Mechanical Stability of a Small, Highly-Luminescent Engineered Protein NanoLuc
Abstract
1. Introduction
2. Results and Discussion
2.1. NanoLuc Protein for Force Spectroscopy
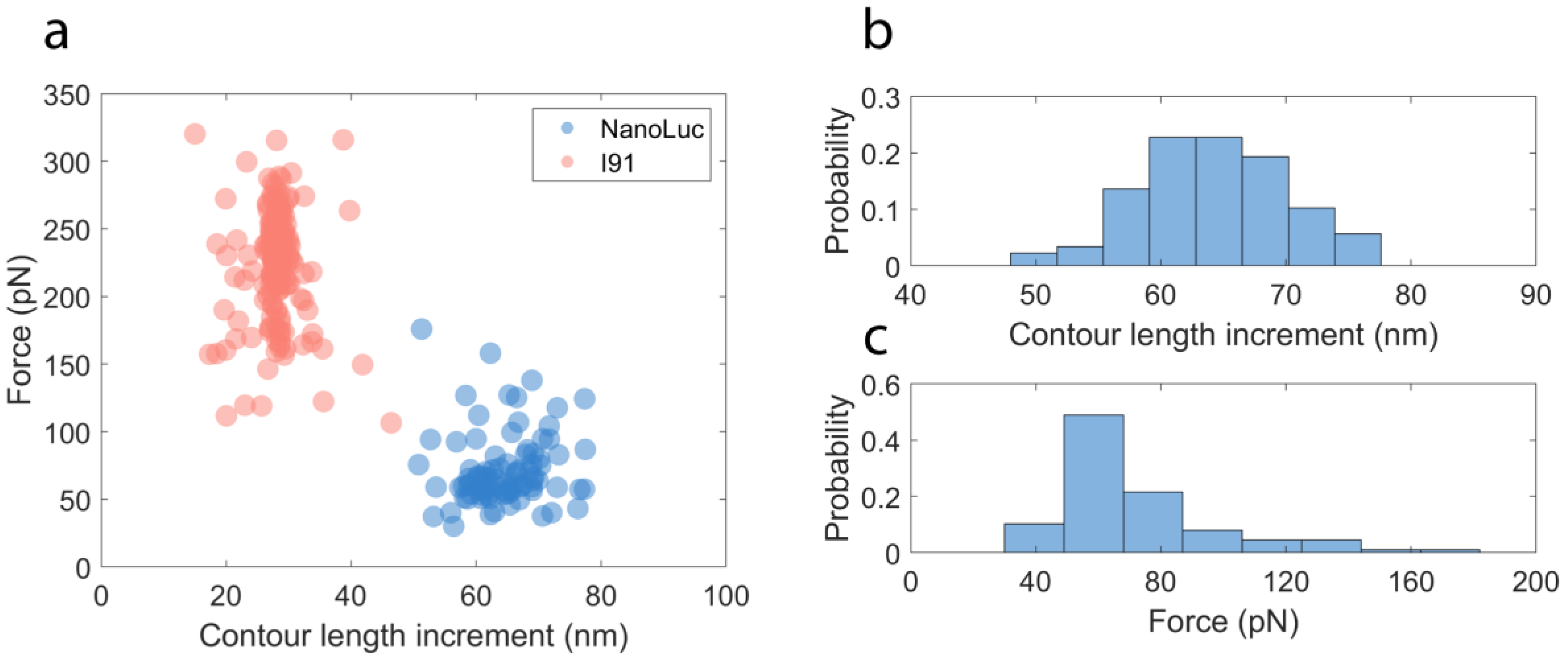
2.2. Mechanical Unfolding of NanoLuc
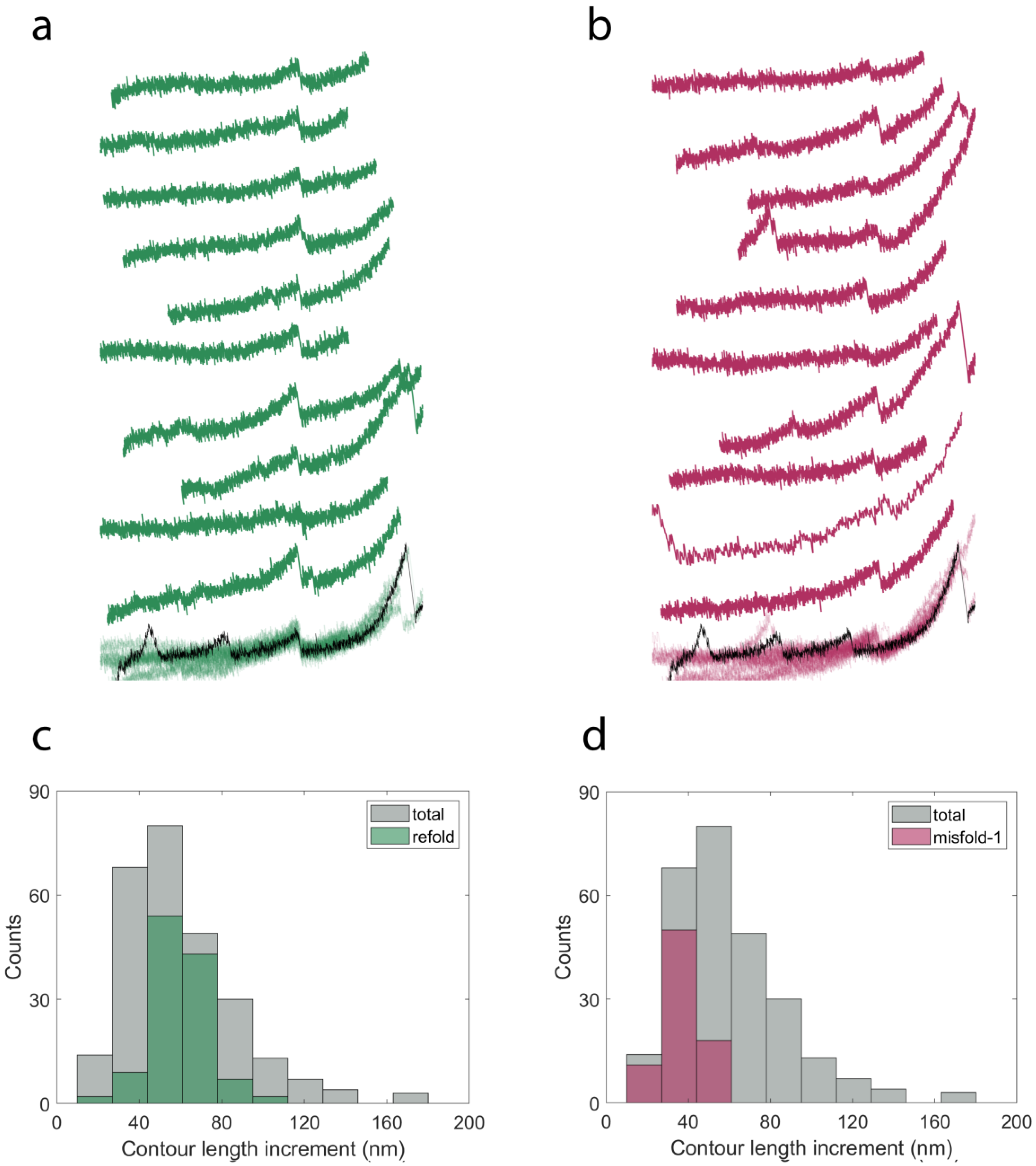
2.3. Cyclic Measurements of NanoLuc Refolding and Misfolding
3. Materials and Methods
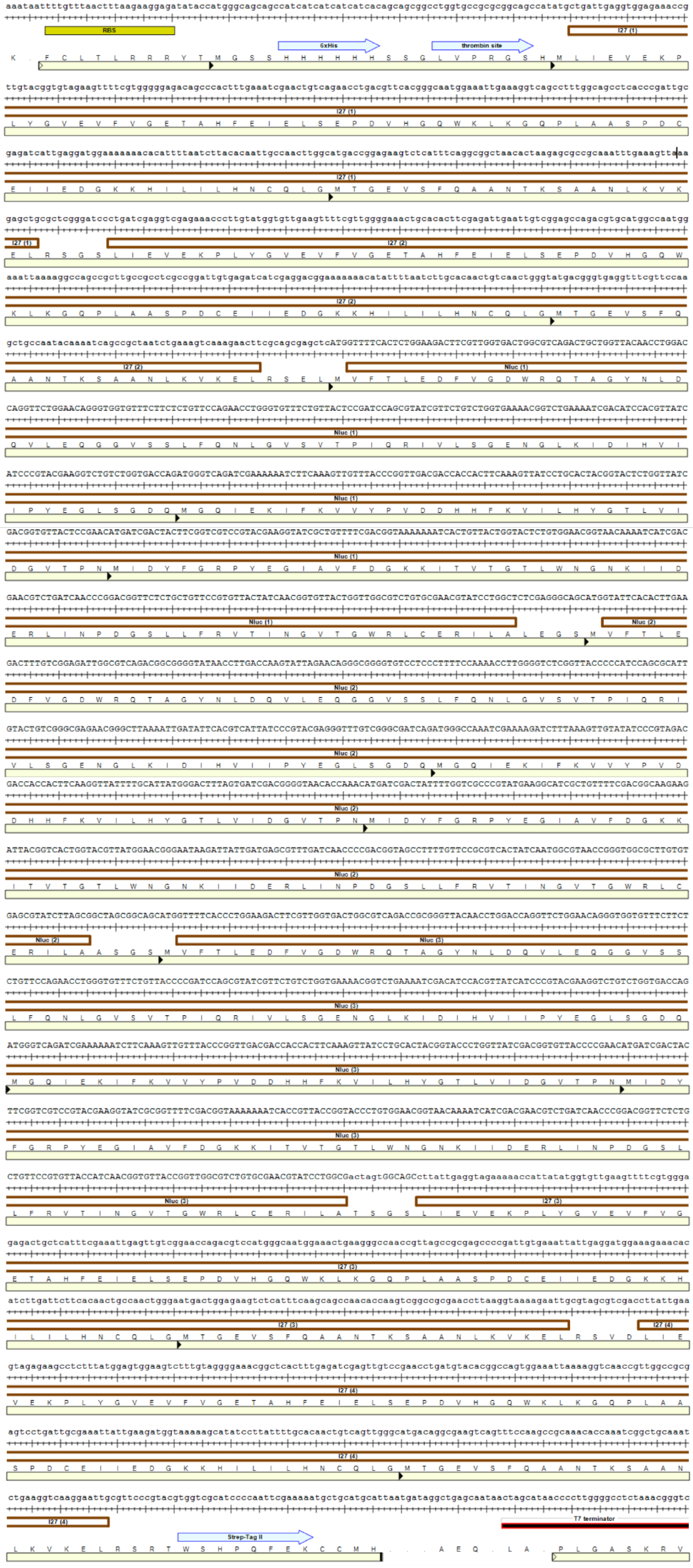
3.1. Protein Purification
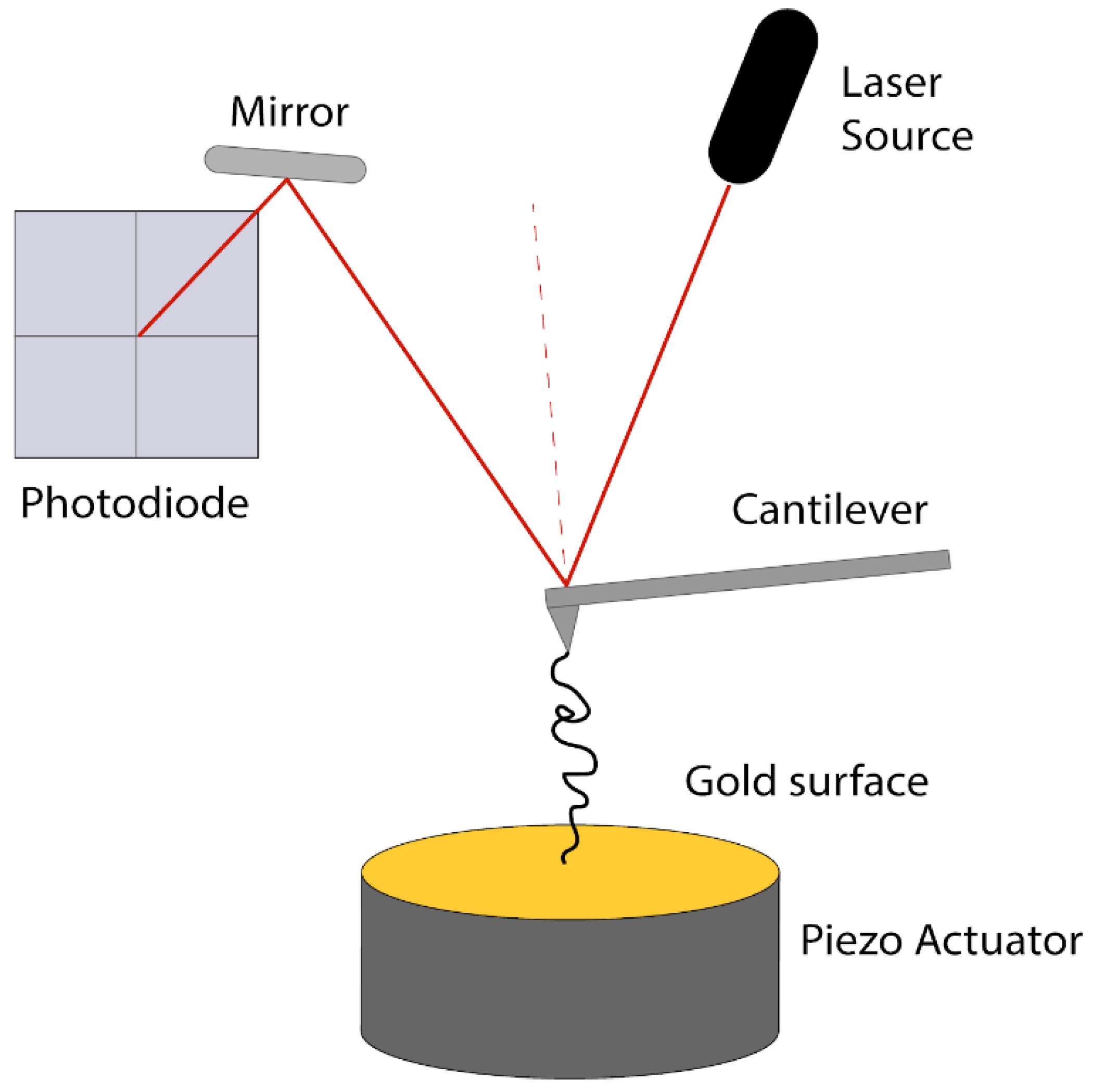
3.2. Atomic Force Microscopy
3.3. Bioluminescence
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AFM | Atomic Force Microscopy |
| SMFS | Single Molecule Force Spectroscopy |
| FLuc | Firefly Luciferase |
References
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Anfinsen, C.B. Principles that govern the folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A.; Chan, H.S. From Levinthal to Pathways to Funnels: The “New View” of Protein Folding Kinetics. Nat. Struct. Biol. 1997, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M.; Evans, P.A.; Radford, S.E. Understanding how proteins fold: The lysozyme story so far. Trends Biochem. Sci. 1994, 19, 31–37. [Google Scholar] [CrossRef]
- Frydman, J.; Erdjument-bromage, H.; Tempst, P.; Hartl, F.U. Co-translational domain folding as the structural basis for the rapid. America (N. Y.) 1999, 6, 697–705. [Google Scholar]
- Jackson, S.E. How do small single-domain proteins fold? Fold. Des. 1998, 3, 81–91. [Google Scholar] [CrossRef]
- Herbst, R.; Schäfer, U.; Seckler, R. Equilibrium intermediates in the reversible unfolding of firefly (Photinus pyralis) luciferase. J. Biol. Chem. 1997, 272, 7099–7105. [Google Scholar] [CrossRef]
- Herbst, R.; Gast, K.; Seckler, R. Folding of firefly (Photinus pyralis) luciferase: Aggregation and reactivation of unfolding intermediates. Biochemistry 1998, 37, 6586–6597. [Google Scholar] [CrossRef]
- Hartl, F.U. Cellular Homeostasis and Aging. Annu. Rev. Biochem. 2016, 85, 1–4. [Google Scholar] [CrossRef]
- Karve, T.M.; Cheema, A.K. Small Changes Huge Impact: The Role of Protein Posttranslational Modifications in Cellular Homeostasis and Disease. J. Amino Acids 2011, 2011, 1–13. [Google Scholar] [CrossRef]
- Cecarini, V.; Gee, J.; Fioretti, E.; Amici, M.; Angeletti, M.; Eleuteri, A.M.; Keller, J.N. Protein oxidation and cellular homeostasis: Emphasis on metabolism. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1773, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Koga, H.; Kaushik, S.; Cuervo, A.M. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res. Rev. 2011, 10, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Villanueva, J.F.; Díaz-Molina, R.; García-González, V. Protein folding and mechanisms of proteostasis. Int. J. Mol. Sci. 2015, 16, 17193–17230. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G. Targeting cancer metabolism: A therapeutic window opens. Nat. Rev. Drug Discov. 2011, 10, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.D.; Shao, S.X.; Jiang, H.P.; Cao, Y.W.; Wang, Y.H.; Yang, X.C.; Wang, Y.L.; Wang, X.S.; Niu, H.T. Warburg effect or reverse warburg effect? a review of cancer metabolism. Oncol. Res. Treat. 2015, 38, 117–122. [Google Scholar] [CrossRef]
- Martinez-Outschoorn, U.E.; Peiris-Pagés, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2017, 14, 11–31. [Google Scholar] [CrossRef]
- Johnson, C.; Warmoes, M.O.; Shen, X.; Locasale, J.W. Epigenetics and cancer metabolism. Cancer Lett. 2015, 356, 309–314. [Google Scholar] [CrossRef]
- Thompson, A.D.; Bernard, S.M.; Skiniotis, G.; Gestwicki, J.E. Visualization and functional analysis of the oligomeric states of Escherichia coli heat shock protein 70 (Hsp70/DnaK). Cell Stress Chaperones 2012, 17, 313–327. [Google Scholar] [CrossRef]
- Chaudhuri, T.K.; Paul, S. Protein-misfolding diseases and chaperone-based therapeutic approaches. FEBS J. 2006, 273, 1331–1349. [Google Scholar] [CrossRef]
- Sherman, M.Y.; Goldberg, A.L. Cellular defenses against unfolded proteins: A cell biologist thinks about neurodegenerative diseases. Neuron 2001, 29, 15–32. [Google Scholar] [CrossRef]
- Carrion-Vazquez, M.; Marszalek, P.E.; Oberhauser, A.F.; Fernandez, J.M. Atomic force microscopy captures length phenotypes in single proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 11288–11292. [Google Scholar] [CrossRef] [PubMed]
- Rief, M.; Grubmüller, H. Force Spectroscopy of Single Biomolecules. ChemPhysChem 2002, 3, 255. [Google Scholar] [CrossRef][Green Version]
- Carrion-Vazquez, M. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog. Biophys. Mol. Biol. 2000, 74, 63–91. [Google Scholar] [CrossRef]
- Eckels, E.C.; Tapia-Rojo, R.; Rivas-Pardo, J.A.; Fernández, J.M. The Work of Titin Protein Folding as a Major Driver in Muscle Contraction. Annu. Rev. Physiol. 2018, 80, 327–351. [Google Scholar] [CrossRef]
- Fernandez, J.M. Force-Clamp Spectroscopy Monitors the Folding Trajectory of a Single Protein. Science 2004, 303, 1674–1678. [Google Scholar] [CrossRef]
- Giganti, D.; Yan, K.; Badilla, C.L.; Fernandez, J.M.; Alegre-Cebollada, J. Disulfide isomerization reactions in titin immunoglobulin domains enable a mode of protein elasticity. Nat. Commun. 2018, 9, 185. [Google Scholar] [CrossRef]
- Dietz, H.; Rief, M. Protein structure by mechanical triangulation. Proc. Natl. Acad. Sci. USA 2006, 103, 1244–1247. [Google Scholar] [CrossRef]
- Haldar, S.; Tapia-Rojo, R.; Eckels, E.C.; Valle-Orero, J.; Fernandez, J.M. Trigger factor chaperone acts as a mechanical foldase. Nat. Commun. 2017, 8, 668. [Google Scholar] [CrossRef]
- Valle-Orero, J.; Rivas-Pardo, J.A.; Tapia-Rojo, R.; Popa, I.; Echelman, D.J.; Haldar, S.; Fernández, J.M. Mechanical Deformation Accelerates Protein Ageing. Angew. Chem. 2017, 129, 9873–9878. [Google Scholar] [CrossRef]
- Popa, I.; Rivas-Pardo, J.A.; Eckels, E.C.; Echelman, D.J.; Badilla, C.L.; Valle-Orero, J.; Fernández, J.M. A HaloTag Anchored Ruler for Week-Long Studies of Protein Dynamics. J. Am. Chem. Soc. 2016, 138, 10546–10553. [Google Scholar] [CrossRef]
- Li, H.; Cao, Y. Protein Mechanics: From Single Molecules to Functional Biomaterials. Acc. Chem. Res. 2010, 43, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Dudek, D.M.; Cao, Y.; Balamurali, M.M.; Gosline, J.; Li, H. Designed biomaterials to mimic the mechanical properties of muscles. Nature 2010, 465, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Milles, L.F.; Schulten, K.; Gaub, H.E.; Bernardi, R.C. Molecular mechanism of extreme mechanostability in a pathogen adhesin. Science 2018, 359, 1527–1533. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Perisic, O.; Peng, Q.; Cao, Y.; Lam, C.; Lu, H.; Li, H. Single-molecule force spectroscopy reveals a mechanically stable protein fold and the rational tuning of its mechanical stability. Proc. Natl. Acad. Sci. USA 2007, 104, 9278–9283. [Google Scholar] [CrossRef] [PubMed]
- Bujalowski, P.J.; Nicholls, P.; Garza, E.; Oberhauser, A.F. The central domain of UNC-45 chaperone inhibits the myosin power stroke. FEBS Open Bio 2018, 8, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Oberhauser, A.F.; Carrión-Vázquez, M. Mechanical Biochemistry of Proteins One Molecule at a Time. J. Biol. Chem. 2008, 283, 6617–6621. [Google Scholar] [CrossRef]
- Oberhauser, A.F.; Hansma, P.K.; Carrion-Vazquez, M.; Fernandez, J.M. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc. Natl. Acad. Sci. USA 2001, 98, 468–472. [Google Scholar] [CrossRef]
- Oberhauser, A.F.; Marszalek, P.E.; Erickson, H.P.; Fernandez, J.M. The molecular elasticity of the extracellular matrix protein tenascin. Nature 1998, 393, 181–185. [Google Scholar] [CrossRef]
- Kim, B.-H.; Palermo, N.Y.; Lovas, S.; Zaikova, T.; Keana, J.F.W.; Lyubchenko, Y.L. Single-Molecule Atomic Force Microscopy Force Spectroscopy Study of Aβ-40 Interactions. Biochemistry 2011, 50, 5154–5162. [Google Scholar] [CrossRef]
- Lv, Z.; Krasnoslobodtsev, A.V.; Zhang, Y.; Ysselstein, D.; Rochet, J.-C.; Blanchard, S.C.; Lyubchenko, Y.L. Direct Detection of α-Synuclein Dimerization Dynamics: Single-Molecule Fluorescence Analysis. Biophys. J. 2015, 108, 2038–2047. [Google Scholar] [CrossRef]
- Zhang, Y.; Lyubchenko, Y.L. The Structure of Misfolded Amyloidogenic Dimers: Computational Analysis of Force Spectroscopy Data. Biophys. J. 2014, 107, 2903–2910. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Galera-Prat, A.; Gómez-Sicilia, A.; Oberhauser, A.F.; Cieplak, M.; Carrión-Vázquez, M. Understanding biology by stretching proteins: Recent progress. Curr. Opin. Struct. Biol. 2010, 20, 63–69. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cecconi, C. Direct Observation of the Three-State Folding of a Single Protein Molecule. Science 2005, 309, 2057–2060. [Google Scholar] [CrossRef] [PubMed]
- Puchner, E.M.; Gaub, H.E. Single-Molecule Mechanoenzymatics. Annu. Rev. Biophys. 2012, 41, 497–518. [Google Scholar] [CrossRef][Green Version]
- Goldman, D.H.; Kaiser, C.M.; Milin, A.; Righini, M.; Tinoco, I.; Bustamante, C. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science 2015, 348, 457–460. [Google Scholar] [CrossRef]
- Kaiser, C.M.; Goldman, D.H.; Chodera, J.D.; Tinoco, I.; Bustamante, C. The Ribosome Modulates Nascent Protein Folding. Science 2011, 334, 1723–1727. [Google Scholar] [CrossRef]
- Kellermayer, M.S.Z.; Smith, S.B.; Granzier, H.L.; Bustamante, C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science 1997, 276, 1112–1116. [Google Scholar] [CrossRef]
- Sen, M.; Maillard, R.A.; Nyquist, K.; Rodriguez-Aliaga, P.; Pressé, S.; Martin, A.; Bustamante, C. The ClpXP Protease Unfolds Substrates Using a Constant Rate of Pulling but Different Gears. Cell 2013, 155, 636–646. [Google Scholar] [CrossRef]
- He, C.; Hu, C.; Hu, X.; Hu, X.; Xiao, A.; Perkins, T.T.; Li, H. Direct Observation of the Reversible Two-State Unfolding and Refolding of an α/β Protein by Single-Molecule Atomic Force Microscopy. Angew. Chem. Int. Ed. 2015, 54, 9921–9925. [Google Scholar] [CrossRef]
- King, G.M.; Carter, A.R.; Churnside, A.B.; Eberle, L.S.; Perkins, T.T. Ultrastable Atomic Force Microscopy: Atomic-Scale Stability and Registration in Ambient Conditions. Nano Lett. 2009, 9, 1451–1456. [Google Scholar] [CrossRef]
- Sullan, R.M.A.; Churnside, A.B.; Nguyen, D.M.; Bull, M.S.; Perkins, T.T. Atomic force microscopy with sub-picoNewton force stability for biological applications. Methods 2013, 60, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Siewny, M.G.W.; Edwards, D.T.; Sanders, A.W.; Perkins, T.T. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science 2017, 355, 945–950. [Google Scholar] [CrossRef] [PubMed]
- Scholl, Z.N.; Yang, W.; Marszalek, P.E. Competing Pathways and Multiple Folding Nuclei in a Large Multidomain Protein, Luciferase. Biophys. J. 2017, 112, 1829–1840. [Google Scholar] [CrossRef] [PubMed]
- Forman, J.R.; Clarke, J. Mechanical unfolding of proteins: Insights into biology, structure and folding. Curr. Opin. Struct. Biol. 2007, 17, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Puchner, E.M.; Gaub, H.E. Force and function: Probing proteins with AFM-based force spectroscopy. Curr. Opin. Struct. Biol. 2009, 19, 605–614. [Google Scholar] [CrossRef]
- Rathore, N.; Yan, Q.; De Pablo, J.J. Molecular simulation of the reversible mechanical unfolding of proteins. J. Chem. Phys. 2004, 120, 5781–5788. [Google Scholar] [CrossRef]
- Chyan, C.L.; Lin, F.C.; Peng, H.; Yuan, J.M.; Chang, C.H.; Lin, S.H.; Yang, G. Reversible mechanical unfolding of single ubiquitin molecules. Biophys. J. 2004, 87, 3995–4006. [Google Scholar] [CrossRef][Green Version]
- Kim, M.; Abdi, K.; Lee, G.; Rabbi, M.; Lee, W.; Yang, M.; Schofield, C.J.; Bennett, V.; Marszalek, P.E. Fast and forceful refolding of stretched α-helical solenoid proteins. Biophys. J. 2010, 98, 3086–3092. [Google Scholar] [CrossRef]
- Popa, I.; Berkovich, R.; Alegre-Cebollada, J.; Badilla, C.L.; Rivas-Pardo, J.A.; Taniguchi, Y.; Kawakami, M.; Fernandez, J.M. Nanomechanics of HaloTag tethers. J. Am. Chem. Soc. 2013, 135, 12762–12771. [Google Scholar] [CrossRef]
- Rief, M.; Gautel, M.; Oesterhelt, F.; Fernandez, J.M.; Gaub, H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 1997, 276, 1109–1112. [Google Scholar] [CrossRef]
- Stigler, J.; Ziegler, F.; Gieseke, A.; Gebhardt, J.C.M.; Rief, M. The complex folding network of single calmodulin molecules. Science 2011, 334, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Žoldák, G.; Rief, M. Force as a single molecule probe of multidimensional protein energy landscapes. Curr. Opin. Struct. Biol. 2013, 23, 48–57. [Google Scholar] [CrossRef]
- Carrion-Vazquez, M.; Li, H.; Lu, H.; Marszalek, P.E.; Oberhauser, A.F.; Fernandez, J.M. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Mol. Biol. 2003, 10, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Carrion-Vazquez, M.; Oberhauser, A.F.; Fowler, S.B.; Marszalek, P.E.; Broedel, S.E.; Clarke, J.; Fernandez, J.M. Mechanical and chemical unfolding of a single protein: A comparison. Proc. Natl. Acad. Sci. USA 1999, 96, 3694–3699. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Haslbeck, M.; Buchner, J. The Heat Shock Response: Life on the Verge of Death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Clerico, E.M.; Meng, W.; Pozhidaeva, A.; Bhasne, K.; Petridis, C.; Gierasch, L.M. Hsp70 molecular chaperones: Multifunctional allosteric holding and unfolding machines. Biochem. J. 2019, 476, 1653–1677. [Google Scholar] [CrossRef]
- Scholl, Z.N.; Yang, W.; Marszalek, P.E. Chaperones rescue luciferase folding by separating its domains. J. Biol. Chem. 2014, 289, 28607–28618. [Google Scholar] [CrossRef]
- Mel’nikov, E.E.; Rotanova, T.V. Molecular chaperones. Bioorg. Khim. 2010, 36, 5–14. [Google Scholar] [CrossRef]
- Hesterkamp, T.; Bukau, B. Role of the DnaK and HscA homologs of Hsp70 chaperones in protein folding in E.coli. EMBO J. 1998, 17, 4818–4828. [Google Scholar] [CrossRef]
- Georgopoulos, C.; Welch, W.J. Role of the major heat shock proteins as molecular chaperones. Annu. Rev. Cell Biol. 1993, 9, 601–634. [Google Scholar] [CrossRef]
- Macario, A.J.L.; Conway de Macario, E. Stress and molecular chaperones in disease. Int. J. Clin. Lab. Res. 2000, 30, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.R.; Gillies, A.T.; Chang, L.; Thompson, A.D.; Gestwicki, J.E. Molecular chaperones DnaK and DnaJ share predicted binding sites on most proteins in the E. coli proteome. Mol. Biosyst. 2012, 8, 2323–2333. [Google Scholar] [CrossRef] [PubMed]
- Nillegoda, N.B.; Wentink, A.S.; Bukau, B. Protein Disaggregation in Multicellular Organisms. Trends Biochem. Sci. 2018, 43, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Wiita, A.P.; Ainavarapu, S.R.K.; Huang, H.H.; Fernandez, J.M. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proc. Natl. Acad. Sci. USA 2006, 103, 7222–7227. [Google Scholar] [CrossRef] [PubMed]
- Wiita, A.P.; Perez-Jimenez, R.; Walther, K.A.; Gräter, F.; Berne, B.J.; Holmgren, A.; Sanchez-Ruiz, J.M.; Fernandez, J.M. Probing the chemistry of thioredoxin catalysis with force. Nature 2007, 450, 124–127. [Google Scholar] [CrossRef]
- Gumpp, H.; Puchner, E.M.; Zimmermann, J.L.; Gerland, U.; Gaub, H.E.; Blank, K. Triggering Enzymatic Activity with Force. Nano Lett. 2009, 9, 3290–3295. [Google Scholar] [CrossRef]
- Puchner, E.M.; Alexandrovich, A.; Kho, A.L.; Hensen, U.; Schafer, L.V.; Brandmeier, B.; Grater, F.; Grubmuller, H.; Gaub, H.E.; Gautel, M. Mechanoenzymatics of titin kinase. Proc. Natl. Acad. Sci. USA 2008, 105, 13385–13390. [Google Scholar] [CrossRef]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; MacHleidt, T.; et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012, 7, 1848–1857. [Google Scholar] [CrossRef]
- Mashaghi, A.; Mashaghi, S.; Tans, S.J. Misfolding of luciferase at the single-molecule level. Angew. Chem. Int. Ed. 2014, 53, 10390–10393. [Google Scholar] [CrossRef]
- England, C.G.; Ehlerding, E.B.; Cai, W. NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjug. Chem. 2016, 27, 1175–1187. [Google Scholar] [CrossRef]
- Schröder, H.; Langer, T.; Hartl, F.U.; Bukau, B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993, 12, 4137–4144. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.; Schroder, H.; Hartl, F.U.; Bukau, B.; Langer, T.; Flanagan, J. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc. Natl. Acad. Sci. USA 2006, 91, 10345–10349. [Google Scholar] [CrossRef] [PubMed]
- Walder, R.; Leblanc, M.A.; Van Patten, W.J.; Edwards, D.T.; Greenberg, J.A.; Adhikari, A.; Okoniewski, S.R.; Sullan, R.M.A.; Rabuka, D.; Sousa, M.C.; et al. Rapid Characterization of a Mechanically Labile α-Helical Protein Enabled by Efficient Site-Specific Bioconjugation. J. Am. Chem. Soc. 2017, 139, 9867–9875. [Google Scholar] [CrossRef] [PubMed]
- Scholl, Z.N.; Li, Q.; Marszalek, P.E. Single molecule mechanical manipulation for studying biological properties of proteins, DNA, and sugars. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2014, 6, 211–229. [Google Scholar] [CrossRef]
- Li, H.; Oberhauser, A.F.; Redick, S.D.; Carrion-Vazquez, M.; Erickson, H.P.; Fernandez, J.M. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. USA 2001, 98, 10682–10686. [Google Scholar] [CrossRef]
- Promega. Chroma-Glo TM Luciferase Assay System; Technical Manual; Promega: Madison, WI, USA, 2015. [Google Scholar]
- Edwards, D.T.; Faulk, J.K.; LeBlanc, M.A.; Perkins, T.T. Force Spectroscopy with 9-μs Resolution and Sub-pN Stability by Tailoring AFM Cantilever Geometry. Biophys. J. 2017, 113, 2595–2600. [Google Scholar] [CrossRef]
- Scott, D.W. Sturges’ rule. WIREs Comput. Stat. 2009, 303–306. [Google Scholar] [CrossRef]
- Probes, C.; Li, Q.; Scholl, Z.N.; Marszalek, P.E. Capturing the Mechanical Unfolding Pathway of a Large Protein with Coiled-Coil Probes**. Angew. Chem. Int. Ed. Engl. 2014, 53, 13429–13433. [Google Scholar] [CrossRef]
- Rief, M.; Pascual, J.; Saraste, M.; Gaub, H.E. Single molecule force spectroscopy of spectrin repeats: Low unfolding forces in helix bundles. J. Mol. Biol. 1999, 286, 553–561. [Google Scholar] [CrossRef]
- Unterauer, E.M.; Nicolaus, T.; Gaub, H.E. Calcium stabilizes the strongest protein fold. Nat. Commun. 2018, 9, 4764. [Google Scholar] [CrossRef]
- Scholl, Z.N.; Li, Q.; Josephs, E.; Apostolidou, D.; Marszalek, P.E. Force Spectroscopy of Single Protein Molecules Using an Atomic Force Microscope. J. Vis. Exp. 2019, 144. [Google Scholar] [CrossRef] [PubMed]
- Scholl, Z.N.; Josephs, E.A.; Marszalek, P.E. Modular, Nondegenerate Polyprotein Scaffolds for Atomic Force Spectroscopy. Biomacromolecules 2016, 17, 2502–2505. [Google Scholar] [CrossRef] [PubMed]
- Florin, E.L.; Rief, M.; Lehmann, H.; Ludwig, M.; Dornmair, C.; Moy, V.T.; Gaub, H.E. Sensing specific molecular interactions with the atomic force microscope. Biosens. Bioelectron. 1995, 10, 895–901. [Google Scholar] [CrossRef]
- Bustamante, C.; Marko, J.F.; Siggia, E.D.; Smith, S. Entropic elasticity of lambda-phage DNA. Science 1994, 265, 1599–1600. [Google Scholar] [CrossRef] [PubMed]
- Marszalek, P.E.; Lu, H.; Li, H.; Carrion-Vazquez, M.; Oberhauser, A.F.; Schulten, K.; Fernandez, J.M. Mechanical unfolding intermediates in titin modules. Nature 1999, 402, 100–103. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Apostolidou, D.; Marszalek, P. Mechanical Stability of a Small, Highly-Luminescent Engineered Protein NanoLuc. Int. J. Mol. Sci. 2021, 22, 55. https://doi.org/10.3390/ijms22010055
Ding Y, Apostolidou D, Marszalek P. Mechanical Stability of a Small, Highly-Luminescent Engineered Protein NanoLuc. International Journal of Molecular Sciences. 2021; 22(1):55. https://doi.org/10.3390/ijms22010055
Chicago/Turabian StyleDing, Yue, Dimitra Apostolidou, and Piotr Marszalek. 2021. "Mechanical Stability of a Small, Highly-Luminescent Engineered Protein NanoLuc" International Journal of Molecular Sciences 22, no. 1: 55. https://doi.org/10.3390/ijms22010055
APA StyleDing, Y., Apostolidou, D., & Marszalek, P. (2021). Mechanical Stability of a Small, Highly-Luminescent Engineered Protein NanoLuc. International Journal of Molecular Sciences, 22(1), 55. https://doi.org/10.3390/ijms22010055




