MD-TSPC4: Computational Method for Predicting the Thermal Stability of I-Motif
Abstract
1. Introduction
2. Results
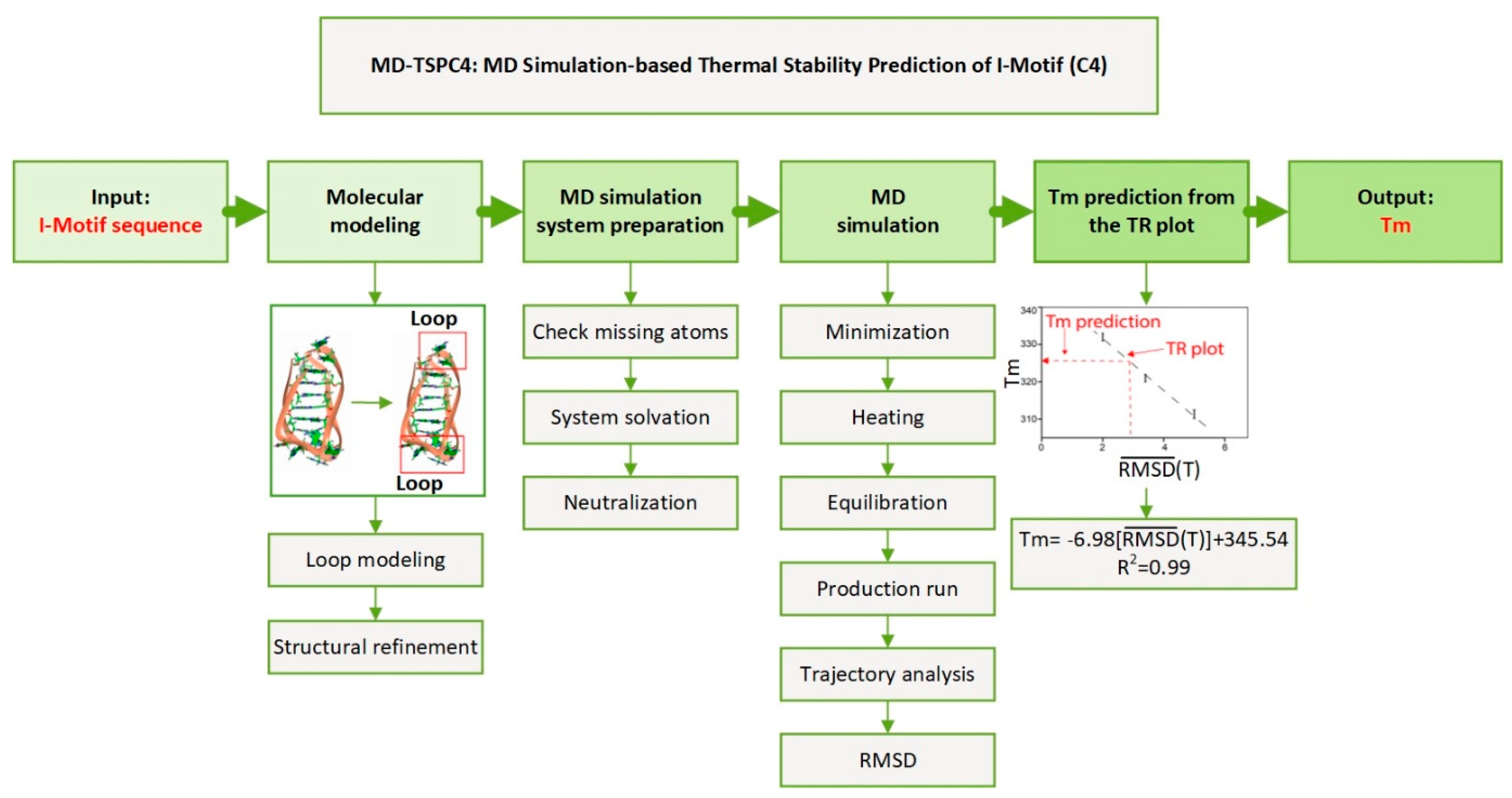
2.1. Research Hypothesis and Molecular Modeling
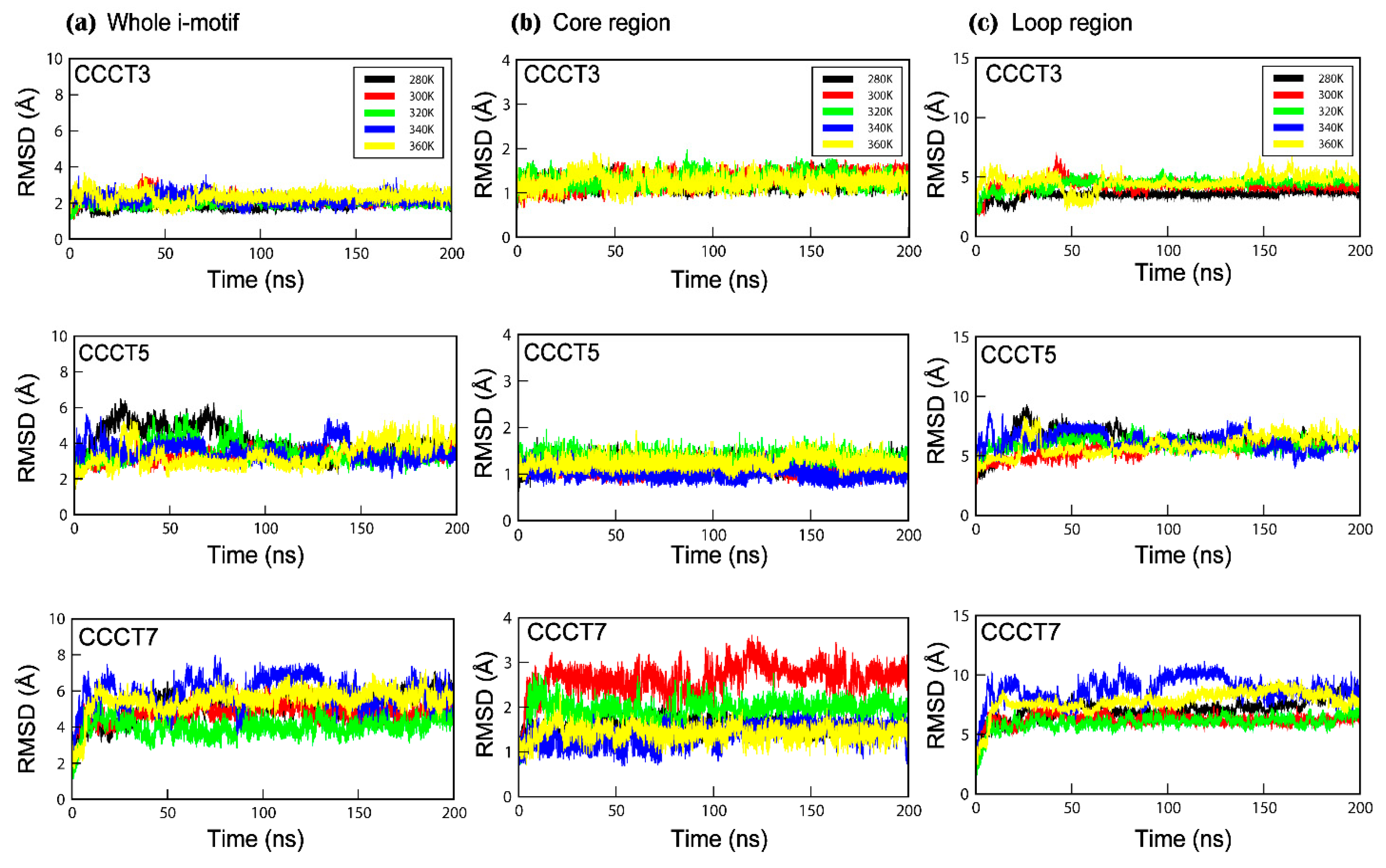
2.2. MD Simulation and Trajectory Analysis
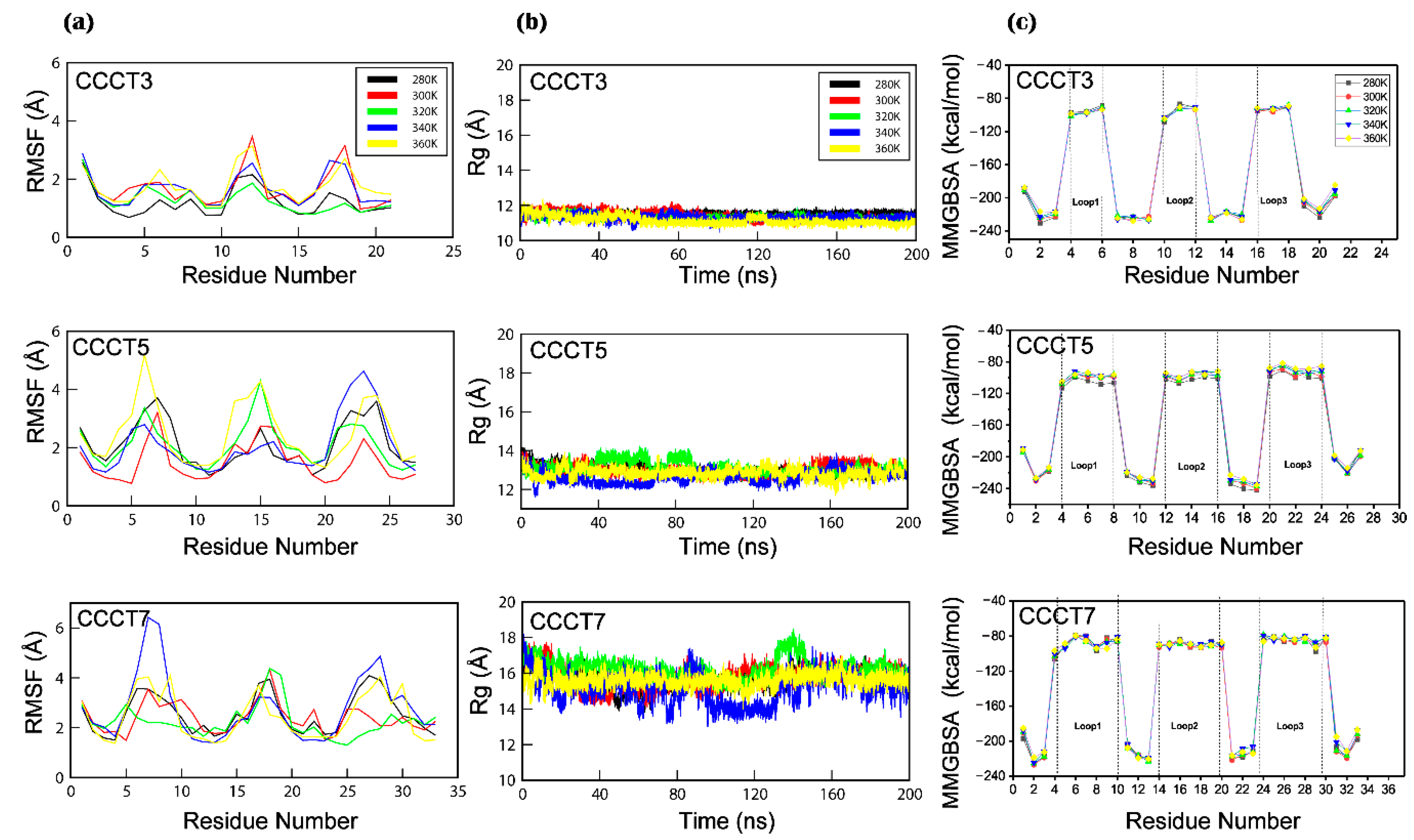
2.3. Conformational Flexibility
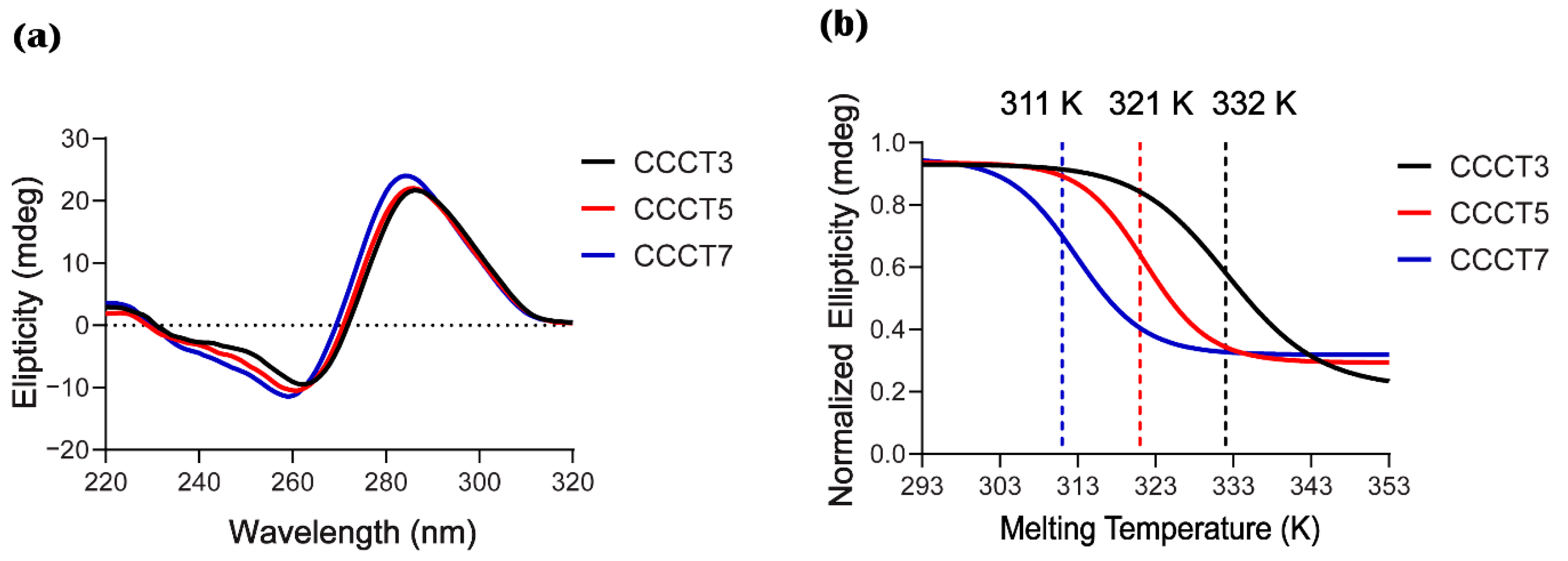
2.4. Experimental Measurement of Thermal Stability
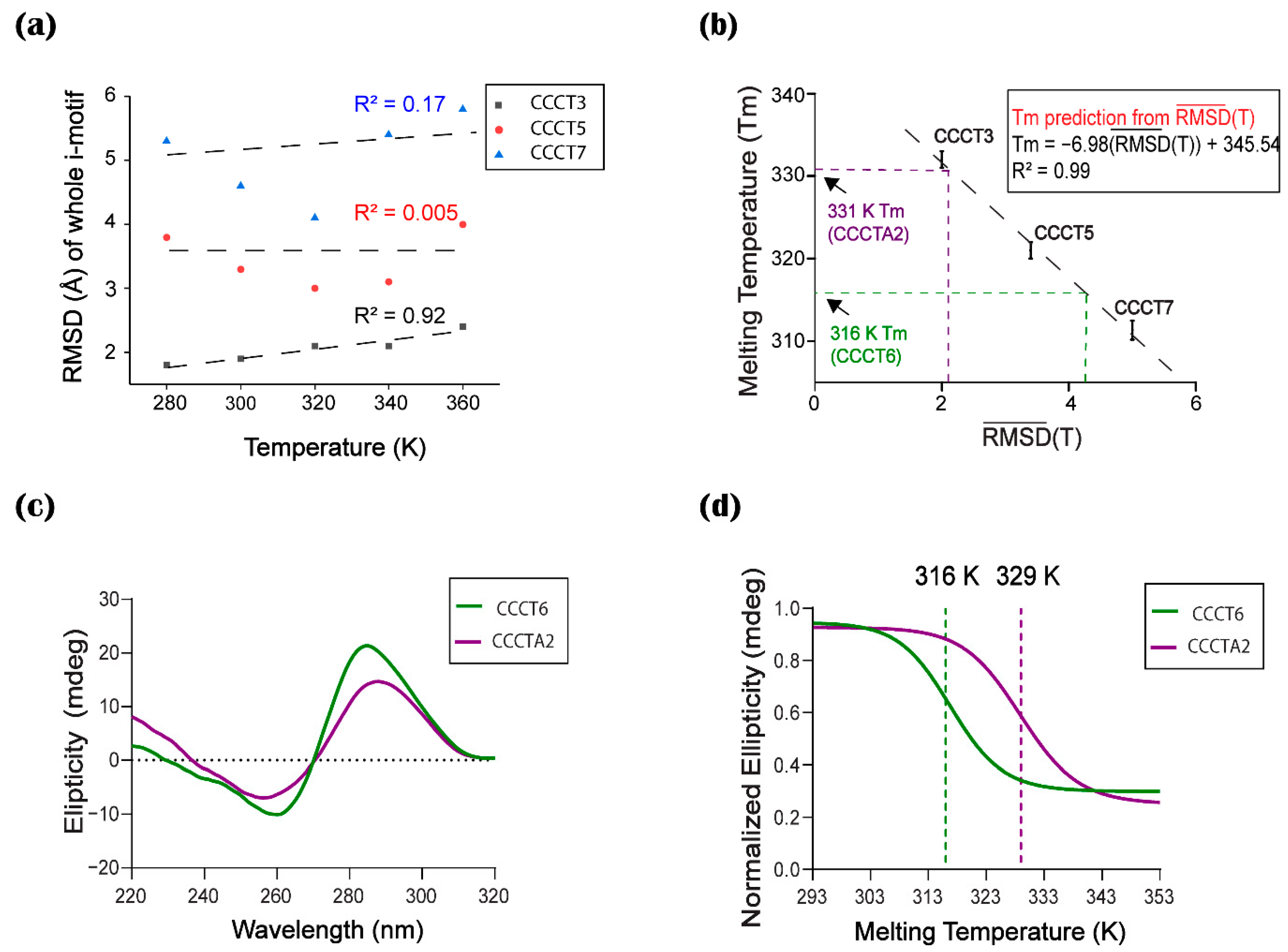
2.5. Correlation between Thermal Stability and Conformational Flexibility
2.6. Validation of the Computational Prediction
2.7. MD Simulation Based Thermal Stability Prediction of I-Motif (C4): MD-TSPC4
3. Discussion
4. Materials and Methods
4.1. Model I-Motif Sequences
4.2. Molecular Modeling
4.3. MD Simulations
4.4. Trajectory Analysis
4.5. Per Residue Binding Free Energy Calculation
4.6. Oligonucleotide Preparation
4.7. CD Spectroscopy
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nat. Cell Biol. 1988, 334, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Gehring, K.; Leroy, J.-L.; Guéron, M. A tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nat. Cell Biol. 1993, 363, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Felsenfeld, G.; Rich, A. Studies on the formation of two- and three-stranded polyribonucleotides. Biochim. Biophys. Acta (BBA) Bioenerg. 1957, 26, 457–468. [Google Scholar] [CrossRef]
- Panayotatos, N.; Wells, R.D. Cruciform structures in supercoiled DNA. Nature 1981, 289, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Subramani, V.; Kim, K.K. Z-DNA in the genome: From structure to disease. Biophys. Rev. 2019, 11, 383–387. [Google Scholar] [CrossRef]
- Dai, X.; Greizerstein, M.B.; Nadas-Chinni, K.; Rothman-Denes, L.B. Supercoil-induced extrusion of a regulatory DNA hairpin. Proc. Natl. Acad. Sci. USA 1997, 94, 2174–2179. [Google Scholar] [CrossRef]
- Thompson, C.A.H.; Wong, J.M.Y. Non-canonical Functions of Telomerase Reverse Transcriptase: Emerging Roles and Biological Relevance. Curr. Top. Med. Chem. 2020, 20, 498–507. [Google Scholar] [CrossRef]
- Tateishi-Karimata, H.; Sugimoto, N. Chemical biology of non-canonical structures of nucleic acids for therapeutic applications. Chem. Commun. 2020, 56, 2379–2390. [Google Scholar] [CrossRef]
- Huppert, J.L.; Bugaut, A.; Kumari, S.; Balasubramanian, S. G-quadruplexes: The beginning and end of UTRs. Nucleic Acids Res. 2008, 36, 6260–6268. [Google Scholar] [CrossRef]
- Beaudoin, J.D.; Perreault, J.P. 5’-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010, 38, 7022–7036. [Google Scholar] [CrossRef] [PubMed]
- Endoh, T.; Kawasaki, Y.; Sugimoto, N. Suppression of gene expression by G-quadruplexes in open reading frames depends on G-quadruplex stability. Angew. Chem. Int. Ed Engl. 2013, 52, 5522–5526. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [PubMed]
- Brooks, T.A.; Hurley, L.H. Targeting MYC Expression through G-Quadruplexes. Genes Cancer 2010, 1, 641–649. [Google Scholar] [CrossRef]
- Guo, K.; Gokhale, V.; Hurley, L.H.; Sun, D. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 2008, 36, 4598–4608. [Google Scholar] [CrossRef]
- Sengar, A.; Vandana, J.J.; Chambers, V.S.; Di Antonio, M.; Winnerdy, F.R.; Balasubramanian, S.; Phan, A.T. Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter. Nucleic Acids Res. 2019, 47, 1564–1572. [Google Scholar] [CrossRef]
- Amato, J.; Pagano, A.; Capasso, D.; Di Gaetano, S.; Giustiniano, M.; Novellino, E.; Randazzo, A.; Pagano, B. Targeting the BCL2 Gene Promoter G-Quadruplex with a New Class of Furopyridazinone-Based Molecules. ChemMedChem 2018, 13, 406–410. [Google Scholar] [CrossRef]
- Cogoi, S.; Xodo, L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006, 34, 2536–2549. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Aggarwal, J.; Thakkar, B. Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants. Sci. Rep. UK 2016, 6, 28211. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Ahn, J.H.; Kim, K.K. Unraveling the Regulatory G-Quadruplex Puzzle: Lessons From Genome and Transcriptome-Wide Studies. Front. Genet 2019, 10, 1002. [Google Scholar] [CrossRef]
- Ravichandran, S.; Kim, Y.E.; Bansal, V.; Ghosh, A.; Hur, J.; Subramani, V.K.; Pradhan, S.; Lee, M.K.; Kim, K.K.; Ahn, J.H. Genome-wide analysis of regulatory G-quadruplexes affecting gene expression in human cytomegalovirus. PLoS Pathog. 2018, 14, e1007334. [Google Scholar] [CrossRef]
- Parveen, N.; Shamim, A.; Cho, S.; Kim, K.K. Computational Approaches to Predict the Non-canonical DNAs. Curr. Bioinform. 2019, 14, 470–479. [Google Scholar] [CrossRef]
- Lombardi, E.P.; Londono-Vallejo, A. A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res. 2020, 48, 1603. [Google Scholar] [CrossRef]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Antonio, M.D.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Yu, Z.; Konik, R.; Cui, Y.; Koirala, D.; Mao, H. G-quadruplex and i-motif are mutually exclusive in ILPR double-stranded DNA. Biophys. J. 2012, 102, 2575–2584. [Google Scholar] [CrossRef]
- Kang, H.J.; Kendrick, S.; Hecht, S.M.; Hurley, L.H. The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 2014, 136, 4172–4185. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Zhang, X.; Deng, H.; Wu, F.; Ren, Y.; Xiang, H.; Zheng, S.; Liu, L.; Huang, L.; Zeng, B.; et al. BmILF and i-motif structure are involved in transcriptional regulation of BmPOUM2 in Bombyx mori. Nucleic Acids Res. 2018, 46, 1710–1723. [Google Scholar] [CrossRef]
- Khan, N.; Avino, A.; Tauler, R.; Gonzalez, C.; Eritja, R.; Gargallo, R. Solution equilibria of the i-motif-forming region upstream of the B-cell lymphoma-2 P1 promoter. Biochimie 2007, 89, 1562–1572. [Google Scholar] [CrossRef]
- Bucek, P.; Jaumot, J.; Avino, A.; Eritja, R.; Gargallo, R. pH-Modulated Watson-Crick duplex-quadruplex equilibria of guanine-rich and cytosine-rich DNA sequences 140 base pairs upstream of the c-kit transcription initiation site. Chemistry 2009, 15, 12663–12671. [Google Scholar] [CrossRef]
- Sun, D.; Hurley, L.H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: Implications for drug targeting and control of gene expression. J. Med. Chem. 2009, 52, 2863–2874. [Google Scholar] [CrossRef]
- Guo, K.; Pourpak, A.; Beetz-Rogers, K.; Gokhale, V.; Sun, D.; Hurley, L.H. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007, 129, 10220–10228. [Google Scholar] [CrossRef]
- Zeraati, M.; Langley, D.B.; Schofield, P.; Moye, A.L.; Rouet, R.; Hughes, W.E.; Bryan, T.M.; Dinger, M.E.; Christ, D. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018, 10, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Abou Assi, H.; Garavis, M.; Gonzalez, C.; Damha, M.J. i-Motif DNA: Structural features and significance to cell biology. Nucleic Acids Res. 2018, 46, 8038–8056. [Google Scholar] [CrossRef] [PubMed]
- McKim, M.; Buxton, A.; Johnson, C.; Metz, A.; Sheardy, R.D. Loop Sequence Context Influences the Formation and Stability of the i-Motif for DNA Oligomers of Sequence (CCCXXX)4, where X = A and/or T, under Slightly Acidic Conditions. J. Phys. Chem. B 2016, 120, 7652–7661. [Google Scholar] [CrossRef] [PubMed]
- Reilly, S.M.; Morgan, R.K.; Brooks, T.A.; Wadkins, R.M. Effect of interior loop length on the thermal stability and pK(a) of i-motif DNA. Biochemistry 2015, 54, 1364–1370. [Google Scholar] [CrossRef]
- Bhavsar-Jog, Y.P.; Van Dornshuld, E.; Brooks, T.A.; Tschumper, G.S.; Wadkins, R.M. Epigenetic Modification, Dehydration, and Molecular Crowding Effects on the Thermodynamics of i-Motif Structure Formation from C-Rich DNA. Biochemistry 2014, 53, 1586–1594. [Google Scholar] [CrossRef]
- Kaiser, C.E.; Van Ert, N.A.; Agrawal, P.; Chawla, R.; Yang, D.; Hurley, L.H. Insight into the Complexity of the i-Motif and G-Quadruplex DNA Structures Formed in the KRAS Promoter and Subsequent Drug-Induced Gene Repression. J. Am. Chem. Soc. 2017, 139, 8522–8536. [Google Scholar] [CrossRef]
- Benabou, S.; Garavis, M.; Lyonnais, S.; Eritja, R.; Gonzalez, C.; Gargallo, R. Understanding the effect of the nature of the nucleobase in the loops on the stability of the i-motif structure. Phys. Chem. Chem. Phys. 2016, 18, 7997–8004. [Google Scholar] [CrossRef]
- Mir, B.; Serrano, I.; Buitrago, D.; Orozco, M.; Escaja, N.; Gonzalez, C. Prevalent Sequences in the Human Genome Can Form Mini i-Motif Structures at Physiological pH. J. Am. Chem. Soc. 2017, 139, 13985–13988. [Google Scholar] [CrossRef]
- Phan, A.T.; Gueron, M.; Leroy, J.L. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000, 299, 123–144. [Google Scholar] [CrossRef]
- Gurung, S.P.; Schwarz, C.; Hall, J.P.; Cardin, C.J.; Brazier, J.A. The importance of loop length on the stability of i-motif structures. Chem. Commun. 2015, 51, 5630–5632. [Google Scholar] [CrossRef]
- Pages, B.J.; Gurung, S.P.; McQuaid, K.; Hall, J.P.; Cardin, C.J.; Brazier, J.A. Stabilization of Long-Looped i-Motif DNA by Polypyridyl Ruthenium Complexes. Front. Chem. 2019, 7, 744. [Google Scholar] [CrossRef] [PubMed]
- Sahakyan, A.B.; Chambers, V.S.; Marsico, G.; Santner, T.; Di Antonio, M.; Balasubramanian, S. Machine learning model for sequence-driven DNA G-quadruplex formation. Sci. Rep. 2017, 7, 14535. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. AMBER 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Ivani, I.; Dans, P.D.; Noy, A.; Perez, A.; Faustino, I.; Hospital, A.; Walther, J.; Andrio, P.; Goni, R.; Balaceanu, A.; et al. Parmbsc1: A refined force field for DNA simulations. Nat. Methods 2016, 13, 55–58. [Google Scholar] [CrossRef]
- Sponer, J.; Burda, J.V.; Mejzlik, P.; Leszczynski, J.; Hobza, P. Hydrogen-bonded trimers of DNA bases and their interaction with metal cations: Ab initio quantum-chemical and empirical potential study. J. Biomol. Struct. Dyn. 1997, 14, 613–628. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Vaught, A. Graphing with Gnuplot and Xmgr: Two graphing packages available under linux. Linux J. 1996, 1996, 7. [Google Scholar]
- Zar, J.H. Biostatistical Analysis, 4th ed.; Prentice Hall, Inc.: Upper Saddle River, NJ, USA, 1999. [Google Scholar]
- Box, G.H.W.; Hunter, J.S. Statistics for Experimenters: An Introduction to Design, Data Analysis, and Model Building; John Wiley & Sons, Inc.: Princeton, NJ, USA, 1978. [Google Scholar]
- Tsui, V.; Case, D.A. Molecular dynamics simulations of nucleic acids with a generalized Born solvation model. J. Am. Chem. Soc. 2000, 122, 2489–2498. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Accounts Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Massova, I.; Kollman, P.A. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect. Drug Discov. Des. 2000, 18, 113–135. [Google Scholar] [CrossRef]

| Name | Sequence (5′-3′) | ** Model |
|---|---|---|
| CCCT3 | * d[(CCCTTT)3CCC] | 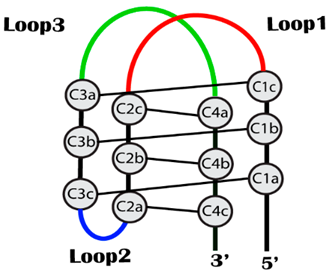 |
| CCCT5 | d[(CCCTTTTT)3CCC] | |
| CCCT6 | d[(CCCTTTTTT)3CCC] | |
| CCCT7 | d[(CCCTTTTTTT)3CCC] | |
| CCCTA2 | d[(CCCTAA)3CCC] |
| Model I-Motifs | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RMSD (Å) | |||||||||||||||
| Temperature | CCCT3 | CCCT5 | CCCT7 | CCCT6 | CCCTA2 | ||||||||||
| Whole I-Motif | Core Region | Loop Region | Whole I-Motif | Core Region | Loop Region | Whole I-Motif | Core Region | Loop Region | Whole I-Motif | Core Region | Loop Region | Whole I-Motif | Core Region | Loop Region | |
| 280 K | 1.8 | 1.2 | 3.5 | 3.8 | 1.2 | 6.1 | 5.3 | 1.5 | 7.0 | 3.5 | 1.5 | 5.3 | 1.9 | 1.4 | 4.0 |
| 300 K | 1.9 | 1.3 | 4.2 | 3.3 | 1.1 | 4.9 | 4.6 | 2.6 | 6.2 | 3.4 | 0.9 | 5.4 | 1.9 | 1.3 | 4.3 |
| 320 K | 2.1 | 1.2 | 4.4 | 3.0 | 1.3 | 5.6 | 4.1 | 1.9 | 5.9 | 4.3 | 1.0 | 5.9 | 2.1 | 1.3 | 4.4 |
| 340 K | 2.1 | 1.2 | 4.5 | 3.1 | 1.0 | 5.9 | 5.4 | 1.3 | 8.7 | 4.6 | 2.3 | 7.2 | 2.5 | 1.3 | 4.8 |
| 360 K | 2.4 | 1.2 | 4.5 | 4.0 | 1.2 | 6.4 | 5.8 | 1.4 | 7.7 | 4.8 | 1.1 | 7.1 | 2.2 | 1.5 | 5.1 |
| 2.0 | 1.2 | 4.2 | 3.4 | 1.2 | 5.8 | 5.0 | 1.7 | 7.1 | 4.1 | 1.3 | 6.2 | 2.1 | 1.4 | 4.5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shamim, A.; Razzaq, M.; Kim, K.K. MD-TSPC4: Computational Method for Predicting the Thermal Stability of I-Motif. Int. J. Mol. Sci. 2021, 22, 61. https://doi.org/10.3390/ijms22010061
Shamim A, Razzaq M, Kim KK. MD-TSPC4: Computational Method for Predicting the Thermal Stability of I-Motif. International Journal of Molecular Sciences. 2021; 22(1):61. https://doi.org/10.3390/ijms22010061
Chicago/Turabian StyleShamim, Amen, Maria Razzaq, and Kyeong Kyu Kim. 2021. "MD-TSPC4: Computational Method for Predicting the Thermal Stability of I-Motif" International Journal of Molecular Sciences 22, no. 1: 61. https://doi.org/10.3390/ijms22010061
APA StyleShamim, A., Razzaq, M., & Kim, K. K. (2021). MD-TSPC4: Computational Method for Predicting the Thermal Stability of I-Motif. International Journal of Molecular Sciences, 22(1), 61. https://doi.org/10.3390/ijms22010061







