OsPP2C09 Is a Bifunctional Regulator in Both ABA-Dependent and Independent Abiotic Stress Signaling Pathways
Abstract
1. Introduction
2. Results
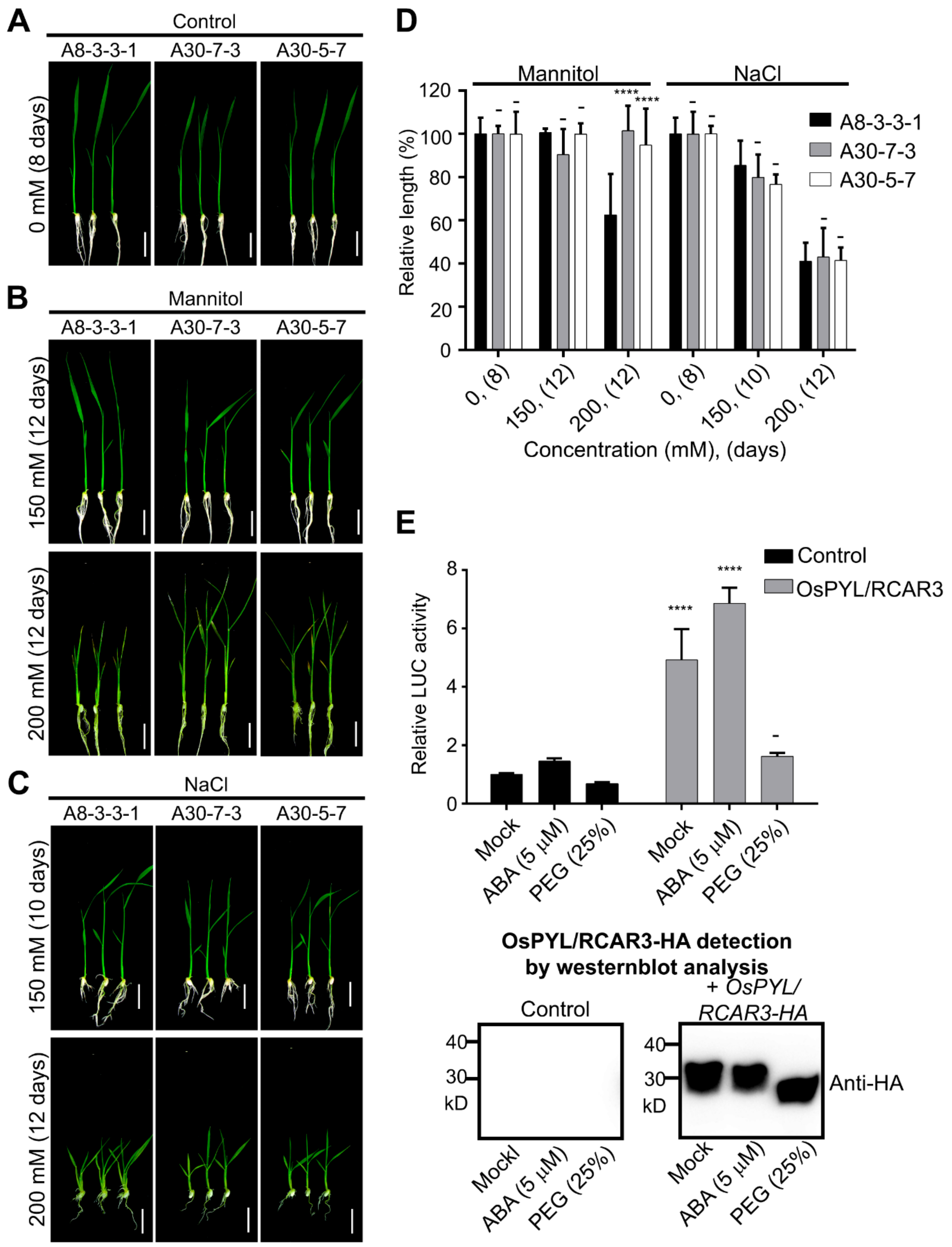
2.1. Transgenic Rice Overexpressing OsPYL/RCAR3 Are Insensitive to Osmotic Stress
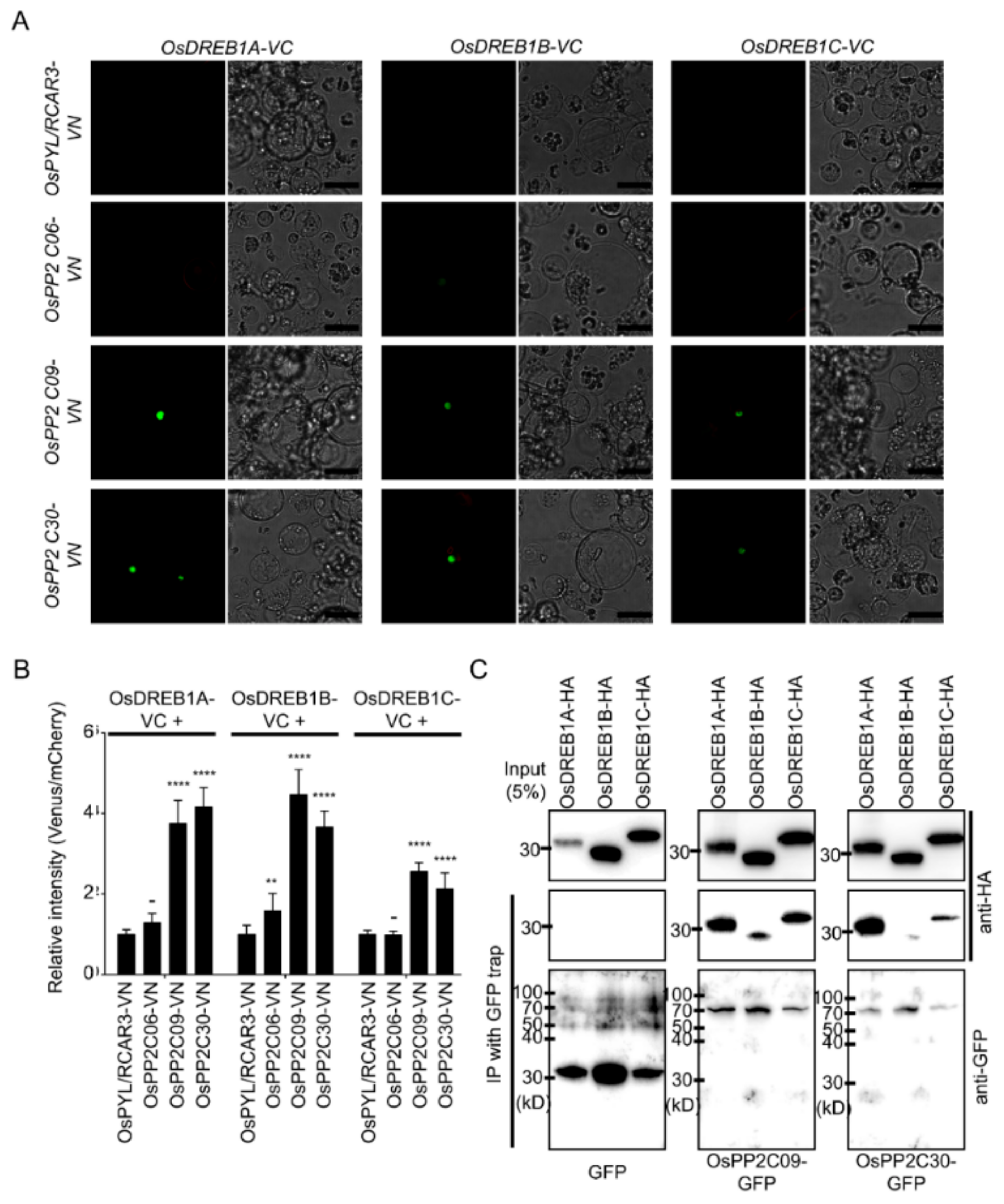
2.2. Stress-Inducible OsPP2CAs Interact with OsPYL/RCAR3 to Activate Abiotic Stress Responses
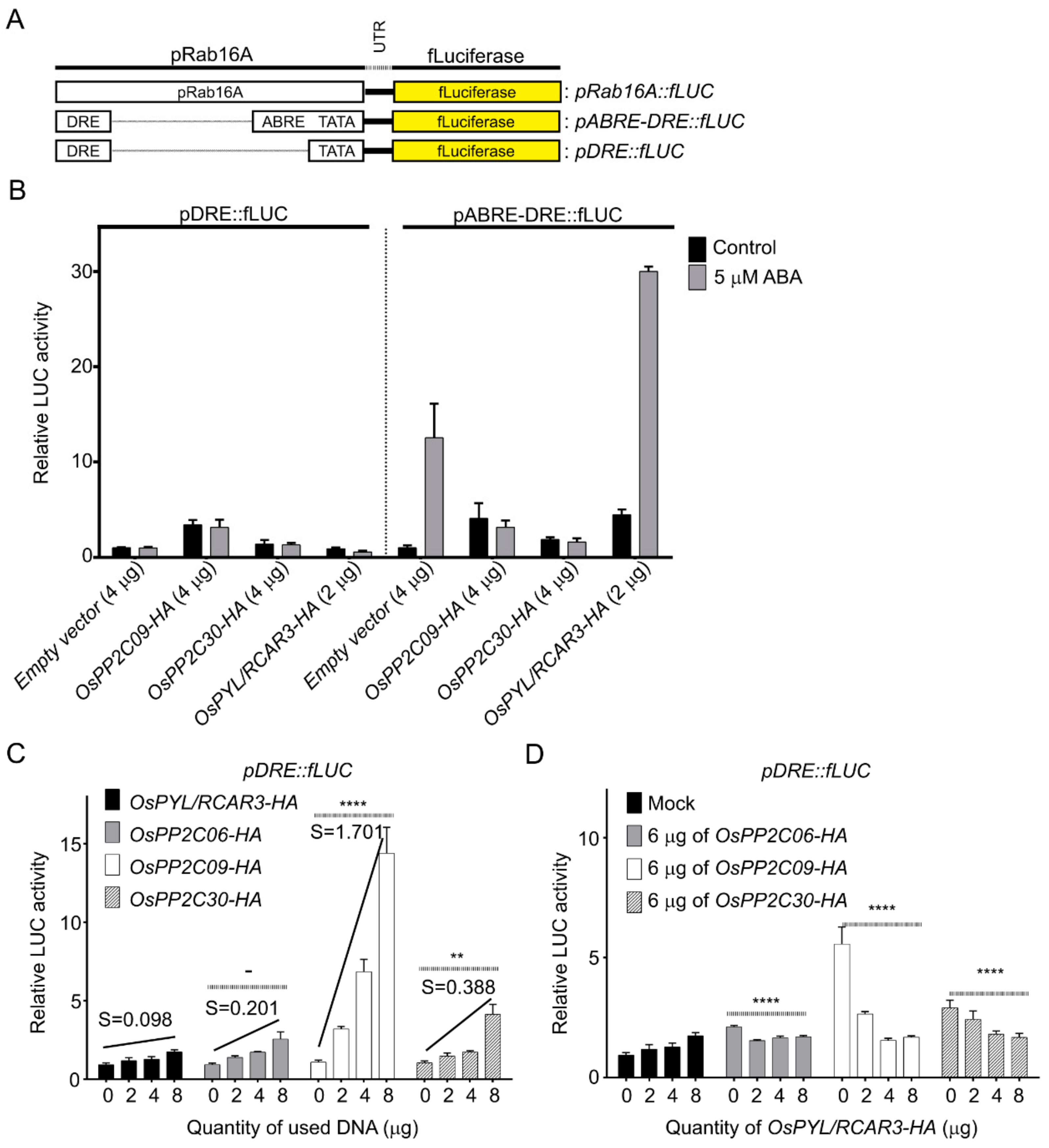
2.3. OsPP2CAs Induce Gene Expression through the DRE Cis-Element
2.4. Transgenic Rice Plants Overexpressing OsPP2C09 Are Hypersensitive to Osmotic Stress
2.5. Transcriptomic Analysis of Rice Overexpressing OsPP2CA09 during Osmotic Stress and ABA Treatment
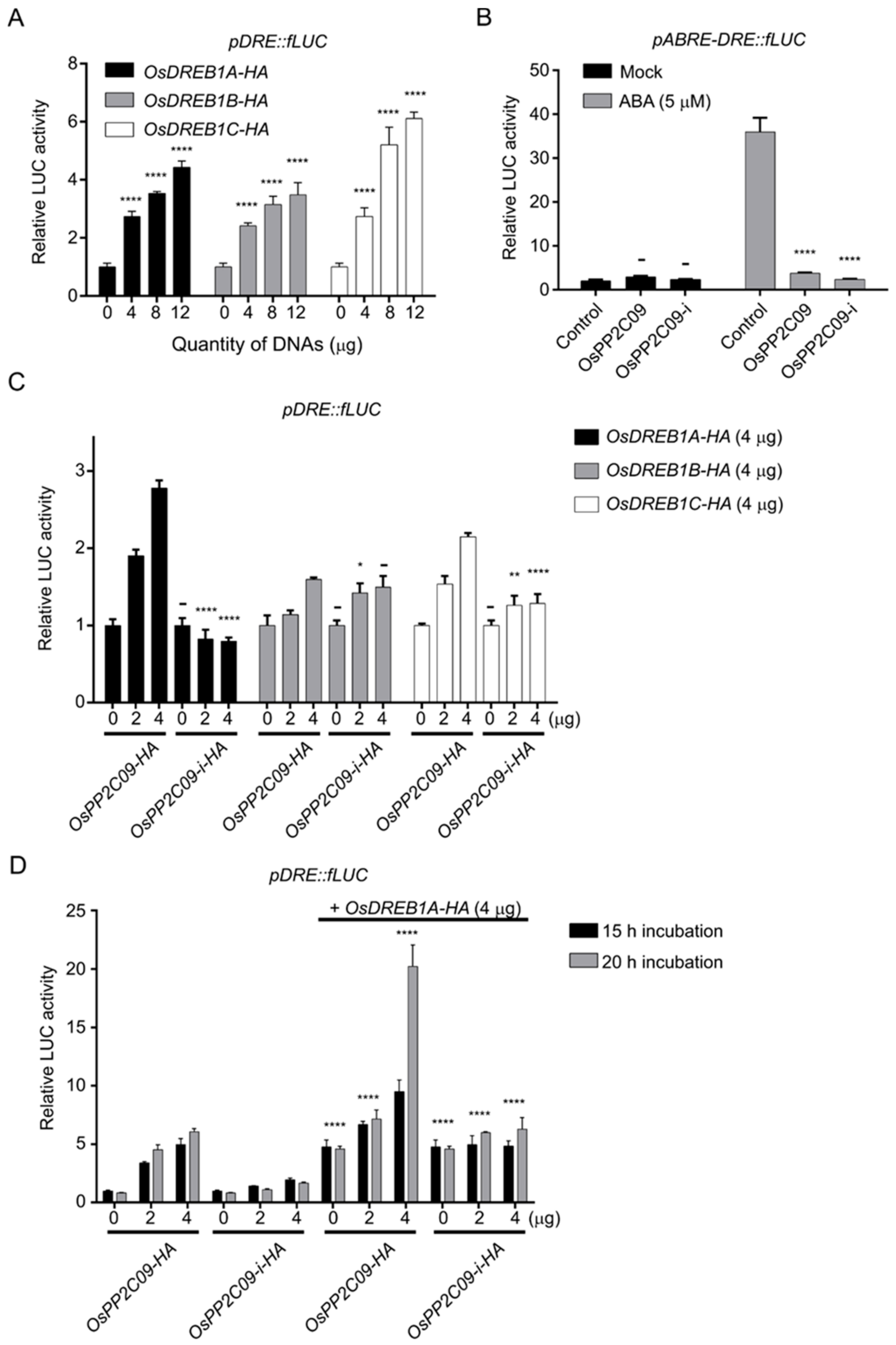
2.6. OsPP2CAs Regulate the Transcriptional Activity of OsDREBs
3. Discussion
3.1. Nucleus-Localized PP2CAs Regulate Diverse Abiotic Stress-Related Transcription Factors
3.2. PP2CAs Might Act as Hubs in ABA-Dependent and -Independent Signaling Crosstalk
4. Materials and Methods
4.1. Plant Materials and Generation of Transgenic Rice
4.2. Post-Germination Assays and Stress Treatment
4.3. RT-PCR and Quantitative PCR
4.4. Luciferase Assay Using Rice Protoplasts
4.5. Subcellular Localization and BiFC
4.6. Co-Immunoprecipitation
4.7. Transcriptome Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Abfs | Aba-responsive element (abre)-binding factors |
| Abi1 | Aba insensitive1 |
| Abre | Aba-responsive element |
| Dre/crt | Dehydration-responsive element/c-repeat |
| Dreb | Dehydration-responsive element binding factor |
| Drip1 | Dreb2a-interacting protein1 |
| Fpkm | Fragments per kilobase million |
| Gfp | Green fluorescent protein |
| Grf7 | Growth-regulating factor7 |
| Hos1 | High expression of osmotically responsive gene1 |
| Ice1 | Inducer of cbf expression1 |
| Pp2cas | Clade a type 2c protein phosphatases |
| Pyr/pyl/rcar | Pyrabactin resistance1/pyr-like/regulatory component of aba receptor |
| Snrk | Snf1-related serine/threonine-protein kinase |
References
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef]
- Kang, J.Y.; Choi, H.I.; Im, M.Y.; Kim, S.Y. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 2002, 14, 343–357. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar]
- Saibo, N.J.; Lourenco, T.; Oliveira, M.M. Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann. Bot. 2009, 103, 609–623. [Google Scholar] [CrossRef]
- Lata, C.; Prasad, M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef]
- Qin, F.; Sakuma, Y.; Tran, L.S.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.; et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef]
- Kim, J.S.; Mizoi, J.; Kidokoro, S.; Maruyama, K.; Nakajima, J.; Nakashima, K.; Mitsuda, N.; Takiguchi, Y.; Ohme-Takagi, M.; Kondou, Y.; et al. Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 2012, 24, 3393–3405. [Google Scholar] [CrossRef]
- Miura, K.; Jin, J.B.; Lee, J.; Yoo, C.Y.; Stirm, V.; Miura, T.; Ashworth, E.N.; Bressan, R.A.; Yun, D.J.; Hasegawa, P.M. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 2007, 19, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Mizoi, J.; Kanazawa, N.; Kidokoro, S.; Takahashi, F.; Qin, F.; Morimoto, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Heat-induced inhibition of phosphorylation of the stress-protective transcription factor DREB2A promotes thermotolerance of Arabidopsis thaliana. J. Biol. Chem. 2019, 294, 902–917. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hwang, H.; Hong, J.W.; Lee, Y.N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.D.; Lee, S.; Lee, S.C.; Kim, B.G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012, 63, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Dupeux, F.; Round, A.; Antoni, R.; Park, S.Y.; Jamin, M.; Cutler, S.R.; Rodriguez, P.L.; Marquez, J.A. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 2009, 462, 665–668. [Google Scholar] [CrossRef]
- Xue, S.; Hu, H.; Ries, A.; Merilo, E.; Kollist, H.; Schroeder, J.I. Central functions of bicarbonate in S-type anion channel activation and OST1 protein kinase in CO2 signal transduction in guard cell. EMBO J. 2011, 30, 1645–1658. [Google Scholar] [CrossRef]
- Tian, X.; Wang, Z.; Li, X.; Lv, T.; Liu, H.; Wang, L.; Niu, H.; Bu, Q. Characterization and Functional Analysis of Pyrabactin Resistance-Like Abscisic Acid Receptor Family in Rice. Rice 2015, 8, 28. [Google Scholar] [CrossRef]
- McLeay, R.C.; Bailey, T.L. Motif Enrichment Analysis: A unified framework and an evaluation on ChIP data. BMC Bioinform. 2010, 11, 165. [Google Scholar] [CrossRef]
- Lu, T.; Zhang, G.; Wang, Y.; He, S.; Sun, L.; Hao, F. Genome-wide characterization and expression analysis of PP2CA family members in response to ABA and osmotic stress in Gossypium. PeerJ 2019, 7, e7105. [Google Scholar] [CrossRef]
- Wang, X.; Guo, C.; Peng, J.; Li, C.; Wan, F.; Zhang, S.; Zhou, Y.; Yan, Y.; Qi, L.; Sun, K.; et al. ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 2019, 221, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Bheri, M.; Mahiwal, S.; Sanyal, S.K.; Pandey, G.K. Plant protein phosphatases: What do we know about their mechanism of action? FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef]
- Min, M.K.; Choi, E.-H.; Kim, J.-A.; Yoon, I.S.; Han, S.; Lee, Y.; Lee, S.; Kim, B.-G. Two Clade A Phosphatase 2Cs Expressed in Guard Cells Physically Interact with Abscisic Acid Signaling Components to Induce Stomatal Closure in Rice. Rice 2019, 12, 37. [Google Scholar] [CrossRef]
- Wang, H.; Tang, J.; Liu, J.; Hu, J.; Liu, J.; Chen, Y.; Cai, Z.; Wang, X. Abscisic Acid Signaling Inhibits Brassinosteroid Signaling through Dampening the Dephosphorylation of BIN2 by ABI1 and ABI2. Mol. Plant 2018, 11, 315–325. [Google Scholar] [CrossRef]
- Bhatnagar, N.; Min, M.K.; Choi, E.H.; Kim, N.; Moon, S.J.; Yoon, I.; Kwon, T.; Jung, K.H.; Kim, B.G. The protein phosphatase 2C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol. Biol. 2017, 93, 389–401. [Google Scholar] [CrossRef]
- Uslu, V.V.; Grossmann, G. The biosensor toolbox for plant developmental biology. Curr. Opin. Plant Biol. 2016, 29, 138–147. [Google Scholar] [CrossRef]
- Durbak, A.; Yao, H.; McSteen, P. Hormone signaling in plant development. Curr. Opin. Plant Biol. 2012, 15, 92–96. [Google Scholar] [CrossRef]
- Nguyen, H.T.K.; Kim, S.Y.; Cho, K.-M.; Hong, J.C.; Shin, J.S.; Kim, H.J. A Transcription Factor γMYB1 Binds to the P1BS cis -Element and Activates PLA 2 -γ Expression with its Co-Activator γMYB2. Plant Cell Physiol. 2016, 57, 784–797. [Google Scholar] [CrossRef][Green Version]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Moon, S.J.; Min, M.K.; Choi, E.H.; Kim, J.A.; Koh, E.Y.; Yoon, I.; Byun, M.O.; Yoo, S.D.; Kim, B.G. Functional characterization and reconstitution of ABA signaling components using transient gene expression in rice protoplasts. Front. Plant Sci. 2015, 6, 614. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A Free, Open-Source System for Microarray Data Management and Analysis. BioTechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Min, M.K.; Kim, R.; Hong, W.-J.; Jung, K.-H.; Lee, J.-Y.; Kim, B.-G. OsPP2C09 Is a Bifunctional Regulator in Both ABA-Dependent and Independent Abiotic Stress Signaling Pathways. Int. J. Mol. Sci. 2021, 22, 393. https://doi.org/10.3390/ijms22010393
Min MK, Kim R, Hong W-J, Jung K-H, Lee J-Y, Kim B-G. OsPP2C09 Is a Bifunctional Regulator in Both ABA-Dependent and Independent Abiotic Stress Signaling Pathways. International Journal of Molecular Sciences. 2021; 22(1):393. https://doi.org/10.3390/ijms22010393
Chicago/Turabian StyleMin, Myung Ki, Rigyeong Kim, Woo-Jong Hong, Ki-Hong Jung, Jong-Yeol Lee, and Beom-Gi Kim. 2021. "OsPP2C09 Is a Bifunctional Regulator in Both ABA-Dependent and Independent Abiotic Stress Signaling Pathways" International Journal of Molecular Sciences 22, no. 1: 393. https://doi.org/10.3390/ijms22010393
APA StyleMin, M. K., Kim, R., Hong, W.-J., Jung, K.-H., Lee, J.-Y., & Kim, B.-G. (2021). OsPP2C09 Is a Bifunctional Regulator in Both ABA-Dependent and Independent Abiotic Stress Signaling Pathways. International Journal of Molecular Sciences, 22(1), 393. https://doi.org/10.3390/ijms22010393





