and associates with transcriptional activator E1A. J. Virol. 2004, 78, 6459–6468. [Google Scholar] [CrossRef]
- Anderson, C.W.; Young, M.E.; Flint, S.J. Characterization of the adenovirus 2 virion protein, mu. Virology 1989, 172, 506–512. [Google Scholar] [CrossRef]
- Vayda, M.E.; Rogers, A.E.; Flint, S.J. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 1983, 11, 441–460. [Google Scholar] [CrossRef]
- Mirza, M.A.; Weber, J. Structure of adenovirus chromatin. Biochim. Biophys. Acta 1982, 696, 76–86. [Google Scholar] [CrossRef]
- Pérez-Berná, A.J.; Marabini, R.; Scheres, S.H.W.; Menéndez-Conejero, R.; Dmitriev, I.P.; Curiel, D.T.; Mangel, W.F.; Flint, S.J.; San Martín, C. Structure and uncoating of immature adenovirus. J. Mol. Biol. 2009, 392, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Martín-González, N.; Hernando-Pérez, M.; Condezo, G.N.; Pérez-Illana, M.; Šiber, A.; Reguera, D.; Ostapchuk, P.; Hearing, P.; San Martín, C.; de Pablo, P.J. Adenovirus major core protein condenses DNA in clusters and bundles, modulating genome release and capsid internal pressure. Nucleic Acids Res. 2019, 47, 9231–9242. [Google Scholar] [CrossRef]
- Ostapchuk, P.; Suomalainen, M.; Zheng, Y.; Boucke, K.; Greber, U.F.; Hearing, P. The adenovirus major core protein VII is dispensable for virion assembly but is essential for lytic infection. PLoS Pathog. 2017, 13, e1006455. [Google Scholar] [CrossRef]
- Ortega-Esteban, A.; Condezo, G.N.; Pérez-Berná, A.J.; Chillón, M.; Flint, S.J.; Reguera, D.; San Martín, C.; de Pablo, P.J. Mechanics of viral chromatin reveals the pressurization of human adenovirus. ACS Nano 2015, 9, 10826–10833. [Google Scholar] [CrossRef]
- Ortega-Esteban, A.; Pérez-Berná, A.J.; Menéndez-Conejero, R.; Flint, S.J.; San Martín, C.; de Pablo, P.J. Monitoring dynamics of human adenovirus disassembly induced by mechanical fatigue. Sci. Rep. 2013, 3, 1434. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Berná, A.J.; Ortega-Esteban, A.; Menéndez-Conejero, R.; Winkler, D.C.; Menéndez, M.; Steven, A.C.; Flint, S.J.; de Pablo, P.J.; San Martín, C. The role of capsid maturation on adenovirus priming for sequential uncoating. J. Biol. Chem. 2012, 287, 31582–31595. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Esteban, A.; Bodensiek, K.; San Martín, C.; Suomalainen, M.; Greber, U.F.; de Pablo, P.J.; Schaap, I.A. Fluorescence tracking of genome release during mechanical unpacking of single viruses. ACS Nano 2015, 9, 10571–10579. [Google Scholar] [CrossRef] [PubMed]
- Condezo, G.N.; Marabini, R.; Ayora, S.; Carazo, J.M.; Alba, R.; Chillón, M.; San Martín, C. Structures of adenovirus incomplete particles clarify capsid architecture and show maturation changes of packaging protein L1 52/55k. J. Virol. 2015, 89, 9653–9664. [Google Scholar] [CrossRef]
- Condezo, G.N.; San Martín, C. Localization of adenovirus morphogenesis players, together with visualization of assembly intermediates and failed products, favor a model where assembly and packaging occur concurrently at the periphery of the replication center. PLoS Pathog. 2017, 13, e1006320. [Google Scholar] [CrossRef]
- Vassal-Stermann, E.; Mottet, M.; Ducournau, C.; Iseni, F.; Vragniau, C.; Wang, H.; Zubieta, C.; Lieber, A.; Fender, P. Mapping of Adenovirus of serotype 3 fibre interaction to desmoglein 2 revealed a novel ‘non-classical’ mechanism of viral receptor engagement. Sci. Rep. 2018, 8, 8381. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, M.A.F.V.; de Vries, A.A.F. Adenovirus: From foe to friend. Rev. Med. Virol. 2006, 16, 167–186. [Google Scholar] [CrossRef] [PubMed]

| Specimen | Virus Type 1 | Genus | PDB 2 ID | Reference |
|---|---|---|---|---|
| Fiber knob | HAdV-C5 | Mastadenovirus | 6HCN | [10] |
| Fiber knob | HAdV-D10 | Mastadenovirus | 6QPM | unpublished |
| Fiber knob | HAdV-D30 | Mastadenovirus | 6STU | [11] |
| Fiber knob | HAdV-D48 | Mastadenovirus | 6FJQ | [10] |
| Fiber knob | HAdV-D49 | Mastadenovirus | 6QPN | [11] |
| Fiber knob | HAdV-D56 | Mastadenovirus | 7AJP | [12] |
| Fiber knob | RAdV-1 | Siadenovirus | 5FJL | [13] |
| Fiber knob | TAdV-3 | Siadenovirus | 4CW8 | [14] |
| Fiber knob | BAdV-4 | Atadenovirus | 4UE0 | [15] |
| Fiber knob | DAdV-1 | Atadenovirus | 6ITX | [16] |
| Fiber knob | SnAdV-1 | Atadenovirus | 4D0V | [17] |
| Fiber knob with CD46 | HAdV-B21 | Mastadenovirus | 3L89 | [18] |
| Fiber knob with sialic acid | HAdV-D26 | Mastadenovirus | 6QU8 | [19] |
| Fiber knob with desmoglein 2 | HAdV-B3 | Mastadenovirus | 6QNT | [20] |
| Fiber knob with desmoglein 2 | HAdV-B7 | Mastadenovirus | 7AGF | [21] |
| Fiber knob with trivalent sialic acid inhibitor | HAdV-D37 | Mastadenovirus | 4XQA | [22] |
| Short fiber knob with 2-O-Methyl-5-N-Acetylneuraminic acid | HAdV-G52 | Mastadenovirus | 4XL8 | [23] |
| Short fiber knob with α-(2,8)-trisialic acid | HAdV-G52 | Mastadenovirus | 6G47 | [24] |
| Fiber knob and shaft with N-acetylglucosamine | MAdV-2 | Mastadenovirus | 5NC1 | [25] |
| Protein LH3 | SnAdV-1 | Atadenovirus | 5G5O | [26] |
| Penton base | HAdV-F41 | Mastadenovirus | 6Z7Q | [27] |
| Viral particle | HAdV-C5 | Mastadenovirus | 6B1T | [28] |
| Viral particle | HAdV-C5 | Mastadenovirus | 6CGV | [8] |
| Viral particle | HAdV-D26 | Mastadenovirus | 5TX1 | [29] |
| Viral particle | HAdV-F41 | Mastadenovirus | 6YBA | [30] |
| Viral particle | HAdV-F41 | Mastadenovirus | 6Z7N | [27] |
| Viral particle | LAdV-2 | Atadenovirus | 6QI5 | [31] |
| Viral particle | BAdV-3 | Mastadenovirus | 3ZIF | [32] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallardo, J.; Pérez-Illana, M.; Martín-González, N.; San Martín, C. Adenovirus Structure: What Is New? Int. J. Mol. Sci. 2021, 22, 5240. https://doi.org/10.3390/ijms22105240
Gallardo J, Pérez-Illana M, Martín-González N, San Martín C. Adenovirus Structure: What Is New? International Journal of Molecular Sciences. 2021; 22(10):5240. https://doi.org/10.3390/ijms22105240
Chicago/Turabian StyleGallardo, José, Marta Pérez-Illana, Natalia Martín-González, and Carmen San Martín. 2021. "Adenovirus Structure: What Is New?" International Journal of Molecular Sciences 22, no. 10: 5240. https://doi.org/10.3390/ijms22105240
APA StyleGallardo, J., Pérez-Illana, M., Martín-González, N., & San Martín, C. (2021). Adenovirus Structure: What Is New? International Journal of Molecular Sciences, 22(10), 5240. https://doi.org/10.3390/ijms22105240